The results section of our study focuses on evaluating the performance of various alignment programs in terms of quality and time efficiency. The tests were performed on a CentOS 7 Linux server with 40 cores (2 × Intel Xeon Gold 6230, 2.20 GHz) and 384 GB of RAM. The workstation also featured four GPUs (Tesla T4 Driver Version 460.27.04, CUDA Version: 11.2) with 16 GB each. In terms of storage, it has four 8 TB SATA HDDs in a RAID 5 configuration for mass storage (where data were stored), two 1 TB SATA SSDs in a RAID 0 configuration for scratch, and two 240 GB SATA SSDs. This machine is housed within the Biocomputing Unit data center at the Spanish National Centre for Biotechnology (CNB-CSIC).
For Warp, we used a different machine with Windows 10 Pro 64-bit (10.0, Build 19045). The desktop has eight cores (AMD Ryzen 7 1700, 3.0 GHz) and 64 GB of RAM (4 × 16 GB). The single RTX 2080 Ti with 11 GB of memory uses driver 536.23. In terms of storage, it has one 500 GB HDD and one 120 GB SSD connected via SATA 600, where the data were copied before the test. This machine is housed within the Sitola laboratory, a joint facility of the Faculty of Informatics and Institute of Computer Science at Masaryk University and the CESNET association at Masaryk University in Brno.
4.1. Quality
The quality assessment of the alignment algorithms involved the analysis of both simulated and real datasets. For the simulated dataset, we evaluated alignment quality by examining the aspect of the signal pattern in the resulting micrographs. This allowed us to assess how well the algorithms handled deformations, noise, and shifts in the simulated data.
In the case of the real dataset, our focus was on measuring the limit resolution criteria of the CTF estimation using two different programs. This criterion provides information about the maximum level of detail that can be resolved in the images. Additionally, we examined the energy decay in the power spectrum density for higher frequencies. This analysis was conducted on three different datasets, each comprising 30 movies. By evaluating these parameters, the study aimed to gain insights into the accuracy and effectiveness of the alignment algorithms in capturing fine details and preserving image quality.
4.1.1. Phantom Movies
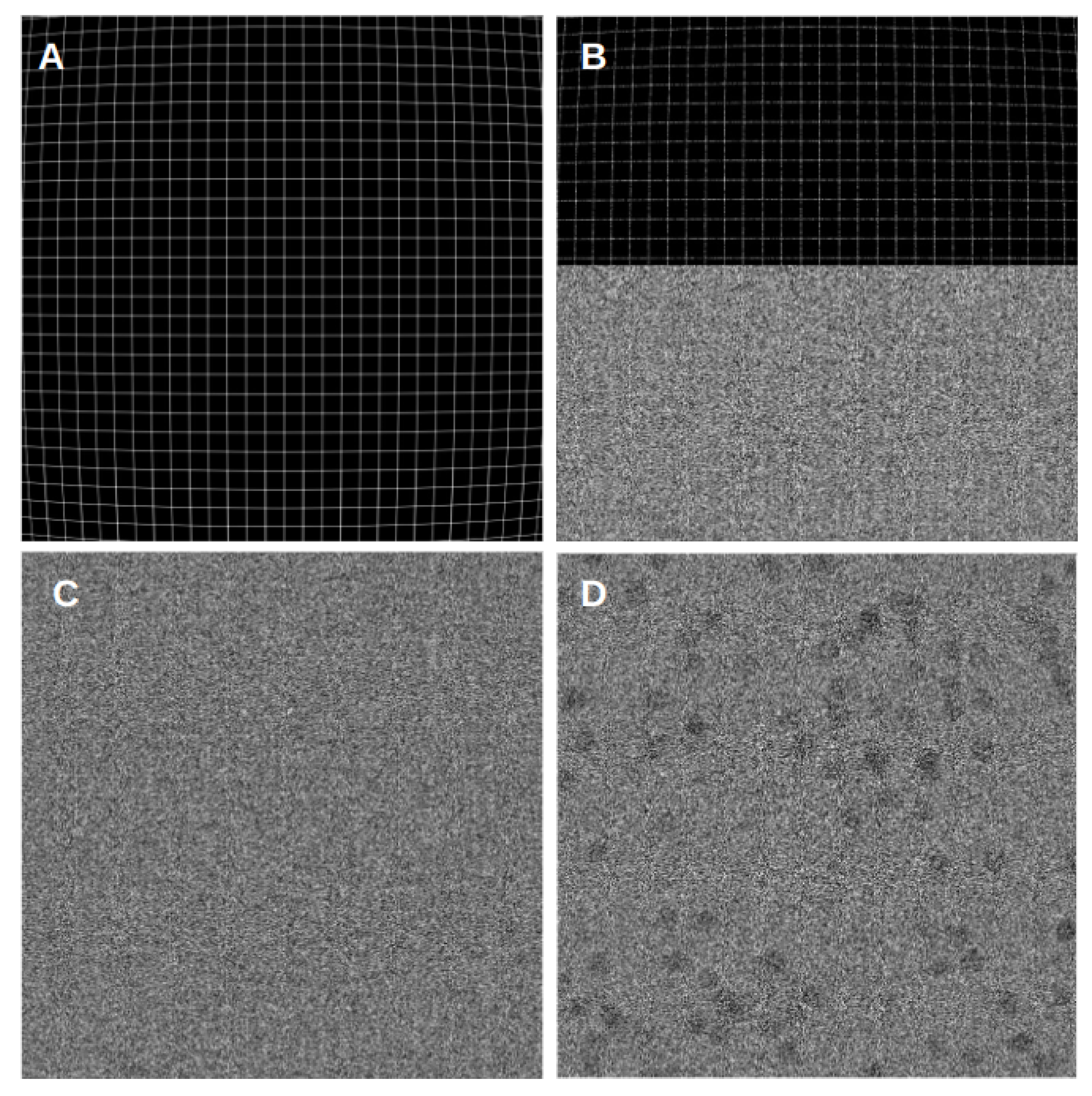
Figure 2 presents examples of simulated aligned phantom movies. We used grid-based phantom movies to examine the grid pattern of the resulting micrographs and evaluate how well the algorithms handle common issues such as noise, deformation, and shifts. For this kind of movie, we have both noiseless and noisy versions, with the noisy ones simulating the ice and dose found in typical cryoEM experiments. Additionally, we included a cryo-EM-based simulated movie that allows for a comparison in a more realistic cryoEM scenario. These types of phantom movies were designed to be a middle ground between particle projections and a regular grid. Furthermore, all types of movies were subject to barrel deformation, which was applied to the movie frames to simulate the common dome effect observed in cryoEM. This results in a more rigid pattern in the center region, gradually curving towards the edges.
By studying the alignment quality of these simulated movies, we aimed to gain valuable insights into the algorithms’ results and their ability to accurately align movies with different characteristics and complexities. However, we cannot reject the possibility that the relative alignment accuracy of the methods may be affected by this particular type of signal.
The visual analysis presented in
Figure 3 provides insights into the quality of the alignment algorithms by comparing the alignment results for pristine movies and their noisy counterparts. This allows for the evaluation of how each algorithm handles deformations, shifts, and noise present in the phantom movies.
In our experiments, we conducted tests on movies of different sizes and observed interesting trends in alignment quality. It became evident that all programs successfully aligned the center region of the movies. However, as we moved towards the edges, the algorithms encountered difficulties and exhibited poor alignments, resulting in a blurring effect in the mesh pattern. This blurring effect was also noticeable in the noisy movies, where incorrect alignment caused the mesh pattern at the edges to appear blurred. This behavior remained consistent across movies of varying sizes and frame counts. When comparing the results of different algorithms, FlexAlign, in particular, excelled at achieving a cleaner pattern in both pristine and noisy images at the edges, indicating superior alignment capabilities.
Furthermore, as we experimented with higher-dimensional images, we simulated a proportional increase in beam-induced movement (BIM) and drift. Consequently, most of the algorithms struggled to align the edges correctly and, in some cases, even struggled to align the central region in the largest dimension. These findings underscore the challenges alignment algorithms encounter when handling larger image deformations, particularly in terms of preserving alignment accuracy at the edges.
The analysis in
Figure 4 allows us to observe the performance of the alignment algorithms when subjected to a 60-pixel shift. By comparing the aligned movies to the pristine and noisy versions, we can assess how effectively each algorithm handles shifts in the phantom movie.
For the experiments, we used movies of the same size (4096 × 4096 × 70) but introduced varying global drift. Specifically, we introduced eight shifts ranging from 50 to 120 pixels, with increments of 10 pixels. Similar to the previous experiment, we observed a consistent pattern where all programs successfully aligned the center region of the movies. However, as we moved toward the edges, the algorithms encountered difficulties, resulting in poor alignment.
It is worth noting that as we increased the drift most of the algorithms struggled to align the edges correctly. FlexAlign, in particular, was sensitive to these changes, as it typically expects frame movement within normal cryoEM conditions. It exhibited alignment failures at shifts over 90 pixels using the default settings. To determine whether FlexAlign could handle larger shifts, we increased the maximum expected shift parameter in the algorithm, leading to improved alignment accuracy.
These findings highlight the challenges alignment algorithms face when confronted with increasing drift levels. They also underscore the importance of parameter optimization and understanding the specific limitations and sensitivities of each algorithm to achieve accurate alignment results, particularly in the presence of significant shifts.
Figure 5 enables a comparison in a more realistic cryoEM scenario. These types of phantom movies were intended to replicate not only cryoEM conditions, such as shifts, dose, noise, and deformations but also to simulate their main signal, which is particle projections. This way, the relative alignment accuracy of the methods is not affected by the particularity of the grid-type signal studied before.
These results corroborated our earlier findings. Despite differences in signal complexity, all programs effectively aligned the central region of the movies. However, as they approached the edges, the algorithms encountered challenges and produced suboptimal alignments, resulting in visible blurring in the particle projections at the micrograph’s periphery. When comparing the outcomes of different algorithms, FlexAlign once again demonstrated its ability to excel by consistently achieving a cleaner pattern, indicating superior alignment capabilities at the edges.
4.1.2. EMPIAR Movies
Table 2 compares the CTF resolution limit, expressed in Angstroms (Å), for each EMPIAR entry using the Gctf and Xmipp methods. The mean value indicates the average CTF resolution limit obtained from each respective method, while the standard deviation represents the variation or dispersion of the CTF criteria around this mean value. By comparing the mean and standard deviation values between the Gctf and Xmipp methods for each EMPIAR entry, we can gain insights into the consistency and accuracy of the CTF estimation provided by these methods.
For consistency and to assess the potentially significant differences in quality performance among different algorithms, a comprehensive statistical study was conducted. This study aimed to evaluate the significant differences in means both collectively using an ANOVA test and individually by comparing all possible program combinations through paired t-tests.
The ANOVA (Analysis of Variance) test was employed to analyze whether there is a statistically significant difference among the means of the various programs. This test allows us to determine if there are significant variations between the groups as a whole and provides an overall assessment of the statistical significance of the observed differences.
If the ANOVA test yielded a significant result, a post hoc analysis was performed to compare the means of each pair of programs individually. This approach enables us to assess the significance of the differences between specific pairs of programs and identify which programs exhibit statistically different performances.
By conducting the ANOVA test and its subsequent post hoc analysis, we can thoroughly investigate the significant differences in means between the programs under consideration. This statistical analysis enhances our understanding of the variations in quality performance among the different movie alignment algorithms in cryoEM.
Figure 6 presents an analysis of the resolution estimation based on the CTF criteria for various EMPIAR entries, facilitating a clear comparison of the resolution performance among different algorithms.
For EMPIAR entry 10,196, the ANOVA test conducted on the group means did not reveal any significant differences.
For EMPIAR entry 10,288, the ANOVA test detected a significant difference in the group means obtained from the Xmipp program at a confidence level of 0.05. Additionally, the post hoc analysis indicated a significant difference between the movie alignment program Warp and the other programs, signifying a poorer quality performance by Warp in this test. Furthermore, CryoSPARC and MotionCor2 exhibited a statistically significant better resolution limit than Relion MotionCor. Regarding the Gctf metric, the ANOVA test did not identify any significant differences.
For EMPIAR entry 10,314, the ANOVA test found a significant difference only for the Xmipp criteria, indicating that different algorithms performed significantly differently for this dataset. The post hoc analysis further revealed a significant difference between all programs and Relion MotionCor and Warp, with the latter two generating micrographs with lower resolutions than the others.
Figure 7 visually represents the observed PSD trends for each program and EMPIAR entry, enabling an analysis of how different alignments affect the PSD pattern. These plots illustrate the distribution of energy across various frequencies. Ideally, with correct alignment, different algorithms should not alter the PSD patterns. The occurrence of such alterations suggests that the algorithm introduces bias to the image.
Regarding the attenuation of PSD energy at higher frequencies, we observed varying degrees of damping among the programs. CryoSPARC and MotionCor2 exhibited minimal damping as they reached higher frequencies. Conversely, FlexAlign and Warp showed slight damping at higher frequencies. It is important to note that this damping occurs close to the Nyquist frequency, at which point the signal’s energy has largely dissipated. Therefore, this energy reduction has a minimal impact on noise reduction. Additionally, it is worth mentioning that for dataset 10,196 Warp exhibited an unusual behavior, displaying different and less favorable energy decay compared to the others. We believe this issue may be associated with Warp’s difficulty in processing this particular dataset.
Finally, Relion MotionCor appeared to lose energy across frequencies. These differences can be attributed to the interpolation function used to generate the output micrograph. CryoSparc and Motioncor2 likely use Fourier cropping, FlexAlign and Warp employ B-spline interpolation, and Relion uses linear interpolation. Relion’s damping begins at lower frequencies than those detected by XMIPP and Gctf. This observation suggests that we may have had sufficient energy to accurately estimate the CTF and extract the resolution limit at higher frequencies. Furthermore, the XMIPP criteria appear to be more consistent with the damping value since its resolution limit is based on the experimental PSD decay rather than the correlation of the theoretical PSD function with the experimental PSD.
To observe how the alignment by various algorithms impacts image content in real space,
Figure 8 visually illustrates the distribution of pixel values in the aligned movies generated by various algorithms, providing a means for comparative analysis of pixel value distribution across different alignment programs. Our experiments revealed variations in the pixel values of the resulting aligned images, depending on the specific algorithm used.
To assess whether there were statistically significant differences in pixel value distribution, we normalized images produced by different alignment algorithms and conducted pairwise Kolmogorov–Smirnov tests to compare them. This non-parametric test evaluates whether two datasets share the same underlying distribution. In most cases, the test rejected the null hypothesis, indicating that the two datasets originated from distinct distributions. This highlights that achieving consistent information content from the alignment of the same image with different algorithms is not as straightforward as image normalization. Instead, it underscores that different alignment algorithms can yield statistically distinct pixel value distributions.
The coefficients of variation (CV) reveal the extent of variability around the mean of all pixel values, even when considering outliers. Across all distributions, we observed low to moderate CV values, indicating that pixel values are relatively close to the mean and exhibit low variability, despite their statistically different distributions.
One possible explanation for these variations is that different algorithms output distinct representations of mean electron impacts or electron impact counts, even after normalization. This can result in differences in observed pixel values in the aligned images and can affect pixel value distribution.
4.2. Performance
The time performance of the programs was evaluated by calculating the mean execution time based on 10 runs of the same simulated movie. Various movie sizes and conditions, including the number of GPUs for parallel computing, multi-threading, batch processing, and SSD storage, were considered to assess the efficiency of the algorithms in terms of processing speed.
Table 3 provides an overview of the mean performance of each algorithm based on 10 trial runs across three different Cryo-EM movie sizes. Please note that Warp was executed on a different machine than the other programs, making an absolute comparison challenging.
For the smallest movie size (4096 × 4096) with 10 frames, suitable for tomographic tilt movies, MotionCor2, FlexAlign, and Warp were the fastest, with MotionCor2 leading. In contrast, Relion MotionCor lagged, and CryoSPARC was the slowest. With 70 frames, which is common for single-particle analysis (SPA), the three fastest algorithms maintained their superiority, with Warp leading, while Relion and CryoSPARC delivered comparable performances.
As the movie size increased to 7676 × 7420, which is typical of larger movies, processing times increased significantly for all programs. In 10-frame tomographic tilt movies, the fastest algorithms’ performances resembled those of the first movie size, with MotionCor2 as the fastest. Relion MotionCor struggled the most with the larger data, while CryoSPARC was less affected but still notably slower. With 70 frames, MotionCor2’s performance declined compared to FlexAlign and Warp, being almost 10 s slower. CryoSPARC also had a notable processing time increase, while Relion MotionCor was the slowest.
For the third movie size, the increase in processing time was not as significant for all programs. In 10-frame movies, MotionCor2, FlexAlign, and Warp remained the fastest, with MotionCor2 slightly ahead. CryoSPARC ranked fourth, and Relion MotionCor was marginally slower. With 70 frames, all programs except FlexAlign and MotionCor2 experienced a substantial loss in performance, indicating difficulty in handling super-resolution-sized movies. FlexAlign consistently outperformed its peers in this scenario.
Figure 9 offers valuable insights into the scalability of alignment algorithms when utilizing GPU parallel processing. The mean processing times for different movie sizes and GPU configurations enable us to assess how efficiently these algorithms make use of parallel computing resources, which is especially important when dealing with large datasets in single-particle analysis (SPA) experiments. Three different movie sizes were tested on a machine with four GPUs.
For movie size 4096 × 4096 × 70, both FlexAlign and MotionCor2 demonstrated excellent scalability. The processing time remained nearly constant as the task was parallelized across multiple GPUs. In essence, processing one movie on one GPU took approximately the same time as processing four movies on four GPUs. However, CryoSPARC exhibited a slight decrease in performance when parallelizing the processing.
Moving to movie size 7676 × 7420 × 70, all algorithms showed a minor trade-off in performance when increasing the number of GPUs for parallel processing. This trade-off implies a small increase in processing time when using multiple GPUs compared to the ideal condition, where processing one movie on one GPU would be as fast as processing two or three movies on two or three GPUs when parallelizing. Therefore, parallelization had a minimal impact on the overall processing time for all algorithms.
For movie size 11,520 × 8184 × 70, CryoSPARC and FlexAlign continued to exhibit a slight trade-off in performance when increasing the number of GPUs, as observed in the previous sizes. However, MotionCor2 displayed a consistent pattern with no trade-off when processing more movies, indicating stable performance with parallelization. These results highlight that the scalability of alignment algorithms can vary depending on the movie size, and some algorithms may exhibit minor trade-offs in performance when parallel processing is employed.
Apart from GPU parallel computing, another approach to accelerating processing times is the use of multi-threading, a feature implemented in Relion MotionCor. Multi-threading involves allocating more CPU cores specifically for the alignment task, allowing the algorithm to leverage the computational power of multiple cores simultaneously. This can lead to improved performance and faster processing times. By optimizing the number of threads to distribute the workload efficiently across the available CPU cores, significant reductions in processing time were achieved for movies of different sizes.
For movie size 4096 × 4096 × 70, the most efficient configuration involved using one process and 36 threads, distributing the workload across 36 out of the 40 available CPU cores. This configuration reduced the processing time from 33.8 ± 5 s to 14.8 ± 0.2 s, more than halving the time required to process a movie of this size.
In the case of movie size 7676 × 7420 × 70, a similar configuration proved to be the most efficient, utilizing one process and 37 threads for workload distribution. This configuration reduced the processing time from 164 ± 7.9 s to 79.3 ± 1.1 s, again cutting the time by more than half.
Finally, for movie size 11,520 × 8184 × 70, a similar optimization was applied, reducing the processing time from 202.6 ± 22.2 s to 110.8 ± 1.2 s. This configuration involved using one process and 35 threads, decreasing the processing time by approximately a minute and a half.
Certainly, optimizing the use of around 35 threads for processing movies with 70 frames aligns well with the manufacturer’s recommendation and system capabilities. Dividing the number of movie frames (70) by the number of threads (35) results in an integer value of 2, indicating that each thread can efficiently process two frames simultaneously. This level of parallelization is the maximum achievable with the available 40 CPU cores in the system, ensuring an efficient utilization of resources.
Fine-tuning the multi-threading option in Relion, specifically for movies with 70 frames, led to a significant improvement in time performance. This optimization brought Relion MotionCor’s processing times into the same range as the faster algorithms for movie size 4096 × 4096 × 70 and significantly reduced the gap for other movie sizes. While it remained slightly slower than the fastest algorithms, this optimization made Relion MotionCor a more competitive choice in terms of processing time. These results underscore the significance of exploring alternative approaches like multi-threading to enhance the efficiency of movie alignment algorithms.
MotionCor2 offers a batch-processing feature, which can be particularly advantageous when dealing with large datasets. For this experiment, we measured the processing time of MotionCor2 for datasets comprising 20 movies of three different sizes (4096 × 4096 × 70, 7676 × 7420 × 70, and 11,520 × 8184 × 70) both using the batch processing flag and without it. Since MotionCor2 is the only algorithm that offers this option, the comparison was limited to MotionCor2 itself.
In
Table 4, we can observe that batch processing significantly improves the alignment time compared to regular processing with MotionCor2.
For movies sized 4096 × 4096 × 70, using batch processing with one GPU reduced the processing time from 170 s to 100 s for the 20-movie dataset. This equates to an alignment pace of approximately 5 s per movie compared to 9 s per movie without batch processing. Further performance gains were achieved by increasing the number of GPUs to two, with a pace of 4 s per movie. However, adding more GPUs beyond this point did not yield further improvements.
As movie sizes grew to 7676 × 7420 × 70, the advantages of batch processing became even more evident. The best-case scenario, using batch processing with three GPUs, halved the pace from 25 s per movie to 12 s per movie, reducing the processing time from 740 s to 245 s. Yet, we reached a machine limit at three GPUs for this size, with no additional gains observed from further GPU additions.
For the largest movie size, 11,520 × 8184 × 70 (super-resolution size), the performance difference was even more substantial. Batch processing reduced the processing time from 38 min to 20 min, lowering the pace from 114 s per movie to 65 s per movie. Remarkably, the machine limit was reached at just one GPU for this size, indicating that the processing time was constrained by the machine’s data reading and writing capabilities, particularly when handling larger movies.
In summary, batch processing clearly accelerates the alignment process, particularly for larger movie sizes. However, it is crucial to account for machine limitations when scaling up the number of GPUs, as there may be no further performance gain beyond a certain point.
Lastly, an additional experiment was conducted involving placing data on a solid-state drive (SSD) for faster reading and writing operations. This approach aimed to further enhance the overall processing speed by leveraging the faster data transfer capabilities of an SSD.
Table 5 clearly illustrates the significant performance improvement gained by storing data on an SSD as opposed to an HDD. The alignment process demonstrated a notable speed increase, approximately 30%, which remained consistently stable. This improvement was most evident for larger movie sizes or those with more frames.
For the 4096 × 4096 movie size with 10 frames, the performance difference between the SSD and HDD was minimal, with differences in the order of tenths of seconds. However, as the number of frames increased to 70, the performance difference became more substantial, with an approximate difference of 5 s.
As the movie size increased, the performance gap between the SSD and HDD became more pronounced. For the 7676 × 7420 movie size with 70 frames, the difference was almost 10 s, highlighting the substantial advantage of SSD storage. The most significant difference was observed with the largest movie size, which is typically used for super-resolution applications. The performance gap reached approximately 20 s, underscoring the substantial impact of the data reading speed on the overall processing time.
These findings emphasize the importance of considering storage options, especially when processing data in real time or at a fast acquisition pace, and prove that utilizing an SSD for data storage can greatly enhance processing efficiency.