On-Line Fluorescence Microscopy for Identification and Imaging of Apoptotic Cell with Synchrotron-Based Soft X-ray Tomography
Abstract
:1. Introduction
2. Materials and Methods
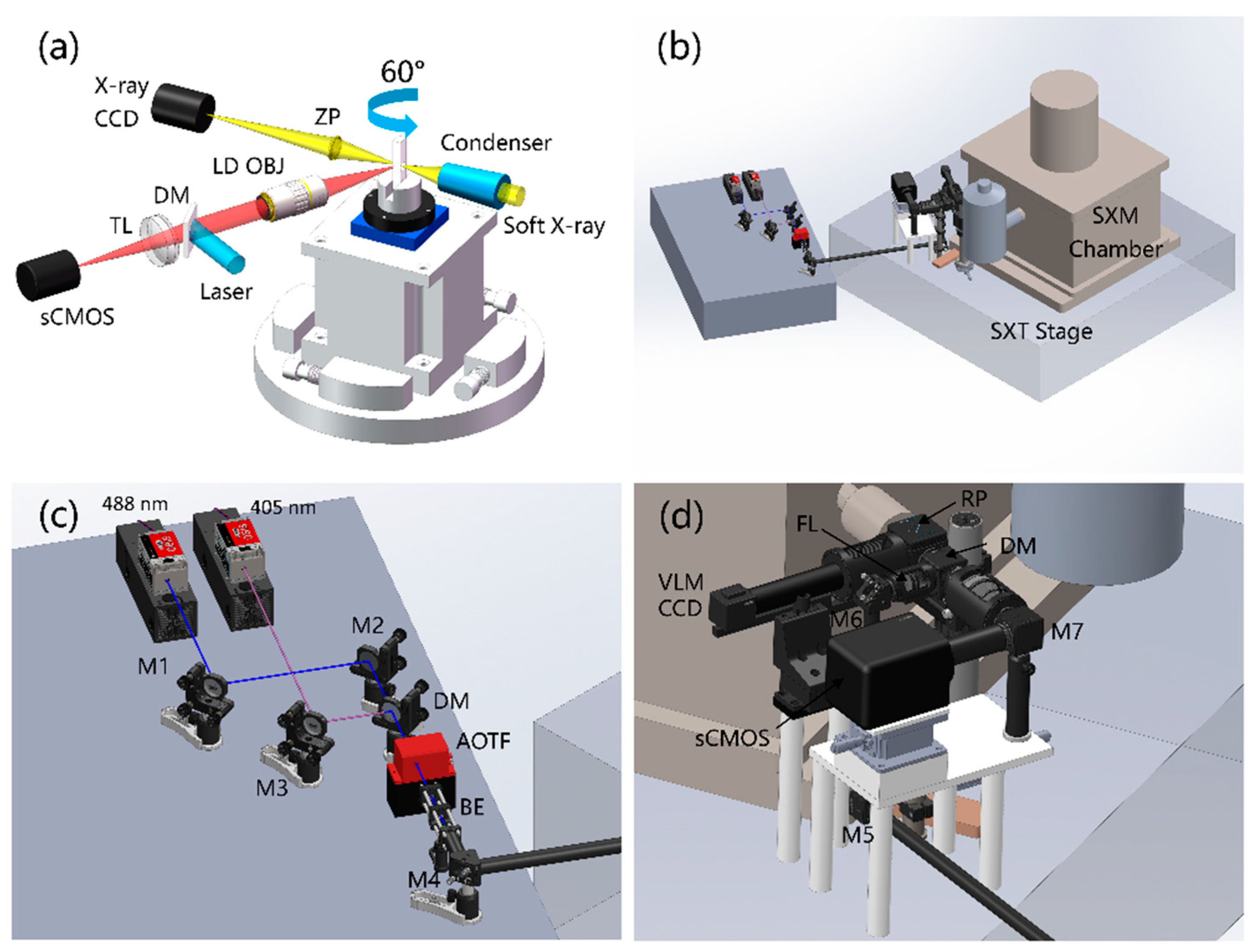
2.1. Optical Design and Implementation
2.2. Sample Preparation and Imaging Workflow
2.3. Reconstruction Algorithms for Linear Absorption Coefficient
3. Results and Discussion
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Zhang, M.M.; Guan, Y.; Dang, Z.; Zhang, P.G.; Zheng, Z.; Chen, L.; Kuang, W.; Wang, C.C.; Liang, G.L. Directly observing intracellular nanoparticle formation with nanocomputed tomography. Sci. Adv. 2020, 6, 6. [Google Scholar] [CrossRef] [PubMed]
- Cao, M.J.; Cai, R.; Zhao, L.N.; Guo, M.Y.; Wang, L.M.; Wang, Y.C.; Zhang, L.L.; Wang, X.F.; Yao, H.D.; Xie, C.Y.; et al. Molybdenum derived from nanomaterials incorporates into molybdenum enzymes and affects their activities in vivo. Nat. Nanotechnol. 2021, 16, 708. [Google Scholar] [CrossRef] [PubMed]
- Dang, Z.; Guan, Y.; Wu, Z.; Tao, X.Y.; Xiong, Y.; Bai, H.B.; Shao, C.S.; Liu, G.; Huang, Q.; Tian, L.J.; et al. Regulating the synthesis rate and yield of bio-assembled FeS nanoparticles for efficient cancer therapy. Nanoscale 2021, 13, 18977. [Google Scholar] [CrossRef]
- Zhou, J.B.; Jiang, Z.H.; Niu, S.W.; Zhu, S.S.; Zhou, J.; Zhu, Y.C.; Liang, J.W.; Han, D.D.; Xu, K.L.; Zhu, L.Q.; et al. Self-Standing Hierarchical P/CNTs@rGO with Unprecedented Capacity and Stability for Lithium and Sodium Storage. Chem 2018, 4, 372–385. [Google Scholar] [CrossRef] [Green Version]
- Zhou, F.; Li, Z.; Lu, Y.Y.; Shen, B.; Guan, Y.; Wang, X.X.; Yin, Y.C.; Zhu, B.S.; Lu, L.L.; Ni, Y.; et al. Diatomite derived hierarchical hybrid anode for high performance all-solid-state lithium metal batteries. Nat. Commun. 2019, 10, 11. [Google Scholar] [CrossRef] [Green Version]
- Bai, H.B.; Guan, Y.; Liu, J.H.; Chen, L.; Wei, W.B.; Liu, G.; Tian, Y.C. Precise correlative method of Cryo-SXT and Cryo-FM for organelle identification. J. Synchrotron Radiat. 2020, 27, 176–184. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bai, H.B.; Dang, Z.; Tao, X.Y.; Guan, Y.; Tian, L.J.; Liu, G.; Wu, Z.; Tian, Y.C. High spatial resolution correlative imaging of Cryo-SXT and GSDIM for identification of three-dimensional subcellular structures. Osa Contin. 2021, 4, 3262–3275. [Google Scholar] [CrossRef]
- Kounatidis, I.; Stanifer, M.L.; Phillips, M.A.; Paul-Gilloteaux, P.; Heiligenstein, X.; Wang, H.; Okolo, C.A.; Fish, T.M.; Spink, M.C.; Stuart, D.I.; et al. 3D Correlative Cryo-Structured Illumination Fluorescence and Soft X-ray Microscopy Elucidates Reovirus Intracellular Release Pathway. Cell 2020, 182, 515–530. [Google Scholar] [CrossRef] [PubMed]
- Le Gros, M.A.; McDermott, G.; Uchida, M.; Knoechel, C.G.; Larabell, C.A. High-aperture cryogenic light microscopy. J. Microsc. 2009, 235, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Smith, E.A.; Cinquin, B.P.; McDermott, G.; Le Gros, M.A.; Parkinson, D.Y.; Kim, H.T.; Larabell, C.A. Correlative microscopy methods that maximize specimen fidelity and data completeness, and improve molecular localization capabilities. J. Struct. Biol. 2013, 184, 12–20. [Google Scholar] [CrossRef]
- Smith, E.A.; McDermott, G.; Do, M.; Leung, K.; Panning, B.; Le Gros, M.A.; Larabell, C.A. Quantitatively Imaging Chromosomes by Correlated Cryo-Fluorescence and Soft X-ray Tomographies. Biophys. J. 2014, 107, 1988–1996. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sorrentino, A.; Nicolas, J.; Valcarcel, R.; Chichon, F.J.; Rosanes, M.; Avila, J.; Tkachuk, A.; Irwin, J.; Ferrer, S.; Pereiro, E. MISTRAL: A transmission soft X-ray microscopy beamline for cryo nano-tomography of biological samples and magnetic domains imaging. J. Synchrotron Radiat. 2015, 22, 1112–1117. [Google Scholar] [CrossRef]
- Varsano, N.; Dadosh, T.; Kapishnikov, S.; Pereiro, E.; Shimoni, E.; Jin, X.T.; Kruth, H.S.; Leiserowitz, L.; Addadi, L. Development of Correlative Cryo-soft X-ray Tomography and Stochastic Reconstruction Microscopy. A Study of Cholesterol Crystal Early Formation in Cells. J. Am. Chem. Soc. 2016, 138, 14931–14940. [Google Scholar] [CrossRef] [PubMed]
- Varsano, N.; Beghi, F.; Elad, N.; Pereiro, E.; Dadosh, T.; Pinkas, I.; Perez-Berna, A.J.; Jin, X.T.; Kruth, H.S.; Leiserowitz, L.; et al. Two polymorphic cholesterol monohydrate crystal structures form in macrophage culture models of atherosclerosis. Proc. Natl. Acad. Sci. USA 2018, 115, 7662–7669. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Groen, J.; Palanca, A.; Aires, A.; Conesa, J.J.; Maestro, D.; Rehbein, S.; Harkiolaki, M.; Villar, A.V.; Cortajarena, A.L.; Pereiro, E. Correlative 3D cryo X-ray imaging reveals intracellular location and effect of designed antifibrotic protein-nanomaterial hybrids. Chem. Sci. 2021, 12, 15090–15103. [Google Scholar] [CrossRef]
- Koronfel, M.; Kounatidis, I.; Mwangangi, D.M.; Vyas, N.; Okolo, C.; Jadhav, A.; Fish, T.; Chotchuang, P.; Schulte, A.; Robinson, R.C.; et al. Correlative cryo-imaging of the cellular universe with soft X-rays and laser light used to track F-actin structures in mammalian cells. Acta Crystallogr. Sect. D-Struct. Biol. 2021, 77, 1479–1485. [Google Scholar] [CrossRef] [PubMed]
- Okolo, C.A.; Jadhav, A.; Phillips, P.; Dumoux, M.; McMurray, A.A.; Joshi, V.D.; Pizzey, C.; Harkiolaki, M. Correlative imaging using super-resolution fluorescence microscopy and soft X-ray tomography at cryogenic temperatures provides a new way to assess virosome solutions for vaccine development. J. Microsc. 2021, 284, 214–232. [Google Scholar] [CrossRef]
- Okolo, C.A. A guide into the world of high-resolution 3D imaging: The case of soft X-ray tomography for the life sciences. Biochem. Soc. Trans. 2022, 50, 649–663. [Google Scholar] [CrossRef]
- Groen, J.; Sorrentino, A.; Aballe, L.; Oliete, R.; Valcarcel, R.; Okolo, C.; Kounatidis, I.; Harkiolaki, M.; Perez-Berna, A.J.; Pereiro, E. A 3D Cartographic Description of the Cell by Cryo Soft X-ray Tomography. J. Vis. Exp. 2021, 17, e62190. [Google Scholar] [CrossRef]
- Zhong, H. Photoactivated localization microscopy (PALM): An optical technique for achieving ~10-nm resolution. Cold Spring Harb. Protoc. 2010, 2010, pdb.top91. [Google Scholar] [CrossRef]
- Lin, Y.; Skaff, H.; Emrick, T.; Dinsmore, A.D.; Russell, T.P. Nanoparticle assembly and transport at liquid-liquid interfaces. Science 2003, 299, 226–229. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chai, Y.; Hasnain, J.; Bahl, K.; Wong, M.; Li, D.; Geissler, P.; Kim, P.Y.; Jiang, Y.F.; Gu, P.Y.; Li, S.Q.; et al. Direct observation of nanoparticle-surfactant assembly and jamming at the water-oil interface. Sci. Adv. 2020, 6, 6. [Google Scholar] [CrossRef] [PubMed]
- HeLa. Available online: https://www.beyotime.com/product/C6330.htm (accessed on 14 December 2022).
- Okolo, C.A.; Kounatidis, I.; Groen, J.; Nahas, K.L.; Balint, S.; Fish, T.M.; Koronfel, M.A.; Cortajarena, A.L.; Dobbie, I.M.; Pereiro, E.; et al. Sample preparation strategies for efficient correlation of 3D SIM and soft X-ray tomography data at cryogenic temperatures. Nat. Protoc. 2021, 16, 2851. [Google Scholar] [CrossRef] [PubMed]
- Liang, Z.T.; Guan, Y.; Liu, G.; Chen, X.Y.; Li, F.H.; Guo, P.F.; Tian, Y.C. A modified discrete algebraic reconstruction technique for multiple grey image reconstruction for limited angle range tomography. J. Synchrotron Radiat. 2016, 23, 606–616. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Guan, Y.; Liang, Z.T.; Guo, L.; Wei, C.X.; Luo, R.H.; Liu, G.; Tian, Y.C. A modified equally sloped algorithm based on the total variation algorithm in computed tomography for insufficient data. J. Synchrotron Radiat. 2017, 24, 490–497. [Google Scholar] [CrossRef]
- Liu, J.H.; Liang, Z.T.; Guan, Y.; Wei, W.B.; Bai, H.B.; Chen, L.; Liu, G.; Tian, Y.C. A modified discrete tomography for improving thereconstruction of unknown multi-gray-level material in the ‘missing wedge’ situation. J. Synchrotron Radiat. 2018, 25, 1847–1859. [Google Scholar] [CrossRef]
- Liu, J.; Li, F.; Chen, L.; Guan, Y.; Tian, L.; Xiong, Y.; Liu, G.; Tian, Y. Quantitative imaging of Candida utilis and its organelles by soft X-ray Nano-CT. J. Microsc. 2018, 270, 64–70. [Google Scholar] [CrossRef]
- Elmore, S. Apoptosis: A review of programmed cell death. Toxicol. Pathol. 2007, 35, 495–516. [Google Scholar] [CrossRef]






| Status | On-Line Fluorescence (Biochemical Information) | Soft X-ray Mosaic (Structural Information) |
|---|---|---|
| Normal | Faint signal | Intact structure |
| Apoptosis | Intense signal indicates caspase 3/7 activated | Cell shrinkage and Intact cell membrane |
| Necrosis | Faint signal | Cell swelling and Cytoplasm released |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, C.; Wu, Z.; Dang, Z.; Tian, L.; Guan, Y.; Liu, G.; Tian, Y. On-Line Fluorescence Microscopy for Identification and Imaging of Apoptotic Cell with Synchrotron-Based Soft X-ray Tomography. Micromachines 2023, 14, 326. https://doi.org/10.3390/mi14020326
Zhang C, Wu Z, Dang Z, Tian L, Guan Y, Liu G, Tian Y. On-Line Fluorescence Microscopy for Identification and Imaging of Apoptotic Cell with Synchrotron-Based Soft X-ray Tomography. Micromachines. 2023; 14(2):326. https://doi.org/10.3390/mi14020326
Chicago/Turabian StyleZhang, Chao, Zhao Wu, Zheng Dang, Lijiao Tian, Yong Guan, Gang Liu, and Yangchao Tian. 2023. "On-Line Fluorescence Microscopy for Identification and Imaging of Apoptotic Cell with Synchrotron-Based Soft X-ray Tomography" Micromachines 14, no. 2: 326. https://doi.org/10.3390/mi14020326





