Controlling the Synchronization of Molecular Oscillators through Indirect Coupling
Abstract
:1. Introduction
2. Method
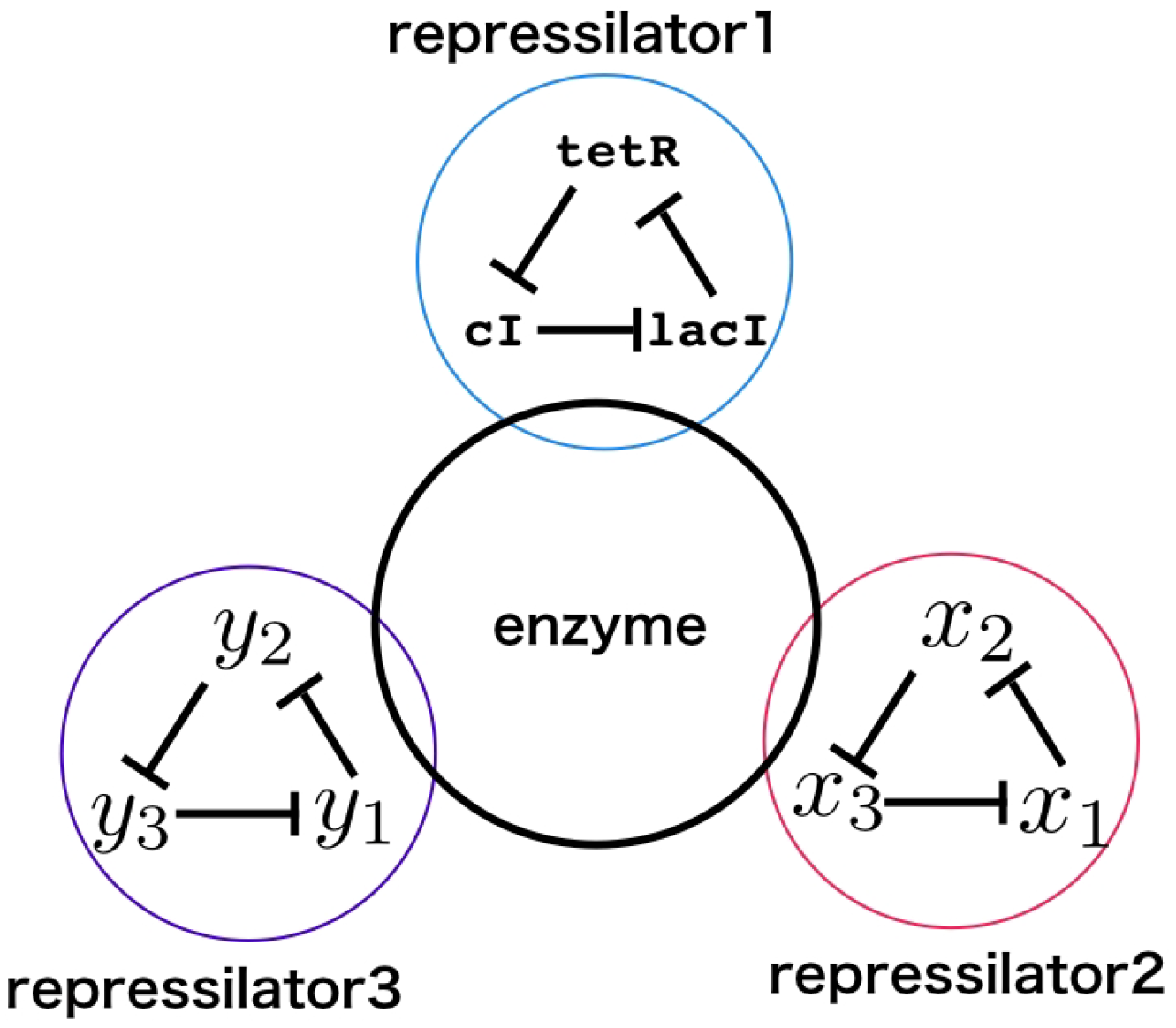
2.1. Model
2.2. Simulation
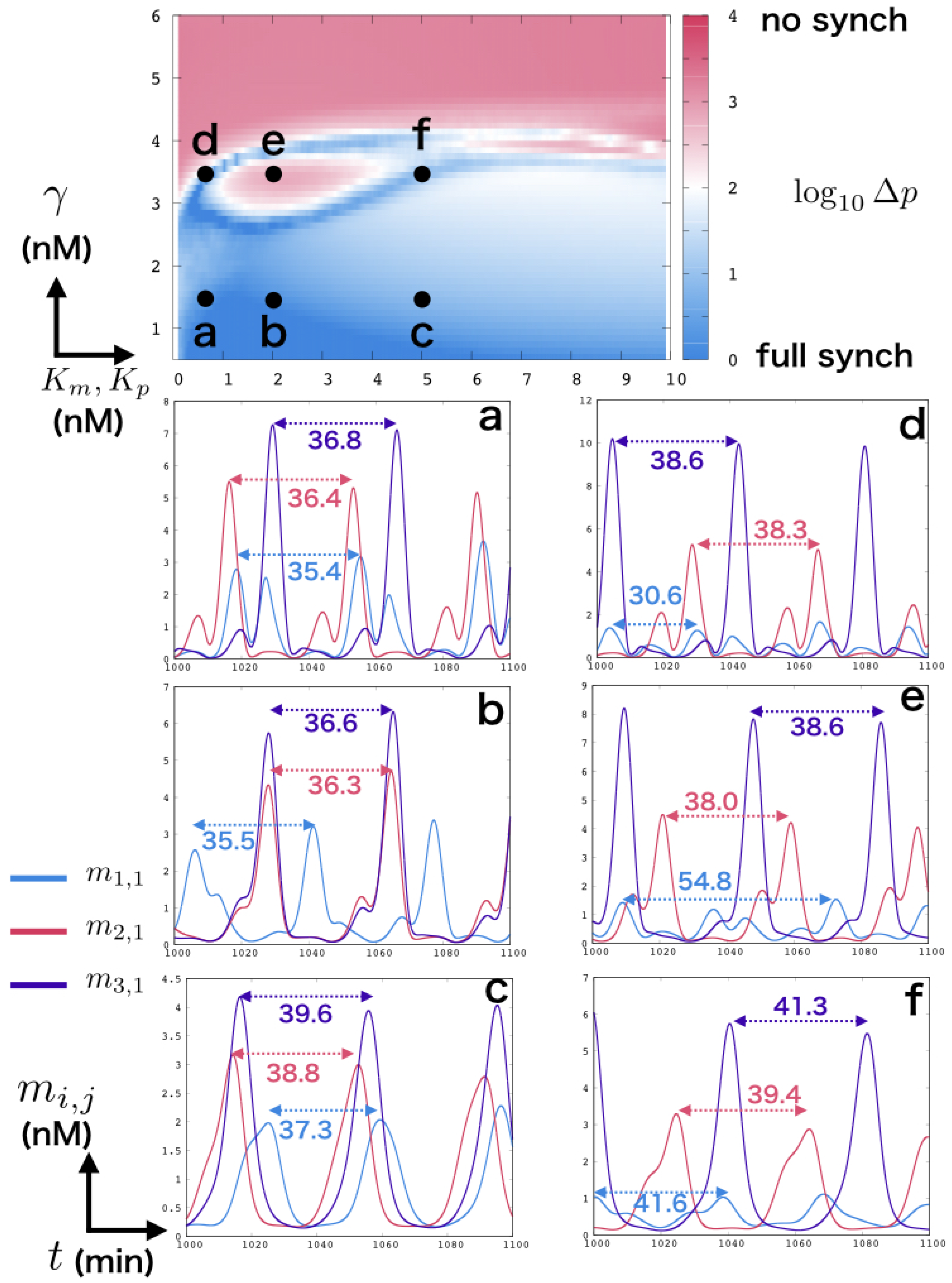
2.3. Measure of Period and Synchronization
3. Result
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Elowitz, M.B.; Leibler, S. A synthetic oscillatory network of transcriptional regulators. Nature 2000, 403, 335–338. [Google Scholar] [CrossRef] [PubMed]
- Gardner, T.S.; Cantor, C.R.; Collins, J.J. Construction of a genetic toggle switch in Escherichia coli. Nature 2000, 403, 339–342. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; White, K.S.; Winfree, E. Construction of an in vitro bistable circuit from synthetic transcriptional switches. Mol. Syst. Biol. 2006, 2, 68. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, J.; Winfree, E. Synthetic in vitro transcriptional oscillators. Mol. Syst. Biol. 2011, 7, 465. [Google Scholar] [CrossRef]
- Montagne, K.; Plasson, R.; Sakai, Y.; Fujii, T.; Rondelez, Y. Programming an in vitro DNA oscillator using a molecular networking strategy. Mol. Syst. Biol. 2011, 7, 466. [Google Scholar] [CrossRef]
- Padirac, A.; Fujii, T.; Rondelez, Y. Bottom-up construction of in vitro switchable memories. Proc. Natl. Acad. Sci. USA 2012, 109, E3212–E3220. [Google Scholar] [CrossRef] [Green Version]
- Stankevich, N.; Volkov, E. Evolution of quasiperiodicity in quorum-sensing coupled identical repressilators. Chaos Interdiscip. J. Nonlinear Sci. 2020, 30, 043122. [Google Scholar] [CrossRef]
- Winfree, A.T. The Geometry of Biological Time; Springer Science & Business Media: New York, NY, USA, 2001; Volume 12. [Google Scholar]
- Irisawa, H.; Brown, H.; Giles, W. Cardiac pacemaking in the sinoatrial node. Physiol. Rev. 1993, 73, 197–227. [Google Scholar] [CrossRef]
- Liu, C.; Weaver, D.R.; Strogatz, S.H.; Reppert, S.M. Cellular construction of a circadian clock: Period determination in the suprachiasmatic nuclei. Cell 1997, 91, 855–860. [Google Scholar] [CrossRef] [Green Version]
- Reppert, S.M.; Weaver, D.R. Forward genetic approach strikes gold: Cloning of a mammalian clock gene. Cell 1997, 89, 487–490. [Google Scholar] [CrossRef] [Green Version]
- Duong, H.A.; Robles, M.S.; Knutti, D.; Weitz, C.J. A molecular mechanism for circadian clock negative feedback. Science 2011, 332, 1436–1439. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garcia-Ojalvo, J.; Elowitz, M.B.; Strogatz, S.H. Modeling a synthetic multicellular clock: Repressilators coupled by quorum sensing. Proc. Natl. Acad. Sci. USA 2004, 101, 10955–10960. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fujii, T.; Rondelez, Y. Predator–prey molecular ecosystems. ACS Nano 2013, 7, 27–34. [Google Scholar] [CrossRef]
- Rondelez, Y. Competition for catalytic resources alters biological network dynamics. PRL 2012, 108, 018102. [Google Scholar] [CrossRef] [PubMed]
- Höll, M.; Kantz, H. The relationship between the detrendend fluctuation analysis and the autocorrelation function of a signal. EPJ B 2015, 88, 327. [Google Scholar] [CrossRef]
- Lobato-Dauzier, N.; Cazenille, L.; Fujii, T.; Genot, A.; Aubert-Kato, N. Temperature-based inputs for molecular reservoir computers. In Proceedings of the Artificial Life Conference, Online, 13–18 July 2020; MIT Press: Cambridge, MA, USA, 2020; pp. 420–422. [Google Scholar]
- Hagiya, M.; Konagaya, A.; Kobayashi, S.; Saito, H.; Murata, S. Molecular robots with sensors and intelligence. Acc. Chem. Res. 2014, 47, 1681–1690. [Google Scholar] [CrossRef]
- Cooke, J.; Zeeman, E.C. A clock and wavefront model for control of the number of repeated structures during animal morphogenesis. J. Theor. Biol. 1976, 58, 455–476. [Google Scholar] [CrossRef]
- Pourquié, O. The segmentation clock: Converting embryonic time into spatial pattern. Science 2003, 301, 328–330. [Google Scholar] [CrossRef] [Green Version]
- Senoussi, A.; Galas, J.C.; Estevez-Torres, A. Programmed mechano-chemical coupling in reaction-diffusion active matter. Sci. Adv. 2021, 7, eabi9865. [Google Scholar] [CrossRef]
- Senoussi, A.; Vyborna, Y.; Berthoumieux, H.; Galas, J.C.; Estevez-Torres, A. Learning from Embryo Development to Engineer Self-organizing Materials. In Out-of-Equilibrium (Supra)molecular Systems and Materials; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2021; Chapter 2; pp. 21–60. [Google Scholar] [CrossRef]
- Buse, O.; Pérez, R.; Kuznetsov, A. Dynamical properties of the repressilator model. Phys. Rev. E 2010, 81, 066206. [Google Scholar] [CrossRef] [Green Version]
- Maša, D.; Hassan, E.; Roman, J.; Tina, L.; Valery G., R.; János, T.; Andreas, W. On three genetic repressilator topologies. React. Kinet. Mech. Catal. 2019, 126, 3–30. [Google Scholar]
- Padirac, A. Tailoring Spatio-Temporal Dynamics with DNA Circuits. Ph.D. Thesis, Université Claude Bernard-Lyon I, Lyon, France, 2012. [Google Scholar]
- Genot, A.; Baccouche, A.; Sieskind, R.; Aubert-Kato, N.; Bredèche, N.; Bartolo, J.; Taly, V.; Fujii, T.; Rondelez, Y. High-resolution mapping of bifurcations in nonlinear biochemical circuits. Nat. Chem. 2016, 8, 760–767. [Google Scholar] [CrossRef] [PubMed]
- Dehne, H.; Reitenbach, A.; Bausch, A. Reversible and spatiotemporal control of colloidal structure formation. Nat. Commun. 2021, 12, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Padirac, A.; Fujii, T.; Estévez-Torres, A.; Rondelez, Y. Spatial waves in synthetic biochemical networks. JACS 2013, 135, 14586–14592. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Inagaki, S.; Aubert-Kato, N. Controlling the Synchronization of Molecular Oscillators through Indirect Coupling. Micromachines 2022, 13, 245. https://doi.org/10.3390/mi13020245
Inagaki S, Aubert-Kato N. Controlling the Synchronization of Molecular Oscillators through Indirect Coupling. Micromachines. 2022; 13(2):245. https://doi.org/10.3390/mi13020245
Chicago/Turabian StyleInagaki, Shiho, and Nathanael Aubert-Kato. 2022. "Controlling the Synchronization of Molecular Oscillators through Indirect Coupling" Micromachines 13, no. 2: 245. https://doi.org/10.3390/mi13020245





