Liposome Deformation Induced by Membrane-Binding Peptides
Abstract
:1. Introduction
2. Materials and Methods
2.1. Experimental Materials
2.2. Preparation of Giant Liposomes Using the Droplet Transfer Method
2.3. Image Analysis of Liposomes
2.4. Measurement of Membrane Capacitance and Subsequent Analysis
3. Results
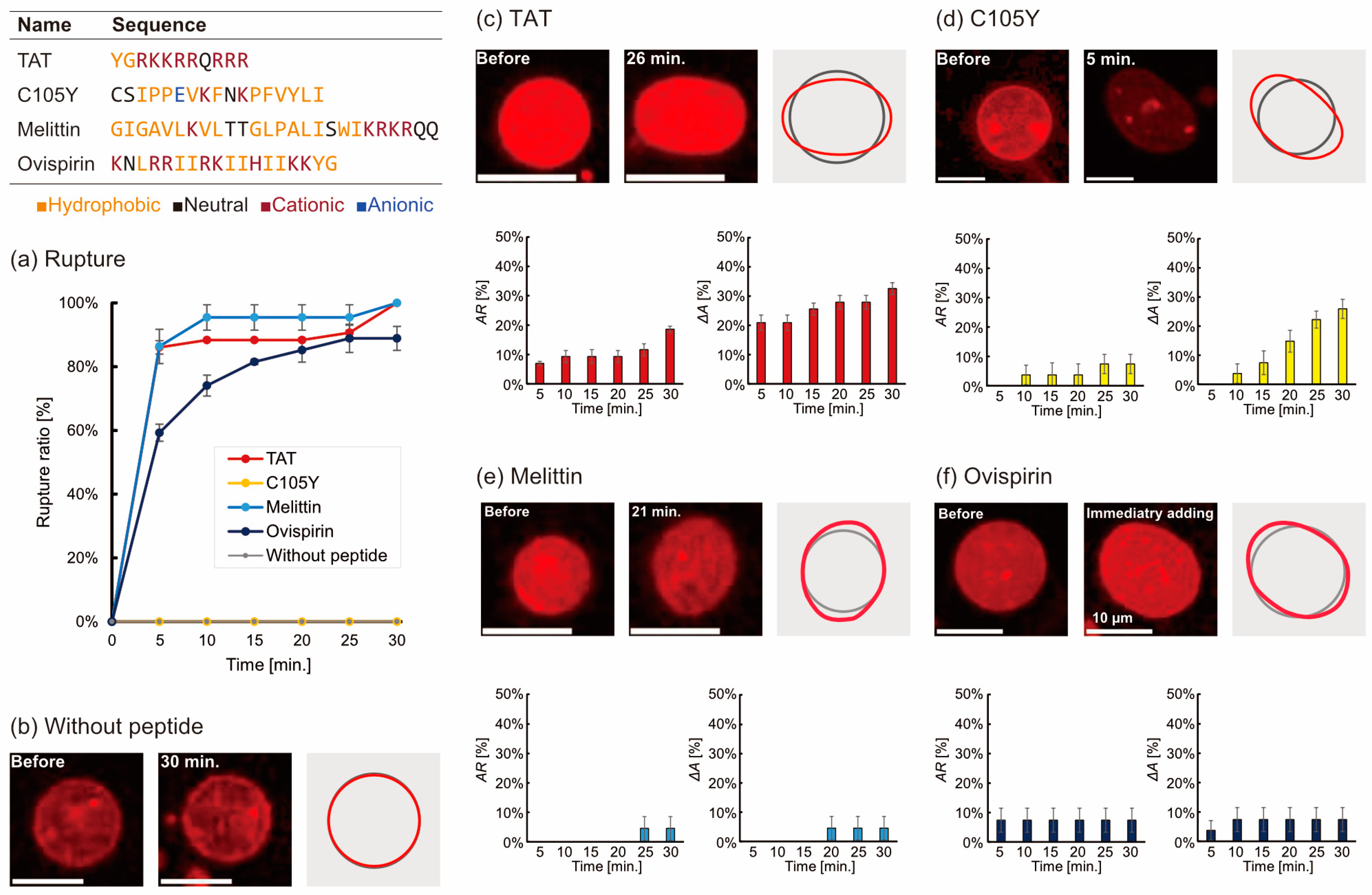
3.1. Microscopic Observation of the Deformation and Rupture of Liposomes by Peptides
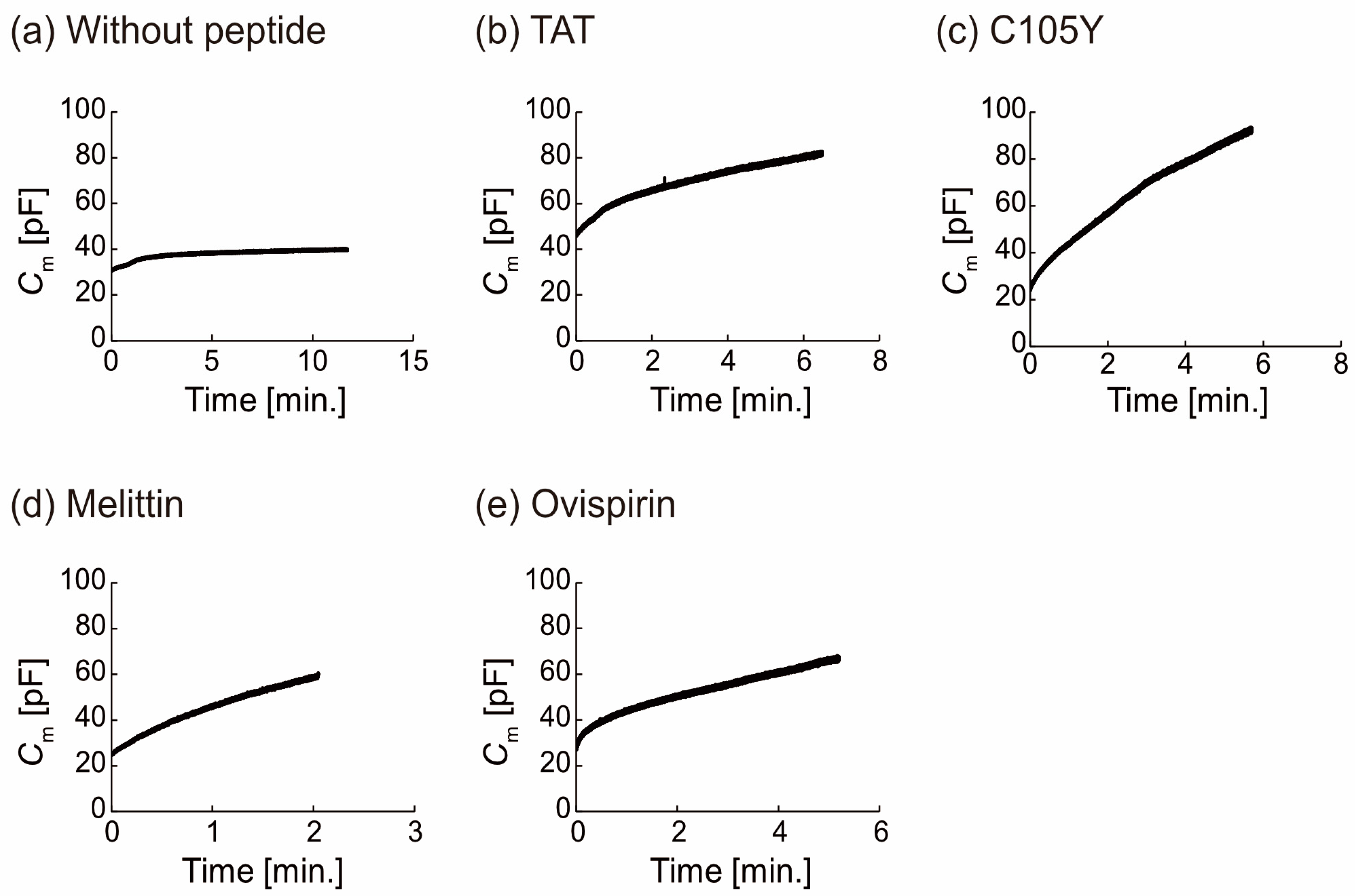
3.2. Binding of Peptide on the Lipid Membrane Estimated by the Membrane Capacitance
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sun, J.; Rijpkema, S.J.; Luan, J.; Zhang, S.; Wilson, D.A. Generating biomembrane-like local curvature in polymersomes via dynamic polymer insertion. Nat. Commun. 2021, 12, 2235. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Wang, J.; Sun, W.; Archibong, E.; Kahkoska, A.R.; Zhang, X.; Lu, Y.; Ligler, F.S.; Buse, J.B.; Gu, Z. Synthetic beta cells for fusion-mediated dynamic insulin secretion. Nat. Chem. Biol. 2018, 14, 86–93. [Google Scholar] [CrossRef]
- Furusato, T.; Horie, F.; Matsubayashi, H.T.; Amikura, K.; Kuruma, Y.; Ueda, T. De Novo Synthesis of Basal Bacterial Cell Division Proteins FtsZ, FtsA, and ZipA Inside Giant Vesicles. ACS Synth. Biol. 2018, 7, 953–961. [Google Scholar] [CrossRef]
- Hagiya, M.; Konagaya, A.; Kobayashi, S.; Saito, H.; Murata, S. Molecular Robots with Sensors and Intelligence. Acc. Chem. Res. 2014, 47, 1681–1690. [Google Scholar] [CrossRef] [PubMed]
- Kawano, R. Synthetic Ion Channels and DNA Logic Gates as Components of Molecular Robots. Chemphyschem 2018, 19, 359–366. [Google Scholar] [CrossRef] [PubMed]
- Shoji, K.; Kawano, R. Recent Advances in Liposome-Based Molecular Robots. Micromachines 2020, 11, 788. [Google Scholar] [CrossRef]
- Sato, Y.; Hiratsuka, Y.; Kawamata, I.; Murata, S.; Nomura, S.-I.M. Micrometer-sized molecular robot changes its shape in response to signal molecules. Sci. Robot. 2017, 2, eaal3735. [Google Scholar] [CrossRef]
- Ha, J.H.; Jeong, Y.J.; Kim, A.Y.; Hong, I.K.; Lee, N.H.; Park, S.N. Preparation and Physicochemical Properties of a Cysteine Derivative-Loaded Deformable Liposomes in Hydrogel for Enhancing Whitening Effects. Eur. J. Lipid Sci. Technol. 2018, 120, 1800125. [Google Scholar] [CrossRef]
- Kim, A.R.; An, H.J.; Jang, E.S.; Lee, J.D.; Park, S.N. Preparation, Physical Characterization, and In Vitro Skin Permeation of Deformable Liposomes Loaded with Taxifolin and Taxifolin Tetraoctanoate. Eur. J. Lipid Sci. Technol. 2019, 121, 1800501. [Google Scholar] [CrossRef]
- Peterlin, P.; Arrigler, V.; Kogej, K.; Svetina, S.; Walde, P. Growth and shape transformations of giant phospholipid vesicles upon interaction with an aqueous oleic acid suspension. Chem. Phys. Lipids 2009, 159, 67–76. [Google Scholar] [CrossRef]
- Sudbrack, T.P.; Archilha, N.L.; Itri, R.; Riske, K.A. Observing the Solubilization of Lipid Bilayers by Detergents with Optical Microscopy of GUVs. J. Phys. Chem. B 2011, 115, 269–277. [Google Scholar] [CrossRef] [PubMed]
- Cortese, D.; Schwab, B., III; Frieden, C.; Elson, E.L. Actin polymerization induces a shape change in actin-containing vesicles. Proc. Natl. Acad. Sci. USA 1989, 86, 5773–5777. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, S.; Takiguchi, K.; Hayashi, M. Repetitive stretching of giant liposomes utilizing the nematic alignment of confined actin. Commun. Phys. 2018, 1, 18. [Google Scholar] [CrossRef]
- Lee, M.-T.; Hung, W.-C.; Chen, F.-Y.; Huang, H.W. Mechanism and kinetics of pore formation in membranes by water-soluble amphipathic peptides. Proc. Natl. Acad. Sci. USA 2008, 105, 5087–5092. [Google Scholar] [CrossRef]
- Lee, M.-T.; Sun, T.-L.; Hung, W.-C.; Huang, H.W. Process of inducing pores in membranes by melittin. Proc. Natl. Acad. Sci. USA 2013, 110, 14243–14248. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Vroman, J.A.; Bae, S.C.; Granick, S. Vesicle Budding Induced by a Pore-Forming Peptide. J. AM. CHEM. SOC. 2010, 132, 195–201. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, T.; Nomura, F.; Yokoyama, Y.; Tanaka-Takiguchi, Y.; Homma, M.; Takiguchi, K. Multiple membrane interactions and versatile vesicle deformations elicited by melittin. Toxins 2013, 5, 637–664. [Google Scholar] [CrossRef]
- Yu, Q.; Sun, J.; Huang, S.; Chang, H.; Bai, Q.; Chen, Y.-X.; Liang, D. Inward Budding and Endocytosis of Membranes Regulated by de Novo Designed Peptides. Langmuir 2018, 34, 6183–6193. [Google Scholar] [CrossRef]
- Mally, M.; Majhenc, J.; Svetina, S.; Zeks, B. The response of giant phospholipid vesicles to pore-forming peptide melittin. Biochim. Biophys. Acta 2007, 1768, 1179–1189. [Google Scholar] [CrossRef]
- Rossman, J.S.; Jing, X.; Leser, G.P.; Lamb, R.A. Influenza virus M2 protein mediates ESCRT-independent membrane scission. Cell 2010, 142, 902–913. [Google Scholar] [CrossRef] [Green Version]
- Shimizu, Y.; Inoue, A.; Tomari, Y.; Suzuki, T.; Yokogawa, T.; Nishikawa, K.; Ueda, T. Cell-free translation reconstituted with purified components. Nat. Biotechnol. 2001, 19, 751–755. [Google Scholar] [CrossRef] [PubMed]
- Stalmans, S.; Bracke, N.; Wynendaele, E.; Gevaert, B.; Peremans, K.; Burvenich, C.; Polis, I.; Spiegeleer, B.D. Cell-Penetrating Peptides Selectively Cross the Blood-Brain Barrier In Vivo. PLoS ONE 2015, 10, e0139652. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Zhang, X.; Guo, C.; Peng, Y.; Liu, X.; Wang, B.; Zhuang, R.; Chang, M.; Wang, R. Hydrocarbon staple constructing highly efficient alpha-helix cell-penetrating peptides for intracellular cargo delivery. Chem. Commun. 2020, 56, 15655–15658. [Google Scholar] [CrossRef] [PubMed]
- Kumara, B.T.; Wijesiri, N.K.; Rathnayake, P.V.G.M.; Ranatunga, R.J.K.U. A Re-evaluation of the Free Energy Profiles for Cell-Penetrating Peptides Across DOPC Membranes. Int. J. Pept. Res. Ther. 2021, 27, 2931–2943. [Google Scholar] [CrossRef]
- Herce, H.D.; Garcia, A.E. Molecular dynamics simulations suggest a mechanism for translocation of the HIV-1 TAT peptide across lipid membranes. Proc. Natl. Acad. Sci. USA 2007, 104, 20805–20810. [Google Scholar] [CrossRef]
- Rhee, M.; Davis, P. Mechanism of uptake of C105Y, a novel cell-penetrating peptide. J. Biol. Chem. 2006, 281, 1233–1240. [Google Scholar] [CrossRef]
- Guha, S.; Ghimire, J.; Wu, E.; Wimley, W.C. Mechanistic Landscape of Membrane-Permeabilizing Peptides. Chem. Rev. 2019, 119, 6040–6085. [Google Scholar] [CrossRef]
- Irudayam, S.J.; Pobandt, T.; Berkowitz, M.L. Free energy barrier for melittin reorientation from a membrane-bound state to a transmembrane state. J. Phys. Chem. B 2013, 117, 13457–13463. [Google Scholar] [CrossRef]
- Krauson, A.J.; He, J.; Wimley, W.C. Gain-of-function analogues of the pore-forming peptide melittin selected by orthogonal high-throughput screening. J. Am. Chem. Soc. 2012, 134, 12732–12741. [Google Scholar] [CrossRef]
- Nguyen, L.T.; Haney, E.F.; Vogel, H.J. The expanding scope of antimicrobial peptide structures and their modes of action. Trends Biotechnol. 2011, 29, 464–472. [Google Scholar] [CrossRef]
- Miyazaki, Y.; Okazaki, S.; Shinoda, W. Free energy analysis of membrane pore formation process in the presence of multiple melittin peptides. Biochim. Biophys. Acta Biomembr. 2019, 1861, 1409–1419. [Google Scholar] [CrossRef] [PubMed]
- Geitani, R.; Moubareck, C.A.; Xu, Z.; Karam Sarkis, D.; Touqui, L. Expression and Roles of Antimicrobial Peptides in Innate Defense of Airway Mucosa: Potential Implication in Cystic Fibrosis. Front. Immunol. 2020, 11, 1198. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, S.; Huster, D.; Waring, A.; Lehrer, R.I.; Kearney, W.; Tack, B.F.; Hong, M. Orientation and Dynamics of an Antimicrobial Peptide in the Lipid Bilayer by Solid-State NMR Spectroscopy. Biophys. J. 2001, 81, 2203–2214. [Google Scholar] [CrossRef] [PubMed]
- Woys, A.M.; Lin, Y.-S.; Reddy, A.S.; Xiong, W.; Pablo, J.J.d.; Skinner, J.L.; Zanni, M.T. 2D IR Line Shapes Probe Ovispirin Peptide Conformation and Depth in Lipid Bilayers. J. Am. Chem. Soc. 2010, 132, 2832–2838. [Google Scholar] [CrossRef] [PubMed]
- Pautot, S.; Frisken, B.J.; Weitz, D.A. Engineering asymmetric vesicles. Proc. Natl. Acad. Sci. USA 2003, 100, 10718–10721. [Google Scholar] [CrossRef]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef]
- Kawano, R.; Tsuji, Y.; Sato, K.; Osaki, T.; Kamiya, K.; Hirano, M.; Ide, T.; Miki, N.; Takeuchi, S. Automated Parallel Recordings of Topologically Identified Single Ion Channels. Sci. Rep. 2013, 3, 1995. [Google Scholar] [CrossRef]
- Sekiya, Y.; Sakashita, S.; Shimizu, K.; Usui, K.; Kawano, R. Channel current analysis estimates the pore-formation and the penetration of transmembrane peptides. Analyst 2018, 143, 3540–3543. [Google Scholar] [CrossRef]
- Sekiya, Y.; Shimizu, K.; Kitahashi, Y.; Ohyama, A.; Kawamura, I.; Kawano, R. Electrophysiological Analysis of Membrane Disruption by Bombinin and Its Isomer Using the Lipid Bilayer System. ACS Appl. Bio. Mater. 2019, 2, 1542–1548. [Google Scholar] [CrossRef]
- Shimizu, K.; Mijiddorj, B.; Usami, M.; Mizoguchi, I.; Yoshida, S.; Akayama, S.; Hamada, Y.; Ohyama, A.; Usui, K.; Kawamura, I.; et al. De novo design of a nanopore for single-molecule detection that incorporates a beta-hairpin peptide. Nat. Nanotechnol. 2022, 17, 67–75. [Google Scholar] [CrossRef]
- Watanabe, H.; Gubbiotti, A.; Chinappi, M.; Takai, N.; Tanaka, K.; Tsumoto, K.; Kawano, R. Analysis of Pore Formation and Protein Translocation Using Large Biological Nanopores. Anal. Chem. 2017, 89, 11269–11277. [Google Scholar] [CrossRef] [PubMed]
- Hirano, M.; Saito, C.; Yokoo, H.; Goto, C.; Kawano, R.; Misawa, T.; Demizu, Y. Development of Antimicrobial Stapled Peptides Based on Magainin 2 Sequence. Molecules 2021, 26, 444. [Google Scholar] [CrossRef] [PubMed]
- Okada, Y. Saishin Pacchikuranpu Zikkengizyutsuhou (How to Use Patch-Clamp Method); Yoshioka Shoten: Kyoto, Japan, 2011. [Google Scholar]
- Hagge, S.O.; Wiese, A.; Seydel, U.; Gutsmann, T. Inner Field Compensation as a Tool for the Characterization of Asymmetric Membranes and Peptide-Membrane Interactions. Biophys. J. 2004, 86, 913–922. [Google Scholar] [CrossRef] [PubMed]
- Funakoshi, K.; Suzuki, H.; Takeuchi, S. Lipid Bilayer Formation by Contacting Monolayers in a Microfluidic Device for Membrane Protein Analysis. Anal. Chem. 2006, 78, 8169–8174. [Google Scholar] [CrossRef] [PubMed]
- Glattli, A.; Chandrasekhar, I.; van Gunsteren, W.F. A molecular dynamics study of the bee venom melittin in aqueous solution, in methanol, and inserted in a phospholipid bilayer. Eur. Biophys. J. 2006, 35, 255–267. [Google Scholar] [CrossRef]
- Valincius, G.; Heinrich, F.; Budvytyte, R.; Vanderah, D.J.; McGillivray, D.J.; Sokolov, Y.; Hall, J.E.; Losche, M. Soluble amyloid beta-oligomers affect dielectric membrane properties by bilayer insertion and domain formation: Implications for cell toxicity. Biophys. J. 2008, 95, 4845–4861. [Google Scholar] [CrossRef] [PubMed]
- Sun, D.; Forsman, J.; Woodward, C.E. Amphipathic membrane-active peptides recognize and stabilize ruptured membrane pores: Exploring cause and effect with coarse-grained simulations. Langmuir 2015, 31, 752–761. [Google Scholar] [CrossRef]
- Van Der Spoel, D.; Lindahl, E.; Hess, B.; Groenhof, G.; Mark, A.E.; Berendsen, H.J. GROMACS: Fast, flexible, and free. J. Comput. Chem. 2005, 26, 1701–1718. [Google Scholar] [CrossRef]
- Huang, J.; Rauscher, S.; Nawrocki, G.; Ran, T.; Feig, M.; de Groot, B.L.; Grubmüller, H.; MacKerell, A.D., Jr. CHARMM36m: An improved force field for folded and intrinsically disordered proteins. Nat. Methods 2017, 14, 71–73. [Google Scholar] [CrossRef]
- Jo, S.; Kim, T.; Iyer, V.G.; Im, W. CHARMM-GUI: A web-based graphical user interface for CHARMM. J. Comput. Chem. 2008, 29, 1859–1865. [Google Scholar] [CrossRef]
- Parrinello, M.; Rahman, A. Polymorphic transitions in single crystals: A new molecular dynamics method. J. Appl. Phys. 1981, 52, 7182–7190. [Google Scholar] [CrossRef]
- Essmann, U.; Perera, L.; Berkowitz, M.L.; Darden, T.; Lee, H.; Pedersen, L.G. A smooth particle mesh Ewald method. J. Chem. Phys. 1995, 103, 8577–8593. [Google Scholar] [CrossRef]
- Hess, B.; Bekker, H.; Berendsen, H.J.C.; Fraaije, J.G.E.M. LINCS: A linear constraint solver for molecular simulations. J. Comput. Chem. 1998, 18, 1463–1472. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Izumi, K.; Saito, C.; Kawano, R. Liposome Deformation Induced by Membrane-Binding Peptides. Micromachines 2023, 14, 373. https://doi.org/10.3390/mi14020373
Izumi K, Saito C, Kawano R. Liposome Deformation Induced by Membrane-Binding Peptides. Micromachines. 2023; 14(2):373. https://doi.org/10.3390/mi14020373
Chicago/Turabian StyleIzumi, Kayano, Chihiro Saito, and Ryuji Kawano. 2023. "Liposome Deformation Induced by Membrane-Binding Peptides" Micromachines 14, no. 2: 373. https://doi.org/10.3390/mi14020373
APA StyleIzumi, K., Saito, C., & Kawano, R. (2023). Liposome Deformation Induced by Membrane-Binding Peptides. Micromachines, 14(2), 373. https://doi.org/10.3390/mi14020373





