Conventional and Microfluidic Methods for the Detection of Nucleic Acid of SARS-CoV-2
Abstract
:1. Introduction
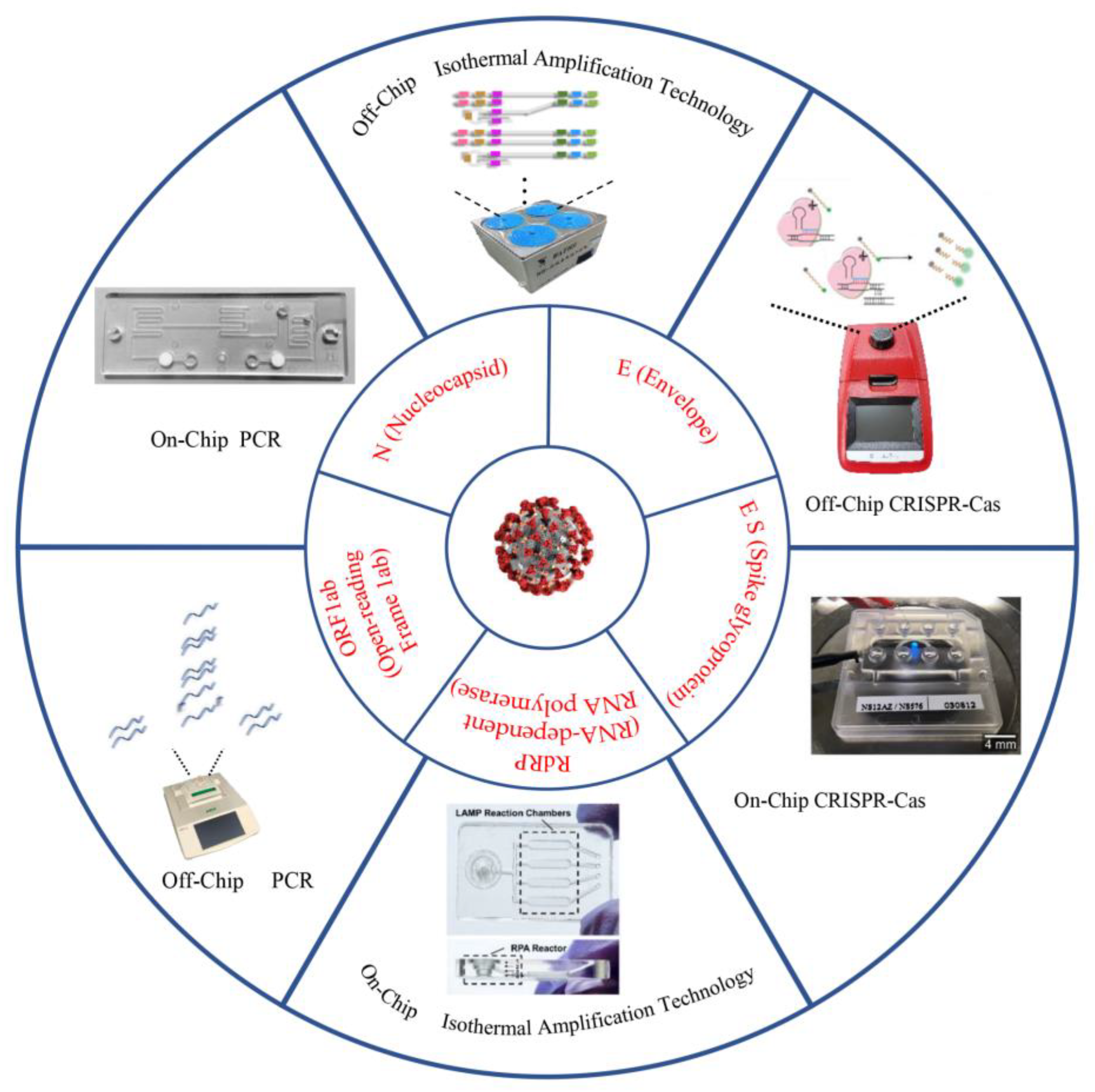
2. Off-Chip Nucleic Acid Detection Method for SARS-CoV-2 Detection
2.1. Nucleic Acid Detection Based on PCR Technology
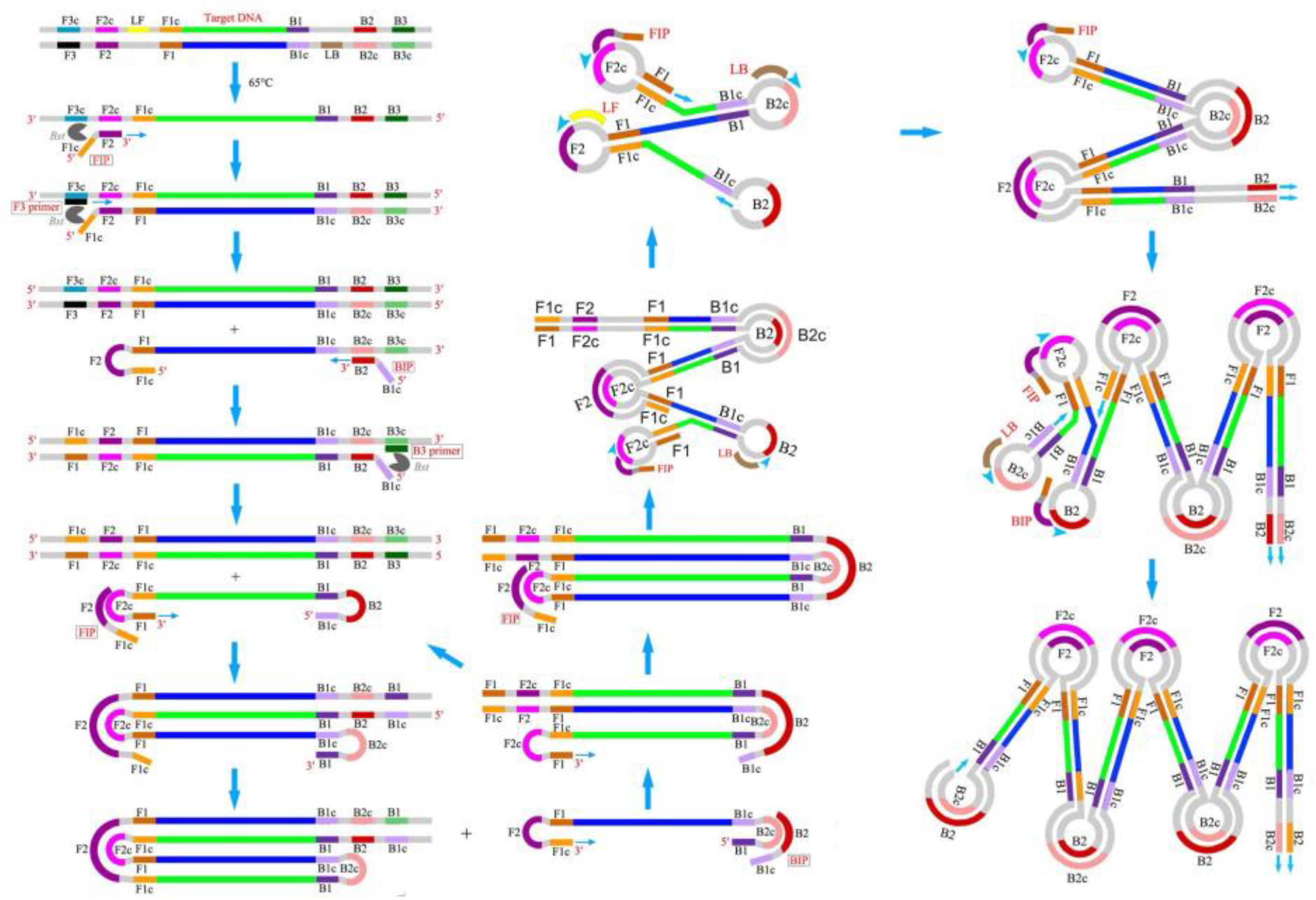
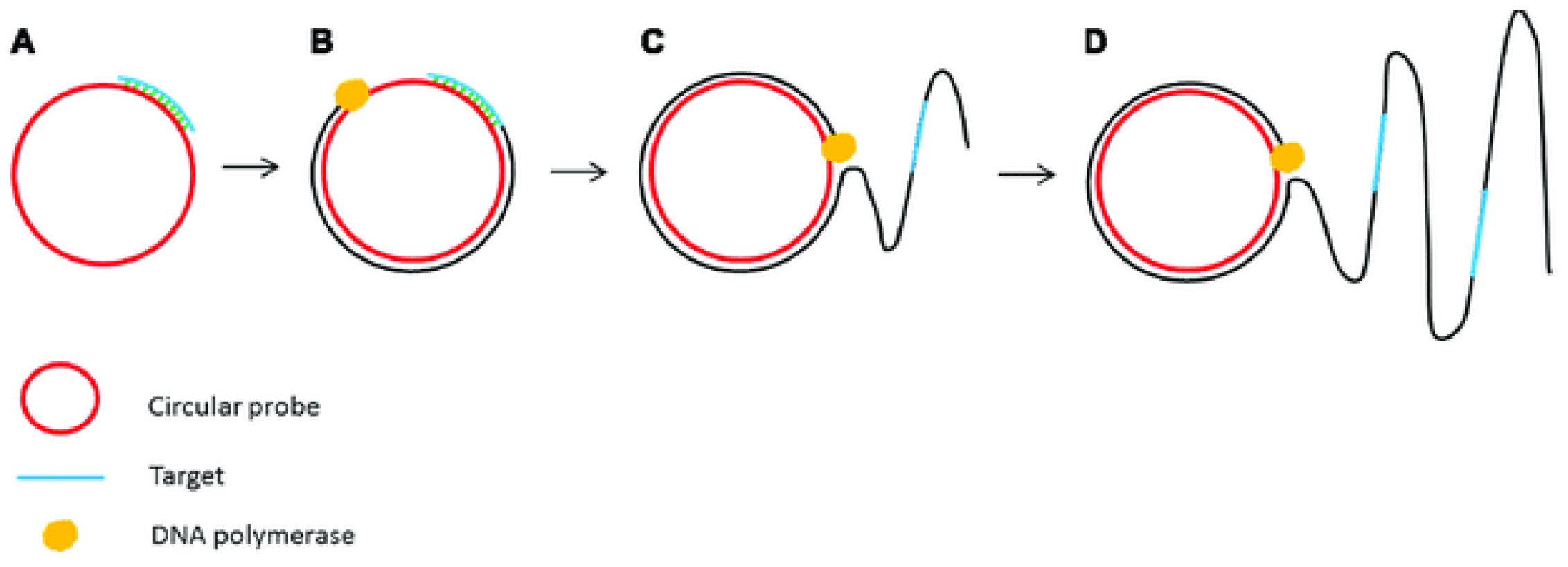
2.2. Nucleic Acid Detection Based on Isothermal Amplification Technology
2.3. Nucleic Acid Detection Based on RNA-Guided CRISPR–Cas System
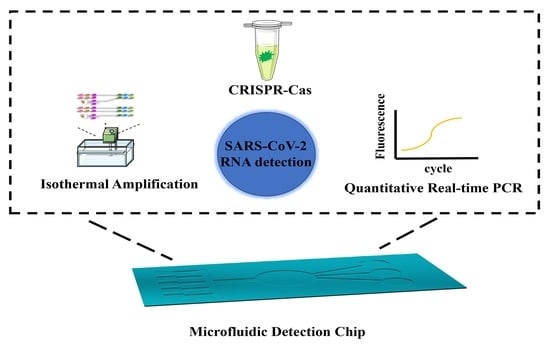
3. Ultrasensitive Microfluidic for SARS-CoV-2 Detection
3.1. Microfluidic Detection Based on PCR Technology
3.2. Microfluidic Detection Based on Isothermal Amplification Technology
3.2.1. LAMP in Microfluidics
3.2.2. Other Isothermal Amplification Techniques in Microfluidics
3.3. Microfluidic Detection Based on CRISPR–Cas Biological Detection Technology
4. Conclusions
4.1. Summary on Traditional and Microfluidic Experiments
4.2. Summary on Nucleic Acid Detection Technology
5. Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wu, F.; Zhao, S.; Yu, B.; Chen, Y.M.; Wang, W.; Song, Z.G.; Hu, Y.; Tao, Z.W.; Tian, J.H.; Pei, Y.Y.; et al. A new coronavirus associated with human respiratory disease in China. Nature 2020, 579, 265–269. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, D.; Hu, B.; Hu, C.; Zhu, F.; Liu, X.; Zhang, J.; Wang, B.; Xiang, H.; Cheng, Z.; Xiong, Y.; et al. Clinical Characteristics of 138 Hospitalized Patients with 2019 Novel Coronavirus-Infected Pneumonia in Wuhan, China. JAMA J. Am. Med. Assoc. 2020, 323, 1061–1069. [Google Scholar] [CrossRef] [PubMed]
- Cevik, M.; Kuppalli, K.; Kindrachuk, J.; Peiris, M. Virology, transmission, and pathogenesis of SARS-CoV-2. BMJ 2020, 371, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Chen, N.; Zhou, M.; Dong, X.; Qu, J.; Gong, F.; Han, Y.; Qiu, Y.; Wang, J.; Liu, Y.; Wei, Y.; et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: A descriptive study. Lancet 2020, 395, 507–513. [Google Scholar] [CrossRef] [Green Version]
- Zhao, J.; Yuan, Q.; Wang, H.; Liu, W.; Liao, X.; Su, Y.; Wang, X.; Yuan, J.; Li, T.; Li, J.; et al. Antibody Responses to SARS-CoV-2 in Patients with Novel Coronavirus Disease 2019. Clin. Infect. Dis. 2020, 71, 2027–2034. [Google Scholar] [CrossRef]
- Long, Q.X.; Liu, B.Z.; Deng, H.J.; Wu, G.C.; Deng, K.; Chen, Y.K.; Liao, P.; Qiu, J.F.; Lin, Y.; Cai, X.F.; et al. Antibody responses to SARS-CoV-2 in patients with COVID-19. Nat. Med. 2020, 26, 845–848. [Google Scholar] [CrossRef]
- Lu, R.; Zhao, X.; Li, J.; Niu, P.; Yang, B.; Wu, H.; Wang, W.; Song, H.; Huang, B.; Zhu, N.; et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: Implications for virus origins and receptor binding. Lancet 2020, 395, 565–574. [Google Scholar] [CrossRef] [Green Version]
- Ren, X.; Liu, Y.; Chen, H.; Liu, W.; Guo, Z.; Zhang, Y.; Chen, C.; Zhou, J.; Xiao, Q.; Jiang, G.-M.; et al. Application and optimization of RT-PCR in diagnosis of SARS-CoV-2 infection. SSRN Electron. J. 2020. [Google Scholar] [CrossRef]
- Zhou, P.; Yang, X.L.; Wang, X.G.; Hu, B.; Zhang, L.; Zhang, W.; Si, H.R.; Zhu, Y.; Li, B.; Huang, C.L.; et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 2020, 579, 270–273. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Wang, Z. Strengths, weaknesses, opportunities and threats (Swot) analysis of china’s prevention and control strategy for the COVID-19 epidemic. Int. J. Environ. Res. Public Health 2020, 17, 2235. [Google Scholar] [CrossRef] [Green Version]
- Berkenbrock, J.A.; Grecco-Machado, R.; Achenbach, S. Arsenal of microfluidic testing devices may combat COVID-19 pandemic. MRS Bull. 2020, 45, 511–514. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Yamamoto, T. Quantification of virus particles using nanopore-based resistive-pulse sensing techniques. Front. Microbiol. 2016, 7, 1500. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, H.; Fohlerová, Z.; Pekárek, J.; Basova, E.; Neužil, P. Recent advances in lab-on-a-chip technologies for viral diagnosis. Biosens. Bioelectron. 2020, 153, 112041. [Google Scholar] [CrossRef] [PubMed]
- Xing, W.; Liu, Y.; Wang, H.; Li, S.; Lin, Y.; Chen, L.; Zhao, Y.; Chao, S.; Huang, X.; Ge, S.; et al. A High-Throughput, Multi-Index Isothermal Amplification Platform for Rapid Detection of 19 Types of Common Respiratory Viruses Including SARS-CoV-2. Engineering 2020, 6, 1130–1140. [Google Scholar] [CrossRef] [PubMed]
- Shu, B.; Zhang, C.; Xing, D. Segmented continuous-flow multiplex polymerase chain reaction microfluidics for high-throughput and rapid foodborne pathogen detection. Anal. Chim. Acta 2014, 826, 51–60. [Google Scholar] [CrossRef]
- Yin, L.; Man, S.; Ye, S.; Liu, G.; Ma, L. CRISPR-Cas based virus detection: Recent advances and perspectives. Biosens. Bioelectron. 2021, 193, 113541. [Google Scholar] [CrossRef]
- Sakai, J.; Tarumoto, N.; Orihara, Y.; Kawamura, R.; Kodana, M.; Matsuzaki, N.; Matsumura, R.; Ogane, K.; Kawamura, T.; Takeuchi, S.; et al. Evaluation of a high-speed but low-throughput RT-qPCR system for detection of SARS-CoV-2. J. Hosp. Infect. 2020, 105, 615–618. [Google Scholar] [CrossRef]
- Yin, K.; Ding, X.; Xu, Z.; Li, Z.; Wang, X.; Zhao, H.; Otis, C.; Li, B.; Liu, C. Multiplexed colorimetric detection of SARS-CoV-2 and other pathogens in wastewater on a 3D printed integrated microfluidic chip. Sens. Actuators B Chem. 2021, 344, 130242. [Google Scholar] [CrossRef]
- Ramachandran, A.; Huyke, D.A.; Sharma, E.; Sahoo, M.K.; Huang, C.; Banaei, N.; Pinsky, B.A.; Santiago, J.G. Electric field-driven microfluidics for rapid CRISPR-based diagnostics and its application to detection of SARS-CoV-2. Proc. Natl. Acad. Sci. USA 2020, 117, 29518–29525. [Google Scholar] [CrossRef]
- Chen, L.; Liu, W.; Zhang, Q.; Xu, K.; Ye, G.; Wu, W.; Sun, Z.; Liu, F.; Wu, K.; Zhong, B.; et al. RNA based mNGS approach identifies a novel human coronavirus from two individual pneumonia cases in 2019 Wuhan outbreak. Emerg. Microbes Infect. 2020, 9, 313–319. [Google Scholar] [CrossRef]
- Kilic, T.; Weissleder, R.; Lee, H. Molecular and Immunological Diagnostic Tests of COVID-19: Current Status and Challenges. iScience 2020, 23, 101406. [Google Scholar] [CrossRef] [PubMed]
- Deng, H.; Gao, Z. Bioanalytical applications of isothermal nucleic acid amplification techniques. Anal. Chim. Acta 2015, 853, 30–45. [Google Scholar] [CrossRef] [PubMed]
- Suo, T.; Liu, X.; Feng, J.; Guo, M.; Hu, W.; Guo, D.; Ullah, H.; Yang, Y.; Zhang, Q.; Wang, X.; et al. ddPCR: A more accurate tool for SARS-CoV-2 detection in low viral load specimens. Emerg. Microbes Infect. 2020, 9, 1259–1268. [Google Scholar] [CrossRef] [PubMed]
- Jayamohan, H.; Lambert, C.J.; Sant, H.J.; Jafek, A.; Patel, D.; Feng, H.; Beeman, M.; Mahmood, T.; Nze, U.; Gale, B.K. SARS-CoV-2 pandemic: A review of molecular diagnostic tools including sample collection and commercial response with associated advantages and limitations. Anal. Bioanal. Chem. 2021, 413, 49–71. [Google Scholar] [CrossRef]
- Fomsgaard, A.S.; Rosenstierne, M.W. An alternative workflow for molecular detection of SARS-CoV-2—Escape from the NA extraction kit-shortage, Copenhagen, Denmark, March 2020. Eurosurveillance 2020, 25, 2000398. [Google Scholar] [CrossRef] [Green Version]
- Williams, E.; Bond, K.; Zhang, B.; Putland, M.; Williamson, D.A. Saliva as a noninvasive specimen for detection of SARS-CoV-2. J. Clin. Microbiol. 2020, 58, e00776-20. [Google Scholar] [CrossRef] [Green Version]
- La Rosa, G.; Iaconelli, M.; Mancini, P.; Bonanno Ferraro, G.; Veneri, C.; Bonadonna, L.; Lucentini, L.; Suffredini, E. First detection of SARS-CoV-2 in untreated wastewaters in Italy. Sci. Total Environ. 2020, 736, 139652. [Google Scholar] [CrossRef]
- Bivins, A.; Greaves, J.; Fischer, R.; Yinda, K.C.; Ahmed, W.; Kitajima, M.; Munster, V.J.; Bibby, K. Persistence of SARS-CoV-2 in Water and Wastewater. Environ. Sci. Technol. Lett. 2020, 7, 937–942. [Google Scholar] [CrossRef]
- Razzini, K.; Castrica, M.; Menchetti, L.; Maggi, L.; Negroni, L.; Orfeo, N.V.; Pizzoccheri, A.; Stocco, M.; Muttini, S.; Balzaretti, C.M. SARS-CoV-2 RNA detection in the air and on surfaces in the COVID-19 ward of a hospital in Milan, Italy. Sci. Total Environ. 2020, 742, 140540. [Google Scholar] [CrossRef]
- Zanoli, L.M.; Spoto, G. Isothermal amplification methods for the detection of nucleic acids in microfluidic devices. Biosensors 2013, 3, 18–43. [Google Scholar] [CrossRef] [Green Version]
- Asadi, R.; Mollasalehi, H. The mechanism and improvements to the isothermal amplification of nucleic acids, at a glance. Anal. Biochem. 2021, 631, 114260. [Google Scholar] [CrossRef] [PubMed]
- Panno, S.; Matić, S.; Tiberini, A.; Caruso, A.G.; Bella, P.; Torta, L.; Stassi, R.; Davino, S. Loop mediated isothermal amplification: Principles and applications in plant virology. Plants 2020, 9, 461. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, J.J.; Xiong, C.; Liu, Y.; Liang, J.S.; Zhou, X.W. Loop-mediated isothermal amplification (LAMP): Emergence as an alternative technology for herbal medicine identification. Front. Plant Sci. 2016, 7, 1956. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pang, B.; Yao, S.; Xu, K.; Wang, J.; Song, X.; Mu, Y.; Zhao, C.; Li, J. A novel visual-mixed-dye for LAMP and its application in the detection of foodborne pathogens. Anal. Biochem. 2019, 574, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Feng, J.; Tian, X. Application of loop-mediated isothermal amplification (LAMP) for rapid detection of Atlantic cod (Gadus morhua), Pacific cod (Gadus macrocephalus) and haddock (Melanogrammus aeglefinus). Mol. Cell. Probes 2019, 47, 101420. [Google Scholar] [CrossRef] [PubMed]
- Hongwarittorrn, I.; Chaichanawongsaroj, N.; Laiwattanapaisal, W. Semi-quantitative visual detection of loop mediated isothermal amplification (LAMP)-generated DNA by distance-based measurement on a paper device. Talanta 2017, 175, 135–142. [Google Scholar] [CrossRef] [PubMed]
- Meena, P.N.; Kharbikar, L.L.; Rana, R.S.; Satpathy, S.; Shanware, A.; Sivalingam, P.N.; Nandanwar, S. Detection of Mesta yellow vein mosaic virus (MeYVMV) in field samples by a loop-mediated isothermal amplification reaction. J. Virol. Methods 2019, 263, 81–87. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Wan, Z.; Yang, S.; Li, Y.; Li, M.; Wang, B.; Hu, Y.; Xia, X.; Jin, X.; Yu, N.; et al. A mismatch-tolerant reverse transcription loop-mediated isothermal amplification method and its application on simultaneous detection of all four serotype of dengue viruses. Front. Microbiol. 2019, 10, 1056. [Google Scholar] [CrossRef] [Green Version]
- Lu, R.; Wu, X.; Wan, Z.; Li, Y.; Zuo, L.; Qin, J.; Jin, X.; Zhang, C. Development of a Novel Reverse Transcription Loop-Mediated Isothermal Amplification Method for Rapid Detection of SARS-CoV-2. Virol. Sin. 2020, 35, 344–347. [Google Scholar] [CrossRef] [Green Version]
- Haque, M.F.U.; Bukhari, S.S.; Ejaz, R.; Zaman, F.U.; Sreejith, K.R.; Rashid, N.; Umer, M.; Shahzad, N. A novel RdRp-based colorimetric RT-LAMP assay for rapid and sensitive detection of SARS-CoV-2 in clinical and sewage samples from Pakistan. Virus Res. 2021, 302, 198484. [Google Scholar] [CrossRef]
- Mohon, A.N.; Oberding, L.; Hundt, J.; van Marle, G.; Pabbaraju, K.; Berenger, B.M.; Lisboa, L.; Griener, T.; Czub, M.; Doolan, C.; et al. Optimization and clinical validation of dual-target RT-LAMP for SARS-CoV-2. J. Virol. Methods 2020, 286, 113972. [Google Scholar] [CrossRef] [PubMed]
- Lau, H.Y.; Botella, J.R. Advanced DNA-based point-of-care diagnostic methods for plant diseases detection. Front. Plant Sci. 2017, 8, 2016. [Google Scholar] [CrossRef] [PubMed]
- Tian, B.; Gao, F.; Fock, J.; Dufva, M.; Hansen, M.F. Homogeneous circle-to-circle amplification for real-time optomagnetic detection of SARS-CoV-2 RdRp coding sequence. Biosens. Bioelectron. 2020, 165, 112356. [Google Scholar] [CrossRef] [PubMed]
- Jiao, J.; Duan, C.; Xue, L.; Liu, Y.; Sun, W.; Xiang, Y. DNA nanoscaffold-based SARS-CoV-2 detection for COVID-19 diagnosis. Biosens. Bioelectron. 2020, 167, 112479. [Google Scholar] [CrossRef] [PubMed]
- Haes, A.J.; Giordano, B.C.; Collins, G.E. Aptamer-based detection and quantitative analysis of ricin using affinity probe capillary electrophoresis. Anal. Chem. 2006, 78, 3758–3764. [Google Scholar] [CrossRef] [PubMed]
- Proske, D.; Blank, M.; Buhmann, R.; Resch, A. Aptamers—Basic research, drug development, and clinical applications. Appl. Microbiol. Biotechnol. 2005, 69, 367–374. [Google Scholar] [CrossRef] [PubMed]
- Platella, C.; Riccardi, C.; Montesarchio, D.; Roviello, G.N.; Musumeci, D. G-quadruplex-based aptamers against protein targets in therapy and diagnostics. Biochim. Biophys. Acta Gen. Subj. 2017, 1861, 1429–1447. [Google Scholar] [CrossRef] [PubMed]
- Woo, C.H.; Jang, S.; Shin, G.; Jung, G.Y.; Lee, J.W. Sensitive fluorescence detection of SARS-CoV-2 RNA in clinical samples via one-pot isothermal ligation and transcription. Nat. Biomed. Eng. 2020, 4, 1168–1179. [Google Scholar] [CrossRef]
- Gootenberg, J.S.; Abudayyeh, O.O.; Lee, J.W.; Essletzbichler, P.; Dy, A.J.; Joung, J.; Verdine, V.; Donghia, N.; Daringer, N.M.; Freije, C.A.; et al. Nucleic acid detection with CRISPR-Cas13a/C2c2. Science 2017, 356, 438–442. [Google Scholar] [CrossRef] [Green Version]
- Ding, X.; Yin, K.; Li, Z.; Lalla, R.V.; Ballesteros, E.; Sfeir, M.M.; Liu, C. Ultrasensitive and visual detection of SARS-CoV-2 using all-in-one dual CRISPR-Cas12a assay. Nat. Commun. 2020, 11, 4711. [Google Scholar] [CrossRef]
- Myhrvold, C.; Freije, C.A.; Gootenberg, J.S.; Abudayyeh, O.O.; Metsky, H.C.; Durbin, A.F.; Kellner, M.J.; Tan, A.L.; Paul, L.M.; Parham, L.A.; et al. Field-deployable viral diagnostics using CRISPR-Cas13. Science 2018, 360, 444–448. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Broughton, J.P.; Deng, X.; Yu, G.; Fasching, C.L.; Servellita, V.; Singh, J.; Miao, X.; Streithorst, J.A.; Granados, A.; Sotomayor-Gonzalez, A.; et al. CRISPR–Cas12-based detection of SARS-CoV-2. Nat. Biotechnol. 2020, 38, 870–874. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Patchsung, M.; Jantarug, K.; Pattama, A.; Aphicho, K.; Suraritdechachai, S.; Meesawat, P.; Sappakhaw, K.; Leelahakorn, N.; Ruenkam, T.; Wongsatit, T.; et al. Clinical validation of a Cas13-based assay for the detection of SARS-CoV-2 RNA. Nat. Biomed. Eng. 2020, 4, 1140–1149. [Google Scholar] [CrossRef] [PubMed]
- Microbiol, C.; Rabaan, A.A.; Ahmed, S.H.A.; Sah, R.; Tiwari, R.; Yatoo, M.I.; Patel, S.K.; Pathak, M.; Malik, Y.S.; Dhama, K.; et al. SARS-CoV-2/COVID-19 and advances in developing potential therapeutics and vaccines to counter this emerging pandemic. Ann. Clin. Microbiol. Antimicrob. 2020, 19, 1–37. [Google Scholar] [CrossRef]
- Wu, D.; Wu, T.; Liu, Q.; Yang, Z. The SARS-CoV-2 outbreak: What we know. Int. J. Infect. Dis. 2020, 94, 44–48. [Google Scholar] [CrossRef]
- Michael-Kordatou, I.; Karaolia, P.; Fatta-Kassinos, D. Sewage analysis as a tool for the COVID-19 pandemic response and management: The urgent need for optimised protocols for SARS-CoV-2 detection and quantification. J. Environ. Chem. Eng. 2020, 8, 104306. [Google Scholar] [CrossRef]
- Ghodake, G.S.; Shinde, S.K.; Kadam, A.A.; Saratale, R.G.; Saratale, G.D.; Syed, A.; Elgorban, A.M.; Marraiki, N.; Kim, D.Y. Biological characteristics and biomarkers of novel SARS-CoV-2 facilitated rapid development and implementation of diagnostic tools and surveillance measures. Biosens. Bioelectron. 2021, 177, 112969. [Google Scholar] [CrossRef]
- Ben-Ami, R.; Klochendler, A.; Seidel, M.; Sido, T.; Gurel-Gurevich, O.; Yassour, M.; Meshorer, E.; Benedek, G.; Fogel, I.; Oiknine-Djian, E.; et al. Large-scale implementation of pooled RNA extraction and RT-PCR for SARS-CoV-2 detection. Clin. Microbiol. Infect. 2020, 26, 1248–1253. [Google Scholar] [CrossRef]
- Ren, K.; Zhou, J.; Wu, H. Materials for microfluidic chip fabrication. Acc. Chem. Res. 2013, 46, 2396–2406. [Google Scholar] [CrossRef]
- Jiang, X.; Shao, N.; Jing, W.; Tao, S.; Liu, S.; Sui, G. Microfluidic chip integrating high throughput continuous-flow PCR and DNA hybridization for bacteria analysis. Talanta 2014, 122, 246–250. [Google Scholar] [CrossRef]
- He, S.; Joseph, N.; Feng, S.; Jellicoe, M.; Raston, C.L. Application of microfluidic technology in food processing. Food Funct. 2020, 11, 5726–5737. [Google Scholar] [CrossRef] [PubMed]
- Park, S.; Zhang, Y.; Lin, S.; Wang, T.H.; Yang, S. Advances in microfluidic PCR for point-of-care infectious disease diagnostics. Biotechnol. Adv. 2011, 29, 830–839. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ji, M.; Xia, Y.; Loo, J.F.C.; Li, L.; Ho, H.P.; He, J.; Gu, D. Automated multiplex nucleic acid tests for rapid detection of SARS-CoV-2, influenza A and B infection with direct reverse-transcription quantitative PCR (dirRT-qPCR) assay in a centrifugal microfluidic platform. RSC Adv. 2020, 10, 34088–34098. [Google Scholar] [CrossRef]
- Trinh, T.N.D.; La, H.C.; Lee, N.Y. Fully Integrated and Foldable Microdevice Encapsulated with Agarose for Long-Term Storage Potential for Point-of-Care Testing of Multiplex Foodborne Pathogens. ACS Sens. 2019, 4, 2754–2762. [Google Scholar] [CrossRef]
- Mitsakakis, K.; D’Acremont, V.; Hin, S.; von Stetten, F.; Zengerle, R. Diagnostic tools for tackling febrile illness and enhancing patient management. Microelectron. Eng. 2018, 201, 26–59. [Google Scholar] [CrossRef]
- Ducrée, J.; Haeberle, S.; Lutz, S.; Pausch, S.; Von Stetten, F.; Zengerle, R. The centrifugal microfluidic Bio-Disk platform. J. Micromechanics Microengineering 2007, 17, S103. [Google Scholar] [CrossRef]
- Kang, B.H.; Lee, Y.; Yu, E.S.; Na, H.; Kang, M.; Huh, H.J.; Jeong, K.H. Ultrafast and Real-Time Nanoplasmonic On-Chip Polymerase Chain Reaction for Rapid and Quantitative Molecular Diagnostics. ACS Nano 2021, 15, 10194–10202. [Google Scholar] [CrossRef]
- Soares, R.R.G.; Akhtar, A.S.; Pinto, I.F.; Lapins, N.; Barrett, D.; Sandh, G.; Yin, X.; Pelechano, V.; Russom, A. Sample-to-answer COVID-19 nucleic acid testing using a low-cost centrifugal microfluidic platform with bead-based signal enhancement and smartphone read-out. Lab Chip 2021, 21, 2932–2944. [Google Scholar] [CrossRef]
- Rodriguez-Mateos, P.; Ngamsom, B.; Walter, C.; Dyer, C.E.; Gitaka, J.; Iles, A.; Pamme, N. A lab-on-a-chip platform for integrated extraction and detection of SARS-CoV-2 RNA in resource-limited settings. Anal. Chim. Acta 2021, 1177, 338758. [Google Scholar] [CrossRef]
- Garneret, P.; Coz, E.; Martin, E.; Manuguerra, J.C.; Brient-Litzler, E.; Enouf, V.; González Obando, D.F.; Olivo-Marin, J.C.; Monti, F.; van der Werf, S.; et al. Performing point-of-care molecular testing for SARS-CoV-2 with RNA extraction and isothermal amplification. PLoS ONE 2021, 16, e0243712. [Google Scholar] [CrossRef]
- Rodriguez-Manzano, J.; Malpartida-Cardenas, K.; Moser, N.; Pennisi, I.; Cavuto, M.; Miglietta, L.; Moniri, A.; Penn, R.; Satta, G.; Randell, P.; et al. Handheld point-of-care system for rapid detection of SARS-CoV-2 extracted RNA in under 20 min. ACS Cent. Sci. 2021, 7, 307–317. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.S.; Abbas, N.; Shin, S. A rapid diagnosis of SARS-CoV-2 using DNA hydrogel formation on microfluidic pores. Biosens. Bioelectron. 2021, 177, 113005. [Google Scholar] [CrossRef] [PubMed]
- Park, J.S.; Hsieh, K.; Chen, L.; Kaushik, A.; Trick, A.Y.; Wang, T.H. Digital CRISPR/Cas-Assisted Assay for Rapid and Sensitive Detection of SARS-CoV-2. Adv. Sci. 2021, 8, 2003564. [Google Scholar] [CrossRef] [PubMed]
- Burmeister, A.; Grünberger, A. Microfluidic cultivation and analysis tools for interaction studies of microbial co-cultures. Curr. Opin. Biotechnol. 2020, 62, 106–115. [Google Scholar] [CrossRef]
- Wang, X.; Liu, Z.; Pang, Y. Concentration gradient generation methods based on microfluidic systems. RSC Adv. 2017, 7, 29966–29984. [Google Scholar] [CrossRef] [Green Version]
- Wu, J.; Kodzius, R.; Cao, W.; Wen, W. Extraction, amplification and detection of DNA in microfluidic chip-based assays. Microchim. Acta 2014, 181, 1611–1631. [Google Scholar] [CrossRef]
- Hu, F.; Li, J.; Zhang, Z.; Li, M.; Zhao, S.; Li, Z.; Peng, N. Smartphone-Based Droplet Digital LAMP Device with Rapid Nucleic Acid Isolation for Highly Sensitive Point-of-Care Detection. Anal. Chem. 2020, 92, 2258–2265. [Google Scholar] [CrossRef] [PubMed]
- Ye, X.; Li, Y.; Fang, X.; Kong, J. Integrated Microfluidic Sample-to-Answer System for Direct Nucleic Acid-Based Detection of Group B Streptococci in Clinical Vaginal/Anal Swab Samples. ACS Sens. 2020, 5, 1132–1139. [Google Scholar] [CrossRef]
- Zhong, J.; Zhao, X. Isothermal Amplification Technologies for the Detection of Foodborne Pathogens. Food Anal. Methods 2018, 11, 1543–1560. [Google Scholar] [CrossRef]
- Barreda-García, S.; Miranda-Castro, R.; de-los-Santos-Álvarez, N.; Miranda-Ordieres, A.J.; Lobo-Castañón, M.J. Helicase-dependent isothermal amplification: A novel tool in the development of molecular-based analytical systems for rapid pathogen detection. Anal. Bioanal. Chem. 2018, 410, 679–693. [Google Scholar] [CrossRef]
- Zhao, Y.; Chen, F.; Li, Q.; Wang, L.; Fan, C. Isothermal Amplification of Nucleic Acids. Chem. Rev. 2015, 115, 12491–12545. [Google Scholar] [CrossRef] [PubMed]
- Suea-Ngam, A.; Bezinge, L.; Mateescu, B.; Howes, P.D.; Demello, A.J.; Richards, D.A. Enzyme-Assisted Nucleic Acid Detection for Infectious Disease Diagnostics: Moving toward the Point-of-Care. ACS Sens. 2020, 5, 2701–2723. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Ge, A.; Liu, M.; Ma, B.; Ma, C.; Shi, C. A fully integrated hand-powered centrifugal microfluidic platform for ultra-simple and non-instrumental nucleic acid detection. Talanta 2020, 219, 121221. [Google Scholar] [CrossRef] [PubMed]
- Yu, W.; Chen, Y.; Wang, Z.; Qiao, L.; Xie, R.; Zhang, J.; Bian, S.; Li, H.; Zhang, Y.; Chen, A. Multiple authentications of high-value milk by centrifugal microfluidic chip-based real-time fluorescent LAMP. Food Chem. 2021, 351, 129348. [Google Scholar] [CrossRef] [PubMed]
- Yuan, D.; Kong, J.; Li, X.; Fang, X.; Chen, Q. Colorimetric LAMP microfluidic chip for detecting three allergens: Peanut, sesame and soybean. Sci. Rep. 2018, 8, 8682. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Q.J.; Wang, L.; Chen, J.; Wang, R.N.; Shi, Y.H.; Li, C.H.; Zhang, D.M.; Yan, X.J.; Zhang, Y.J. Development and evaluation of a real-time fluorogenic loop-mediated isothermal amplification assay integrated on a microfluidic disc chip (on-chip LAMP) for rapid and simultaneous detection of ten pathogenic bacteria in aquatic animals. J. Microbiol. Methods 2014, 104, 26–35. [Google Scholar] [CrossRef]
- Dhar, B.C.; Lee, N.Y. Lab-on-a-Chip Technology for Environmental Monitoring of Microorganisms. Biochip J. 2018, 12, 173–183. [Google Scholar] [CrossRef]
- Zhou, X.; Liu, D.; Zhong, R.; Dai, Z.; Wu, D.; Wang, H.; Du, Y.; Xia, Z.; Zhang, L.; Mei, X.; et al. Determination of SARS-coronavirus by a microfluidic chip system. Electrophoresis 2004, 25, 3032–3039. [Google Scholar] [CrossRef]

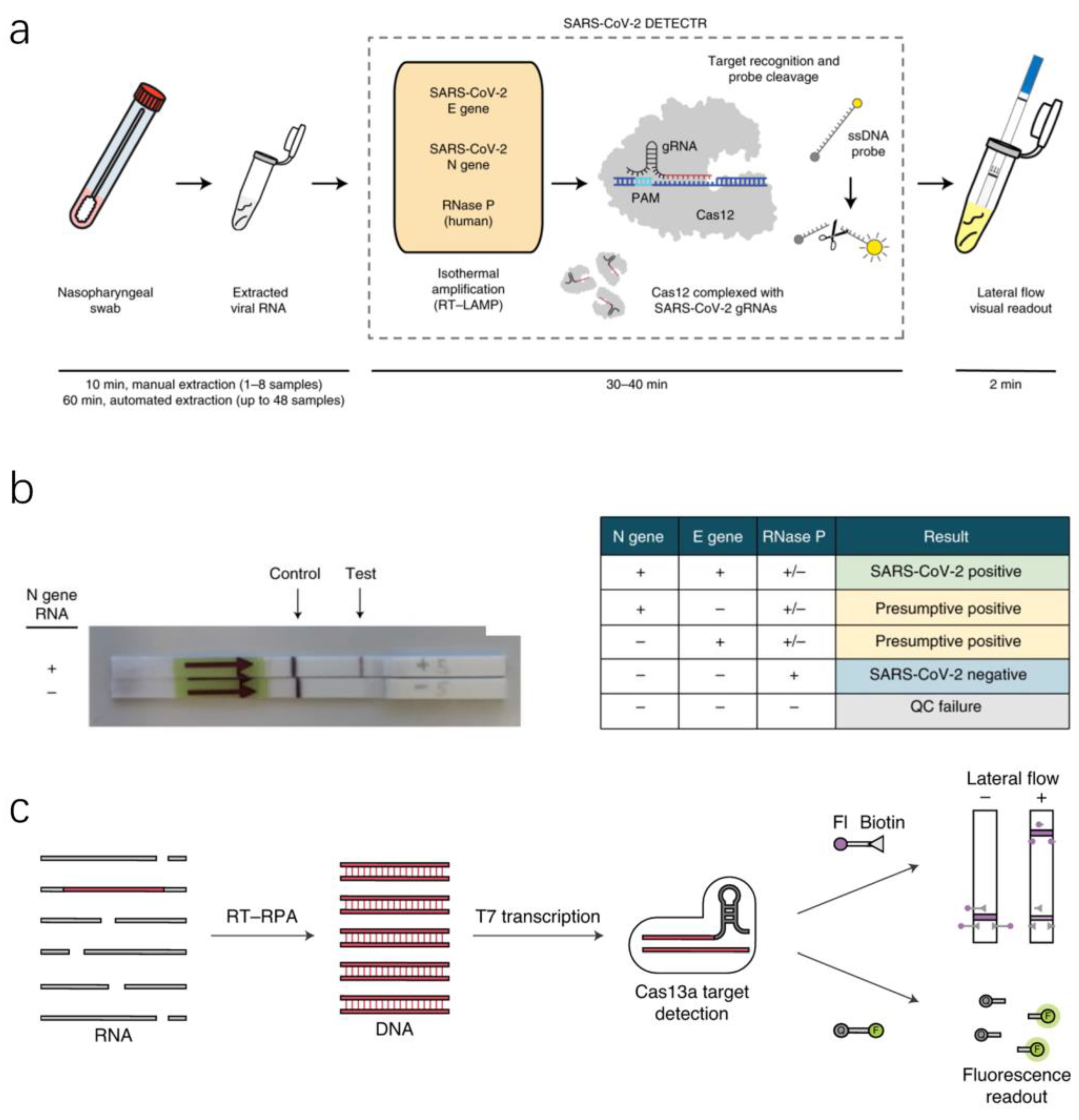

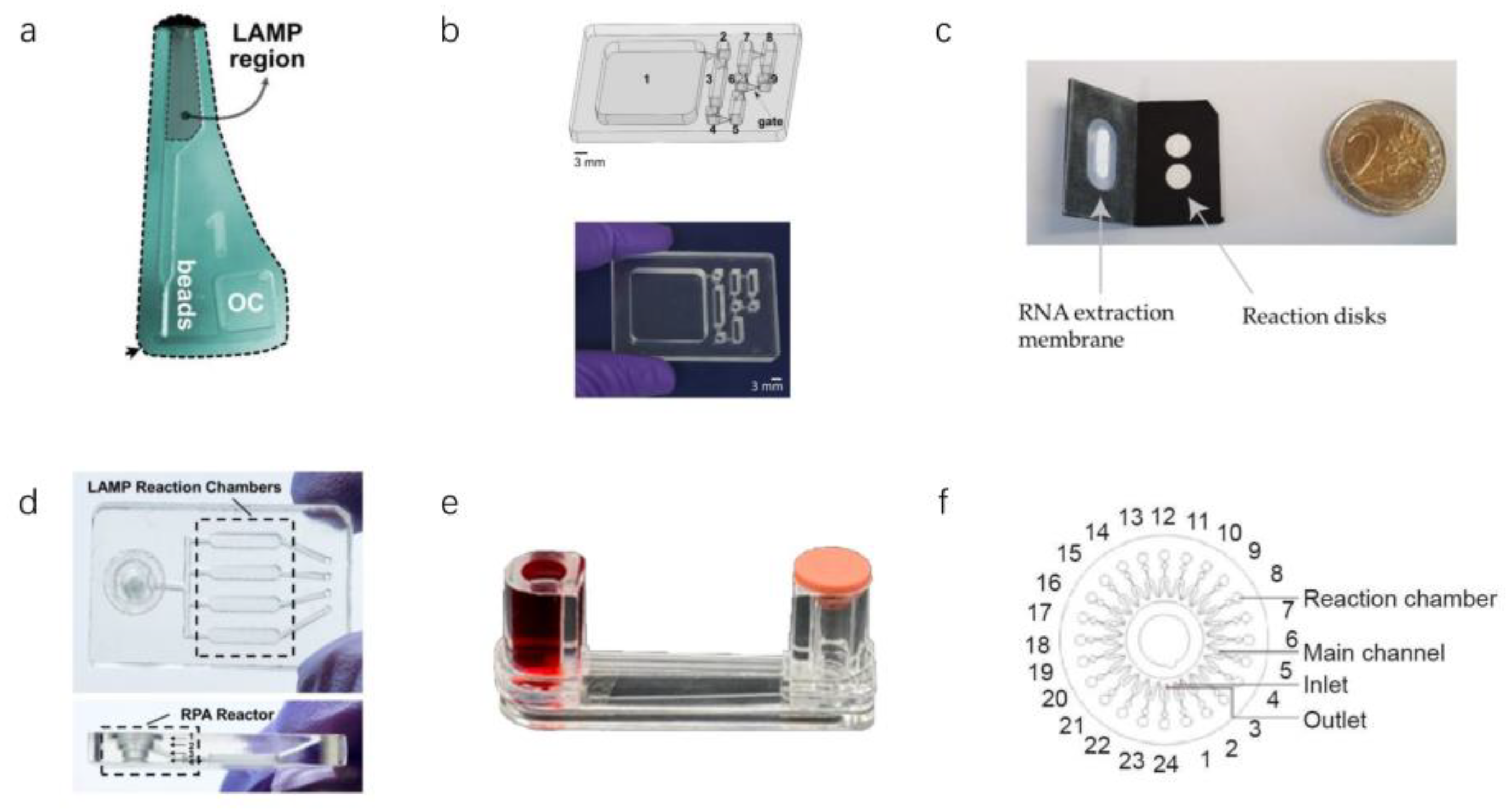
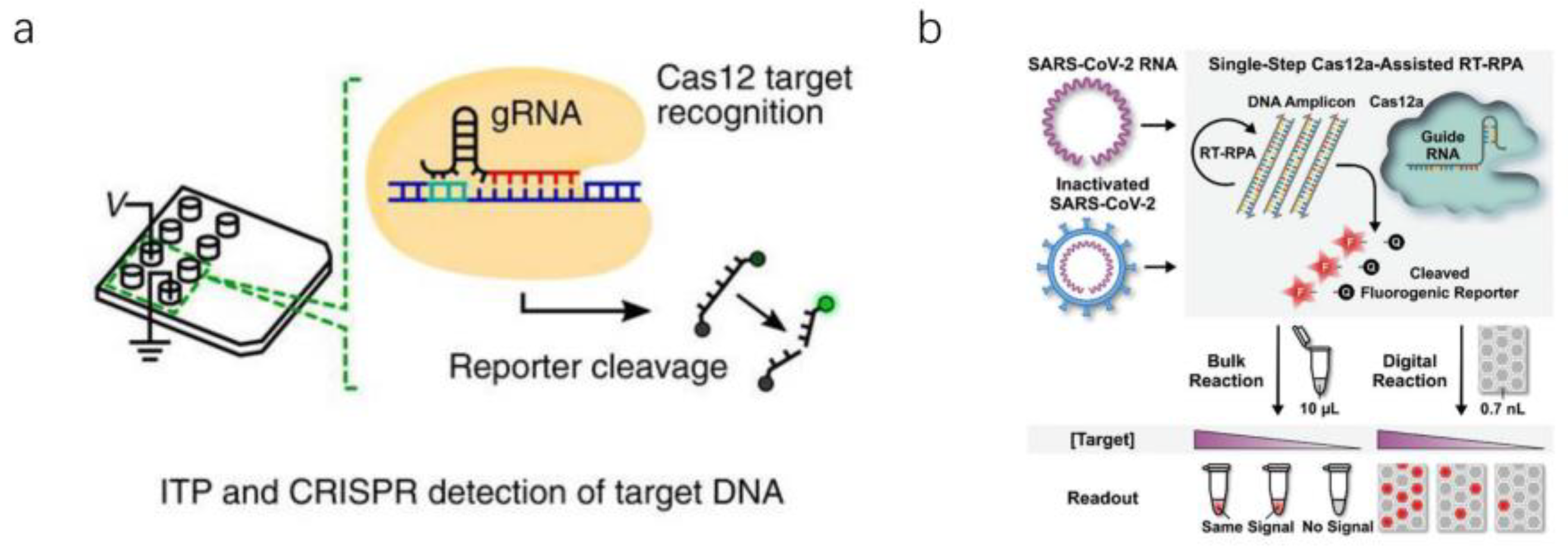
| Sample Type | Sensitivity | Target Genes | Application Scenario | Assay Time | Reference |
|---|---|---|---|---|---|
| Oropharyngeal swabs (OPS) | 97.4% | - | Lack of nucleic acid extraction reagent | PCR45 cycles | [25] |
| Oropharyngeal swabs (OPS) | 94% | N | Laboratory testing | PCR40 cycles | [23] |
| Saliva | 95% | RdRP | Throat swabs and protective equipment are in short supply | PCR30 cycles | [26] |
| Wastewater | - | E | Outdoor water source | 40 cycles | [28] |
| Air | - | - | hospital | 45 cycles | [29] |
| Sample Type | Lower Detection Limit | Sensitivity | Target Genes | Application Scenario | Assay Time | Reference |
|---|---|---|---|---|---|---|
| Clinical sample | - | 100% | RdRP | Isothermal heating instrument | 40 min | [39] |
| Synthetic DNA | 0.4 fM | 100% | RdRP | Laboratory | 100 min | [43] |
| Synthetic DNA | 0.96 pM | - | - | Fluorescence detector | 10 min | [44] |
| Clinical sample, nasopharyngeal swabs (NPS) | 0.1 aM | 95% | RdRp | Fluorescence detector | 30–50 min | [48] |
| Sewage and clinical samples | 10 copies | 93% | RdRp | Visual observation | 20 min | [40] |
| Nasopharyngeal swabs (NPS) | 25–50 copies/Reaction | 95.8% | RdRp, S | Fluorescence detector | 30 min | [41] |
| Sample Type | Lower Detection Limit | Sensitivity | Target Genes | Application Scenario | Assay Time | Reference |
|---|---|---|---|---|---|---|
| Clinical sample | - | 100% | N | Hand warmer | 20 min | [50] |
| Oropharyngeal swabs (OPS), nasopharyngeal swabs (NPS) | - | 95% | E+N | Limited medical resources | 40 min | [52] |
| Oropharyngeal swabs (OPS), nasopharyngeal swabs (NPS) | 42/Reaction | 96% | N, Orf1ab | Limited medical resources | 1 h | [53] |
| Sample Type | Lower Detection Limit | Sensitivity | Target Genes | Application Scenario | Assay Time | Reference |
|---|---|---|---|---|---|---|
| Oropharyngeal swabs (OPS), nasopharyngeal swabs (NPS) | 10 copies/μL | 100% | N | Centrifugal microfluidic detection instrument | 1.5 h | [63] |
| Nasopharyngeal swabs (NPS) | 10 copies/20 μL | 92% | N | Centrifugal microfluidic detection instrument | 95 min | [17] |
| Synthesis Plasmid | - | - | E | Lighting | 306 s | [67] |
| Pretreatment (On-Chip and Off-Chip) | Sample Motion (Centrifugation, Capillary Force) | Lower Detection Limit | Target Genes | Test Method | Assay Time | Reference |
|---|---|---|---|---|---|---|
| On-chip | Magnetic force | 470 copies/mL | ORF1a, N | RT-LAMP | 30–45 min | [69] |
| Off-chip | - | 10 copies/reaction | N | RT-LAMP | 20 min | [71] |
| On-chip | Fold | 1 copy/μL | ORF1ab | RT-LAMP | 20–60 min | [70] |
| On-chip | Propulsive force | 100 (GE)/mL | E, N ORF1a | RT-LAMP | 60 min | [18] |
| Off-chip | - | 0.7 aM | - | RPA | 15 min | [72] |
| Off-chip | Centrifugal force | 50 copies/μL | S, N | LAMP | 90 min | [68] |
| Off-chip | Centrifugal force | 100 and 1000 copies/10 μL | ORF1ab | NASBA | 60 min | [14] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Song, W.; Zhang, T.; Lin, H.; Yang, Y.; Zhao, G.; Huang, X. Conventional and Microfluidic Methods for the Detection of Nucleic Acid of SARS-CoV-2. Micromachines 2022, 13, 636. https://doi.org/10.3390/mi13040636
Song W, Zhang T, Lin H, Yang Y, Zhao G, Huang X. Conventional and Microfluidic Methods for the Detection of Nucleic Acid of SARS-CoV-2. Micromachines. 2022; 13(4):636. https://doi.org/10.3390/mi13040636
Chicago/Turabian StyleSong, Weidu, Taiyi Zhang, Huichao Lin, Yujing Yang, Gaozhen Zhao, and Xiaowen Huang. 2022. "Conventional and Microfluidic Methods for the Detection of Nucleic Acid of SARS-CoV-2" Micromachines 13, no. 4: 636. https://doi.org/10.3390/mi13040636
APA StyleSong, W., Zhang, T., Lin, H., Yang, Y., Zhao, G., & Huang, X. (2022). Conventional and Microfluidic Methods for the Detection of Nucleic Acid of SARS-CoV-2. Micromachines, 13(4), 636. https://doi.org/10.3390/mi13040636