3D-Printed Dip Slides Miniaturize Bacterial Identification and Antibiotic Susceptibility Tests Allowing Direct Mastitis Sample Analysis
Abstract
:1. Introduction
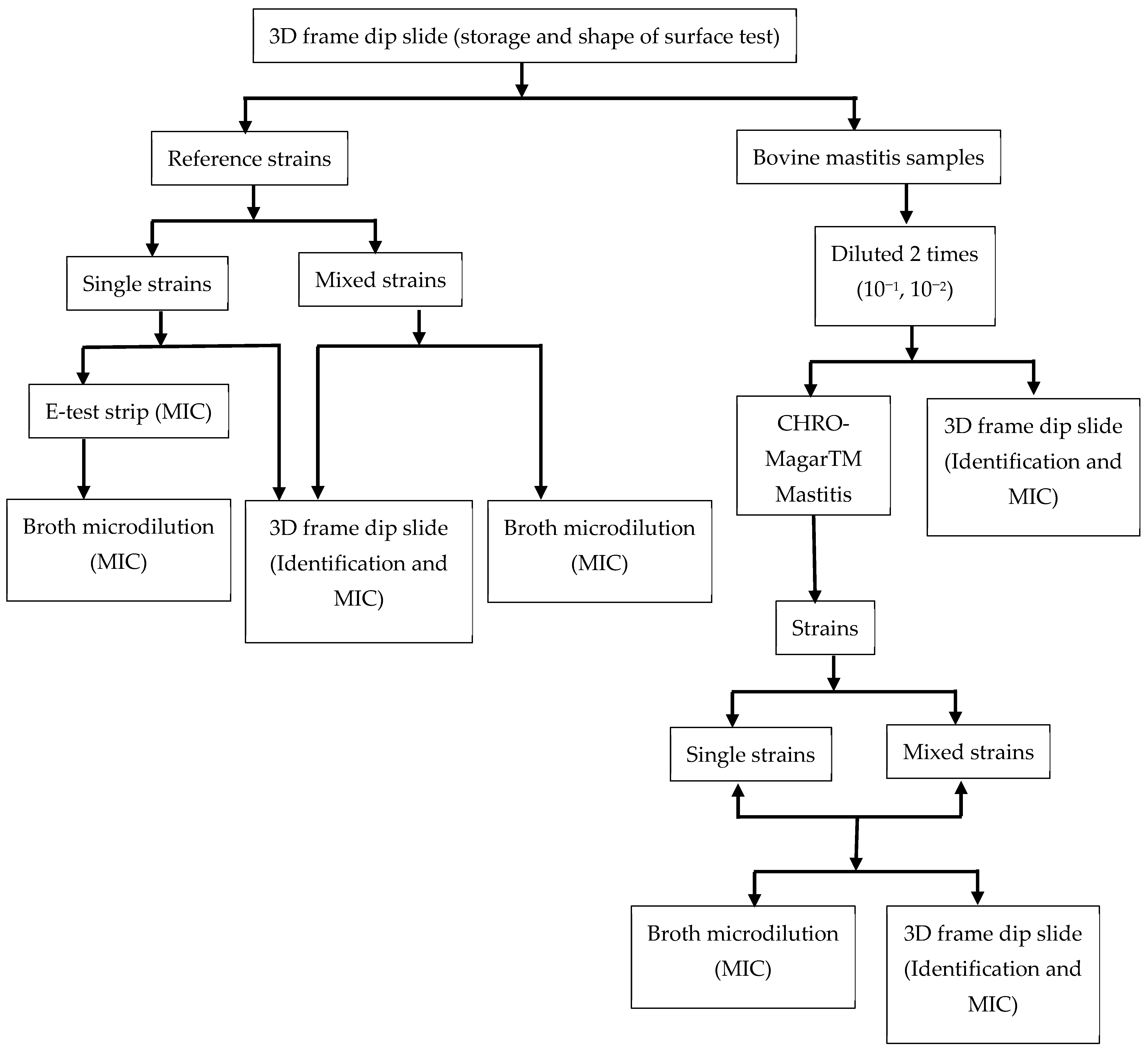
2. Materials and Methods
2.1. Experimental Approach
2.2. Bacterial Strains, Mastitis Samples, Media, and Measurement of MIC
2.3. Bacterial Identification and MIC Measurement Using 3D-Printed Dip Slides
2.3.1. Preparation of 3D Printed Dip-Slide Frame
2.3.2. Detection of Bacterial Species and Determining MIC Using 3D-Printed Dip Slides
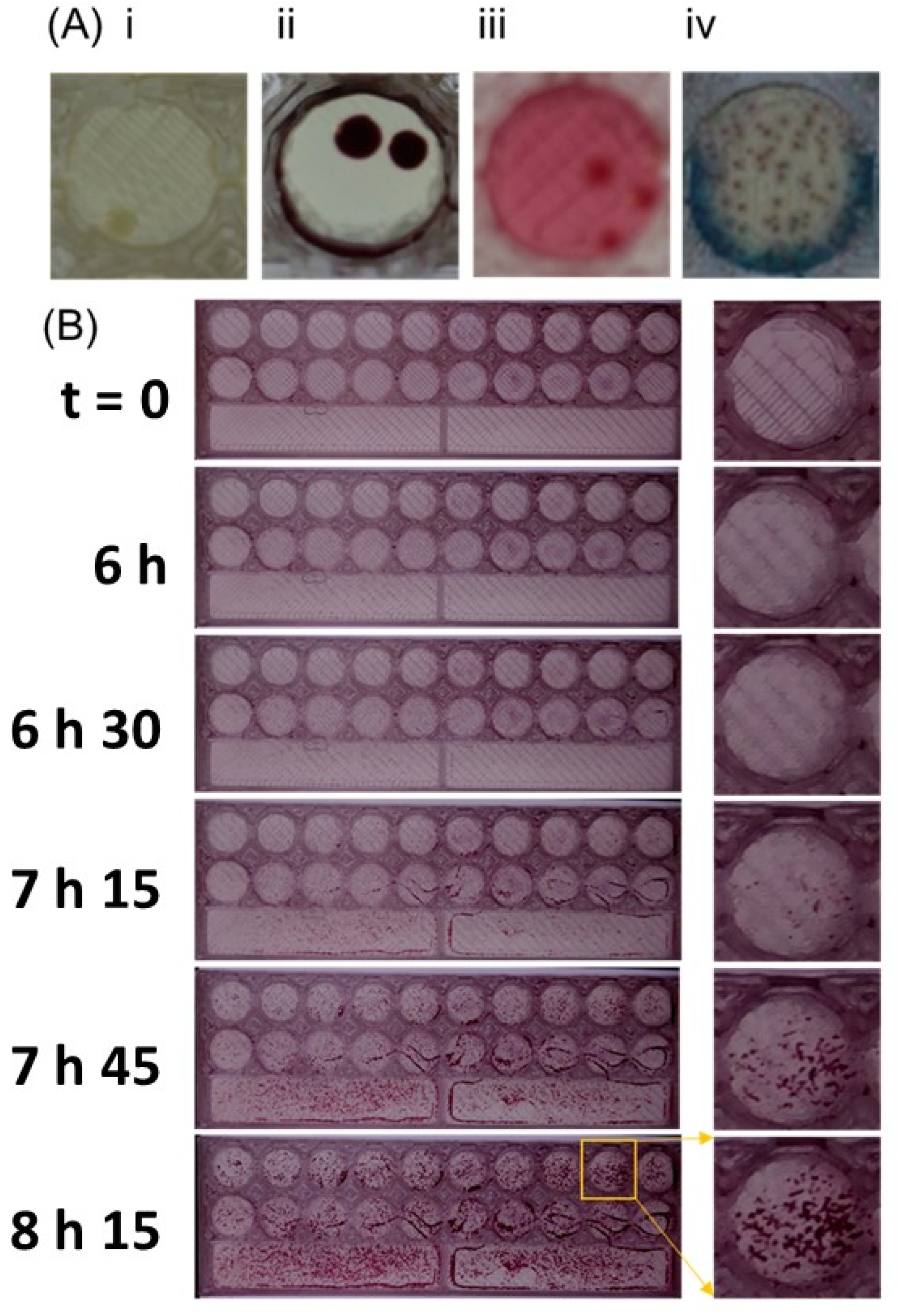
2.4. Time-Lapse Imaging to Check Kinetics of Bacterial Growth on Millifluidic Solid Microbiological Media
2.5. Comparison of MIC Breakpoints with E-Test Trip and Broth Microdilution in a Single Strain and a Mixture of Bacterial Strains
3. Results
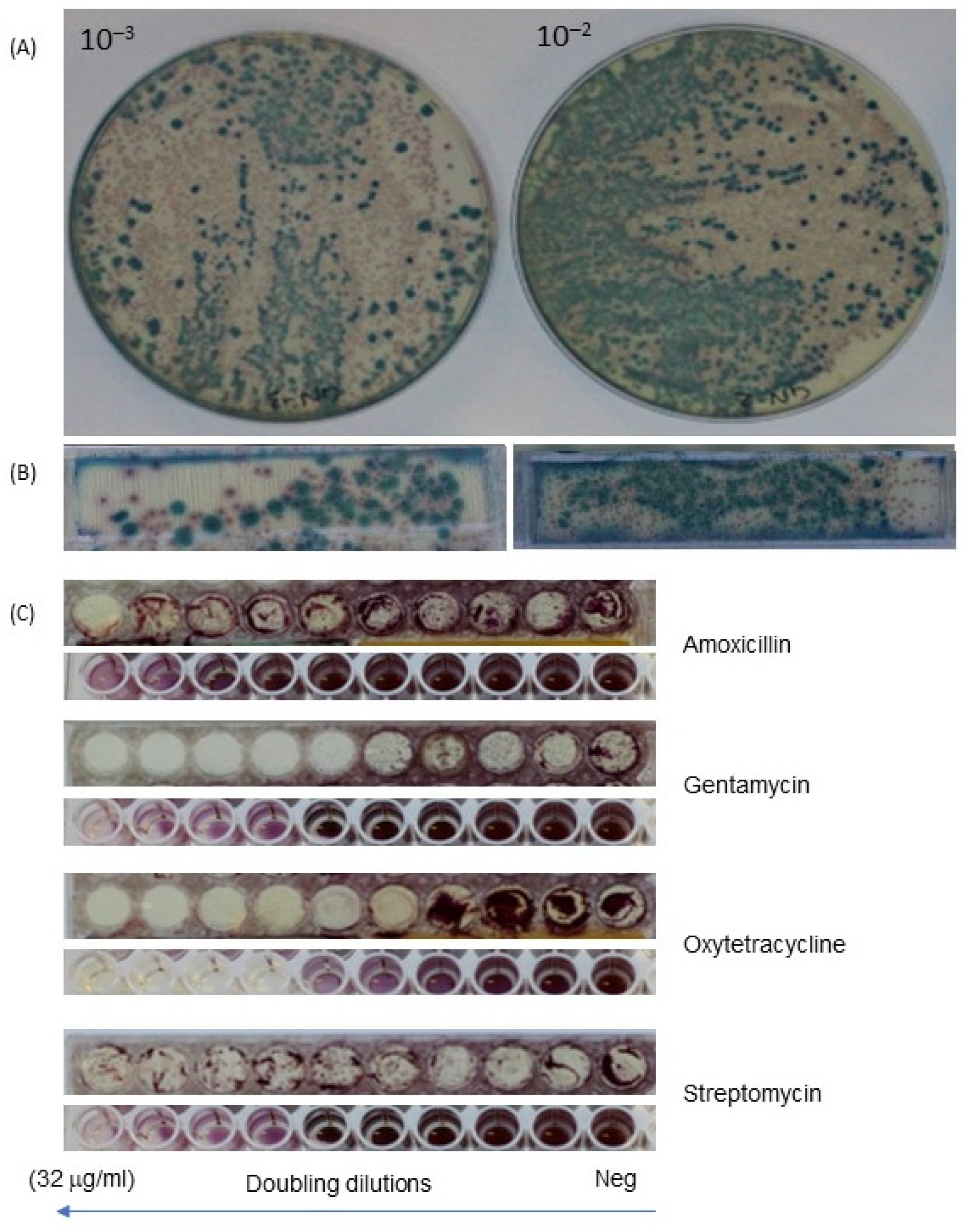
3.1. Use of 3D-Printed Multiplex Dip Slide for Bacterial Identification and Antibiotic Susceptibility Testing
3.2. Characteristics of Multiplex Multifluidic Dip Slides
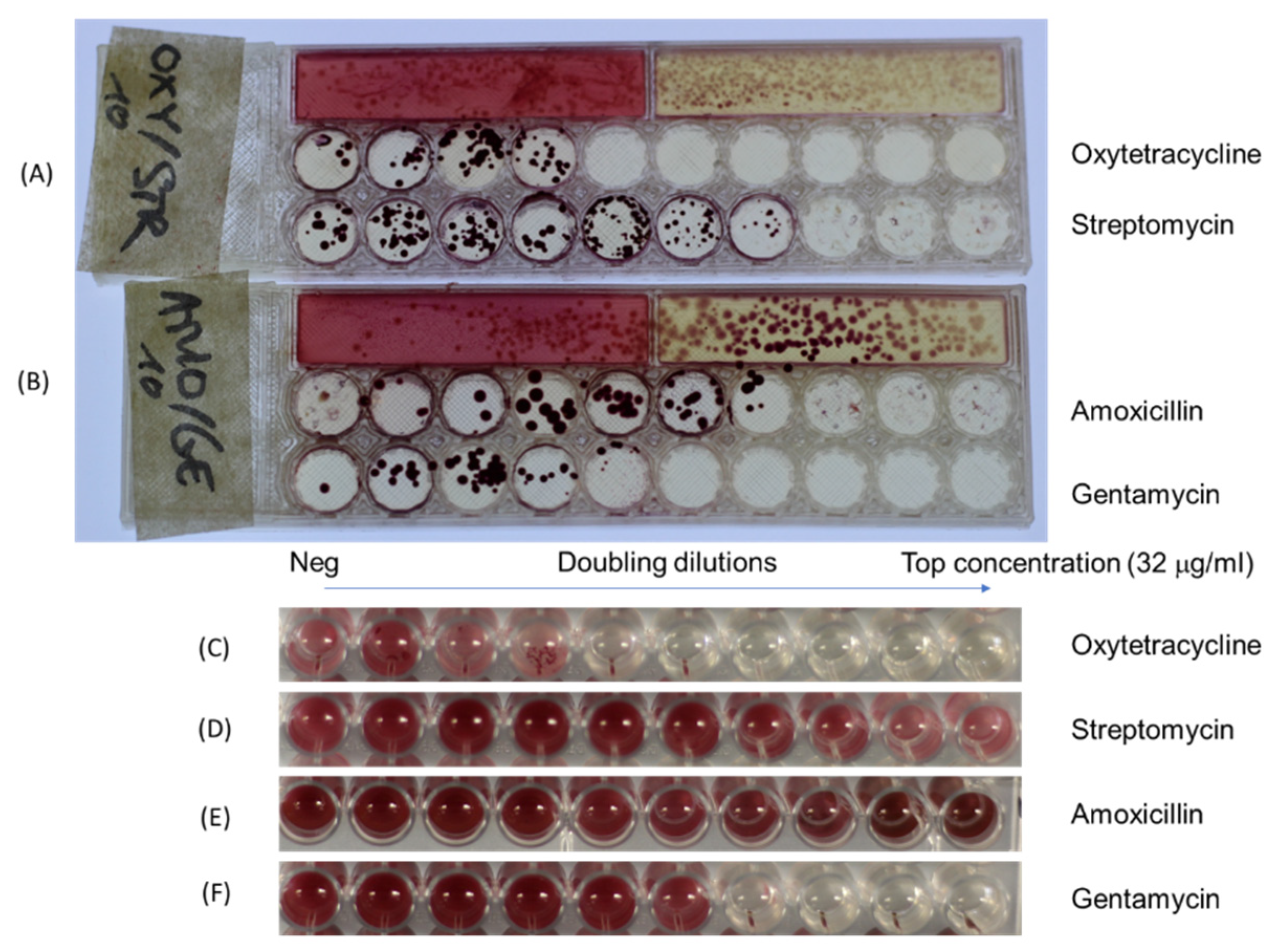
3.3. Detection of Individual Bacteria Isolates and Their Antibiotic Resistance Profile
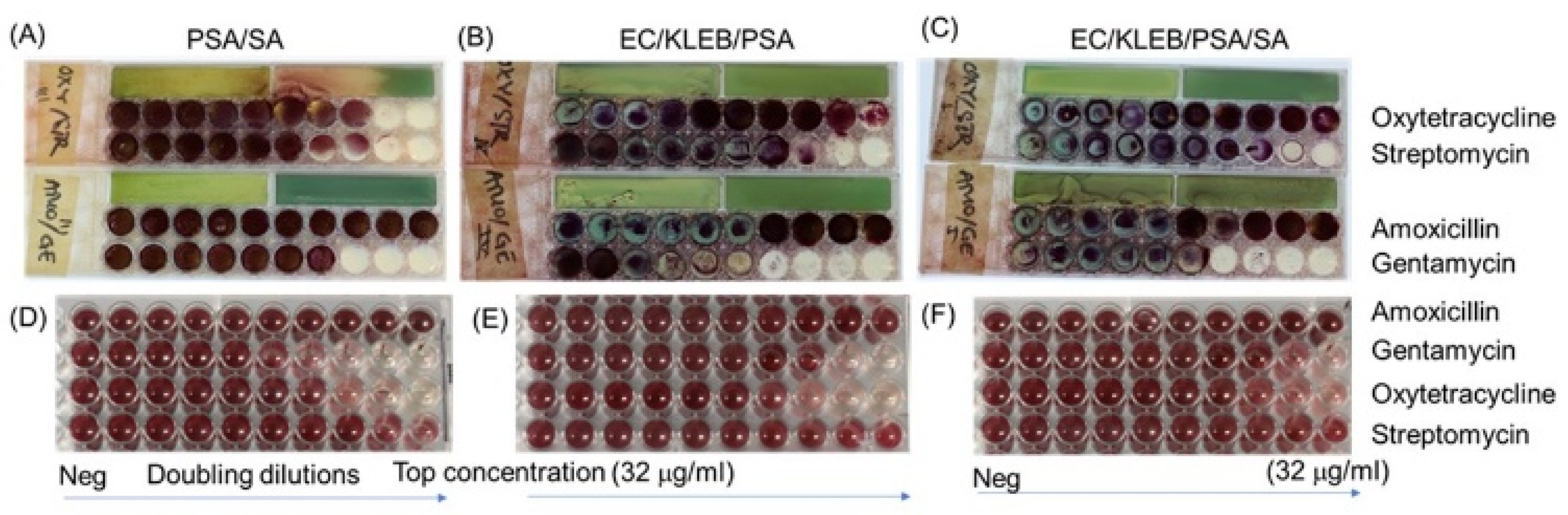
3.4. Direct Testing of Mixtures of Bacteria and Their Antibiotic Resistance Profile
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Hoque, M.N.; Istiaq, A.; Clement, R.A.; Gibson, K.M.; Saha, O.; Islam, O.K.; Abir, R.A.; Sultana, M.; Siddiki, A.Z.; Crandall, K.A.; et al. Insights Into the Resistome of Bovine Clinical Mastitis Microbiome, a Key Factor in Disease Complication. Front. Microbiol. 2020, 11, 860. [Google Scholar] [CrossRef] [PubMed]
- Falentin, H.; Rault, L.; Nicolas, A.; Bouchard, D.S.; Lassalas, J.; Lamberton, P.; Aubry, J.-M.; Marnet, P.-G.; Le Loir, Y.; Even, S. Bovine Teat Microbiome Analysis Revealed Reduced Alpha Diversity and Significant Changes in Taxonomic Profiles in Quarters with a History of Mastitis. Front. Microbiol. 2016, 7, 480. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baumgartner, M.; Bayer, F.; Pfrunder-Cardozo, K.R.; Buckling, A.; Hall, A.R. Resident microbial communities inhibit growth and antibiotic-resistance evolution of Escherichia coli in human gut microbiome samples. PLoS Biol. 2020, 18, e3000465. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Perri, R.; Kolvenbach, B.A.; Corvini, P.F.X. Subsistence and complexity of antimicrobial resistance on a community-wide level. Environ. Microbiol. 2020, 22, 2463–2468. [Google Scholar] [CrossRef] [PubMed]
- Murray, A.K.; Zhang, L.; Yin, X.; Zhang, T.; Buckling, A.; Snape, J.; Gaze, W.H. Novel Insights into Selection for Antibiotic Resistance in Complex Microbial Communities. mBio 2018, 9, e00969-18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ribeiro, C.F.A.; Silveira, G.; Candido, E.S.; Cardoso, M.H.; Espinola Carvalho, C.M.; Franco, O.L. Effects of Antibiotic Treatment on Gut Microbiota and How to Overcome Its Negative Impacts on Human Health. ACS Infect. Dis. 2020, 6, 2544–2559. [Google Scholar] [CrossRef]
- Zhang, L.; Forst, C.V.; Gordon, A.; Gussin, G.; Geber, A.B.; Fernandez, P.J.; Ding, T.; Lashua, L.; Wang, M.; Balmaseda, A.; et al. Characterization of antibiotic resistance and host-microbiome interactions in the human upper respiratory tract during influenza infection. Microbiome 2020, 8, 39. [Google Scholar] [CrossRef] [Green Version]
- Turnidge, J.; Paterson, D.L. Setting and revising antibacterial susceptibility breakpoints. Clin. Microbiol. Rev. 2007, 20, 391–408. [Google Scholar] [CrossRef] [Green Version]
- Mouton, J.W.; Muller, A.E.; Canton, R.; Giske, C.G.; Kahlmeter, G.; Turnidge, J. MIC-based dose adjustment: Facts and fables. J. Antimicrob. Chemother. 2018, 73, 564–568. [Google Scholar] [CrossRef] [Green Version]
- Nielsen, E.I.; Cars, O.; Friberg, L.E. Pharmacokinetic/pharmacodynamic (PK/PD) indices of antibiotics predicted by a semimechanistic PKPD model: A step toward model-based dose optimization. Antimicrob. Agents Chemother. 2011, 55, 4619–4630. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Xie, S.; Ahmed, S.; Wang, F.; Gu, Y.; Zhang, C.; Chai, X.; Wu, Y.; Cai, J.; Cheng, G. Antimicrobial Activity and Resistance: Influencing Factors. Front. Pharmacol. 2017, 8, 364. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- de Boer, M.; Heuer, C.; Hussein, H.; McDougall, S. Minimum inhibitory concentrations of selected antimicrobials against Escherichia coli and Trueperella pyogenes of bovine uterine origin. J. Dairy Sci. 2015, 98, 4427–4438. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Klumper, U.; Recker, M.; Zhang, L.; Yin, X.; Zhang, T.; Buckling, A.; Gaze, W.H. Selection for antimicrobial resistance is reduced when embedded in a natural microbial community. ISME J. 2019, 13, 2927–2937. [Google Scholar] [CrossRef]
- Benkova, M.; Soukup, O.; Marek, J. Antimicrobial susceptibility testing: Currently used methods and devices and the near future in clinical practice. J. Appl. Microbiol. 2020, 129, 806–822. [Google Scholar] [CrossRef] [PubMed]
- Jenkins, S.G.; Schuetz, A.N. Current concepts in laboratory testing to guide antimicrobial therapy. Mayo Clin. Proc. 2012, 87, 290–308. [Google Scholar] [CrossRef] [Green Version]
- Lemos, T.C.; Cogo, L.L.; Maestri, A.C.; Hadad, M.; Nogueira, K.D.S. Is it possible to perform bacterial identification and antimicrobial susceptibility testing with a positive blood culture bottle for quick diagnosis of bloodstream infections? Rev. Soc. Bras. Med. Trop. 2018, 51, 215–218. [Google Scholar] [CrossRef] [Green Version]
- Pereira, D.C.; Goldani, L.Z. Integrating Bacterial Identification and Susceptibility Testing: A Simple and Rapid Approach to Reduce the Turnaround Time in the Management of Blood Cultures. BioMed Res. Int. 2019, 2019, 8041746. [Google Scholar] [CrossRef]
- Choi, J.; Jung, Y.G.; Kim, J.; Kim, S.; Jung, Y.; Na, H.; Kwon, S. Rapid antibiotic susceptibility testing by tracking single cell growth in a microfluidic agarose channel system. Lab Chip 2013, 13, 280–287. [Google Scholar] [CrossRef]
- Choi, J.; Jeong, H.Y.; Lee, G.Y.; Han, S.; Han, S.; Jin, B.; Lim, T.; Kim, S.; Kim, D.Y.; Kim, H.C.; et al. Direct, rapid antimicrobial susceptibility test from positive blood cultures based on microscopic imaging analysis. Sci. Rep. 2017, 7, 1148. [Google Scholar] [CrossRef]
- Toosky, M.N.; Grunwald, J.T.; Pala, D.; Shen, B.; Zhao, W.; D’Agostini, C.; Coghe, F.; Angioni, G.; Motolese, G.; Abram, T.; et al. A rapid, point-of-care antibiotic susceptibility test for urinary tract infections. J. Med. Microbiol. 2020, 69, 52. [Google Scholar]
- Mo, M.; Yang, Y.; Zhang, F.; Jing, W.; Iriya, R.; Popovich, J.; Wang, S.; Grys, T.; Haydel, S.E.; Tao, N. Rapid Antimicrobial Susceptibility Testing of Patient Urine Samples Using Large Volume Free-Solution Light Scattering Microscopy. Anal. Chem. 2019, 91, 10164–10171. [Google Scholar] [CrossRef] [PubMed]
- Bär, J.; Boumasmoud, M.; Kouyos, R.D.; Zinkernagel, A.S.; Vulin, C. Efficient microbial colony growth dynamics quantification with ColTapp, an automated image analysis application. Sci. Rep. 2020, 10, 16084. [Google Scholar] [CrossRef] [PubMed]
- Åkerlund, A.; Jonasson, E.; Matuschek, E.; Serrander, L.; Sundqvist, M.; Kahlmeter, G. EUCAST rapid antimicrobial susceptibility testing (RAST) in blood cultures: Validation in 55 European laboratories. J. Antimicrob. Chemother. 2020, 75, 3230–3238. [Google Scholar] [CrossRef] [PubMed]
- Diep, T.T.; Ray, P.P.; Edwards, A.D. Methods for rapid prototyping novel labware: Using CAD and desktop 3D printing in the microbiology laboratory. Lett. Appl. Microbiol. 2021, 74, 247–257. [Google Scholar] [CrossRef]
- Needs, S.H.; Donmez, S.I.; Bull, S.P.; McQuaid, C.; Osborn, H.M.I.; Edwards, A.D. Challenges in Microfluidic and Point-of-Care Phenotypic Antimicrobial Resistance Tests. Front. Mech. Eng. 2020, 6, 73. [Google Scholar] [CrossRef]
- Michael, A.; Kelman, T.; Pitesky, M. Overview of Quantitative Methodologies to Understand Antimicrobial Resistance via Minimum Inhibitory Concentration. Animals 2020, 10, 1405. [Google Scholar] [CrossRef]
- Flentie, K.; Spears, B.R.; Chen, F.; Purmort, N.B.; DaPonte, K.; Viveiros, E.; Phelan, N.; Krebill, C.; Flyer, A.N.; Hooper, D.C.; et al. Microplate-based surface area assay for rapid phenotypic antibiotic susceptibility testing. Sci. Rep. 2019, 9, 237. [Google Scholar] [CrossRef] [Green Version]
- Avesar, J.; Rosenfeld, D.; Truman-Rosentsvit, M.; Ben-Arye, T.; Geffen, Y.; Bercovici, M.; Levenberg, S. Rapid phenotypic antimicrobial susceptibility testing using nanoliter arrays. Proc. Natl. Acad. Sci. USA 2017, 114, E5787–E5795. [Google Scholar] [CrossRef] [Green Version]
- Wistrand-Yuen, P.; Malmberg, C.; Fatsis-Kavalopoulos, N.; Lubke, M.; Tangden, T.; Kreuger, J. A Multiplex Fluidic Chip for Rapid Phenotypic Antibiotic Susceptibility Testing. mBio 2020, 11, e03109-19. [Google Scholar] [CrossRef] [Green Version]
- Lee, K.; Kim, D.W.; Lee, D.H.; Kim, Y.S.; Bu, J.H.; Cha, J.H.; Thawng, C.N.; Hwang, E.-M.; Seong, H.J.; Sul, W.J.; et al. Mobile resistome of human gut and pathogen drives anthropogenic bloom of antibiotic resistance. Microbiome 2020, 8, 2. [Google Scholar] [CrossRef] [Green Version]
- Letten, A.D.; Baumgartner, M.; Pfrunder-Cardozo, K.R.; Levine, J.M.; Hall, A.R. Human-associated microbiota suppress invading bacteria even under disruption by antibiotics. ISME J. 2021, 15, 2809–2812. [Google Scholar] [CrossRef] [PubMed]
- Diep, T.T.; Bizley, S.; Ray, P.P.; Edwards, A.D. MicroMI: A portable microbiological mobile incubator that uses inexpensive lithium power banks for field microbiology. HardwareX 2021, 10, e00242. [Google Scholar] [CrossRef] [PubMed]


| Testing Samples | Methods | Antibiotic MIC (mg/mL) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Reference Strains | AMP | AMO | AMI | CEF | CEFO | OFL | OXT | STR | GEN | IMI | |
| Escherichia coli ATCC 25922 (EC) | Frame | 4 | >32 | 0.5 | 16 | >32 | 2 | 1 | 0.125 | ||
| MIC broth | 4 | >32 | 0.5 | 16 | >32 | 2 | 1 | 0.125 | |||
| E—strip test | 4 | 1 | 0.125 | ||||||||
| Klebsiella pneuminae ATCC 13883 (KB) | Frame | >32 | >32 | 1 | 16 | >32 | 2 | 1 | 0.5 | ||
| MIC broth | >32 | >32 | 1 | 16 | >32 | 2 | 1 | 0.5 | |||
| E—strip test | >32 | 1 | 0.5 | ||||||||
| Pseudomonas aeruginosa ATCC 10145 (PSA) | Frame | >32 | >32 | 2 | 16 | >32 | 2 | 0.5 | 1 | ||
| MIC broth | >32 | >32 | 2 | 16 | >32 | 2 | 0.5 | 1 | |||
| E—strip test | 0.5 | 1 | |||||||||
| Staphylococcus aureus ATCC 12600 (SA) | Frame | <0.125 | >32 | 1 | 16 | >32 | 2 | ||||
| MIC broth | <0.125 | >32 | 1 | 16 | >32 | 2 | |||||
| E—strip test | 0.032 | ||||||||||
| Direct from milk mastitis samples | |||||||||||
| 19.02 MRF (EC) | Frame | >32 | 1 | >32 | 4 | ||||||
| MIC broth | >32 | 1 | >32 | 4 | |||||||
| 19.03 MRF (Klebsiella sp.—K) | Frame | >32 | 2 | 16 | 4 | ||||||
| MIC broth | >32 | 2 | 16 | 4 | |||||||
| 19.07 MRF (SA) | Frame | >32 | >32 | >32 | 4 | ||||||
| MIC broth | >32 | >32 | >32 | 4 | |||||||
| 19.08 MRF (SA) | Frame | 16 | 8 | ||||||||
| MIC broth | 16 | 8 | |||||||||
| 19.09 MRF (SA) | Frame | >32 | 16 | >32 | 8 | ||||||
| MIC broth | >32 | 16 | >32 | 8 | |||||||
| 19.09 MRF (Klebsiella sp.) | Frame | >32 | >32 | >32 | 16 | 4 | 2 | ||||
| MIC broth | >32 | >32 | >32 | 16 | 4 | 2 | |||||
| 19.10 MRF (EC) | Frame | >32 | 1 | >32 | 2 | ||||||
| MIC broth | >32 | 1 | >32 | 2 | |||||||
| 19.10 MRF (Streptococcus uberis) | Frame | >32 | 16 | ||||||||
| MIC broth | >32 | 16 | |||||||||
| 19.11 MRF (Streptococcus agalactiae) | Frame | 8 | 16 | ||||||||
| MIC broth | 8 | 16 | |||||||||
| 19.14 MRF (PSA) | Frame | >32 | >32 | 1 | |||||||
| MIC broth | >32 | >32 | 1 | ||||||||
| 19.15 MRF (S. aureus—SA) | Frame | 16 | >32 | 2 | |||||||
| MIC broth | 16 | >32 | 2 | ||||||||
| 19.16 MRF (EC) | Frame | 16 | 8 | 2 | >32 | >32 | |||||
| MIC broth | 16 | 8 | 2 | >32 | >32 | ||||||
| 19.16 MRF (SA) | Frame | >32 | 1 | 8 | 16 | ||||||
| MIC broth | >32 | 1 | 8 | 16 | |||||||
| 19.17 MRF (Streptococcus agalactiae) | Frame | 8 | 16 | ||||||||
| MIC broth | 8 | 16 | |||||||||
| 19.17 MRF (SA) | Frame | >32 | 8 | 16 | 4 | ||||||
| MIC broth | >32 | 8 | 16 | 4 | |||||||
| 20. 20 MRF (S. aureus—SA) | Frame | 8 | 32 | ||||||||
| MIC broth | 8 | 32 | |||||||||
| 20. 20 MRF (PSA) | Frame | >32 | >32 | >32 | 8 | ||||||
| MIC broth | >32 | >32 | >32 | 8 | |||||||
| Testing Samples | Methods | Antibiotic MIC (mg/mL) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| AMP | AMO | AMI | CEF | CEFO | OFL | OXT | STR | GEN | IMI | ||
| Reference Strains | |||||||||||
| EC/SA/KB (Reference strains) | Frame | >32 | >32 | 2 | 16 | >32 | 2 | ||||
| MIC broth | >32 | >32 | 2 | 16 | >32 | 2 | |||||
| EC/PSA/KB (Reference strains) | Frame | >32 | >32 | 2 | 16 | >32 | 2 | >32 | >32 | >32 | |
| MIC broth | >32 | >32 | 2 | 16 | >32 | 2 | >32 | >32 | >32 | ||
| EC/SA/PSA/KB (Reference strains) | Frame | >32 | >32 | 2 | 16 | >32 | 2 | >32 | >32 | >32 | |
| MIC broth | >32 | >32 | 2 | 16 | >32 | 2 | >32 | >32 | >32 | ||
| Strains Isolated from mastitis samples | |||||||||||
| 19.15 MRF (SA)/20.20 MRF (PSA) | Frame | >32 | >32 | >32 | 8 | ||||||
| MIC broth | >32 | >32 | >32 | 8 | |||||||
| 19.17 MRF (EC)/19.09 MRF (KB)/20.20 MRF (PSA) | Frame | >32 | >32 | >32 | >32 | ||||||
| MIC broth | >32 | >32 | >32 | >32 | |||||||
| 19.17 MRF (EC)/19.15 MRF (SA)/20.20 MRF (PSA)/19.09 MRF (K) | Frame | >32 | >32 | >32 | >32 | ||||||
| MIC broth | >32 | >32 | >32 | >32 | |||||||
| 19.17 MRF (EC)/19.09 MRF (K) | Frame | >32 | 16 | >32 | >32 | ||||||
| MIC broth | >32 | 16 | >32 | >32 | |||||||
| 19.17 MRF (Streptococcus agalactiae) + 20.20 MRF (S. aureus—SA) | Frame | 8 | >32 | ||||||||
| MIC broth | 8 | >32 | |||||||||
| 19.10 MRF (Streptococcus uberis) + 19.15 MRF (S. aureus—SA) | Frame | >32 | >32 | ||||||||
| MIC broth | >32 | >32 | |||||||||
| Direct from milk mastitis samples | |||||||||||
| 19.07 MRF | Frame | >32 | >32 | >32 | 4 | ||||||
| MIC broth | >32 | >32 | >32 | 4 | |||||||
| 19.09 MRF | Frame | >32 | >32 | >32 | 16 | 4 | 2 | 16 | >32 | 8 | |
| MIC broth | >32 | >32 | >32 | 16 | 4 | 2 | 16 | >32 | 8 | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Diep, T.T.; Bizley, S.; Edwards, A.D. 3D-Printed Dip Slides Miniaturize Bacterial Identification and Antibiotic Susceptibility Tests Allowing Direct Mastitis Sample Analysis. Micromachines 2022, 13, 941. https://doi.org/10.3390/mi13060941
Diep TT, Bizley S, Edwards AD. 3D-Printed Dip Slides Miniaturize Bacterial Identification and Antibiotic Susceptibility Tests Allowing Direct Mastitis Sample Analysis. Micromachines. 2022; 13(6):941. https://doi.org/10.3390/mi13060941
Chicago/Turabian StyleDiep, Tai The, Samuel Bizley, and Alexander Daniel Edwards. 2022. "3D-Printed Dip Slides Miniaturize Bacterial Identification and Antibiotic Susceptibility Tests Allowing Direct Mastitis Sample Analysis" Micromachines 13, no. 6: 941. https://doi.org/10.3390/mi13060941






