A Genosensor Based on the Modification of a Microcantilever: A Review
Abstract
1. Introduction
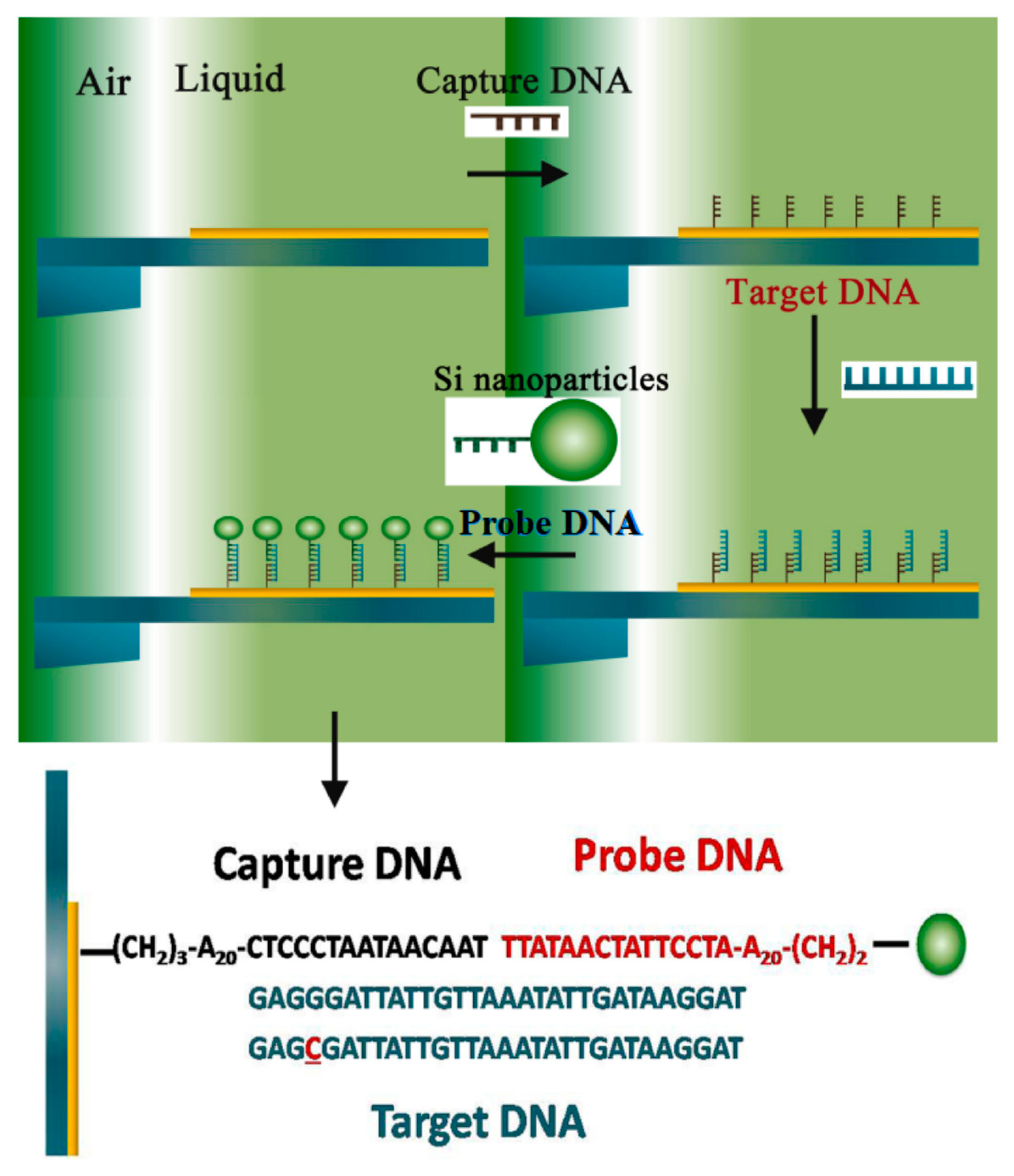
2. Detection Procedure of Genetic-Probe-Modified Microcantilever
- Moderate length. The length of a nucleic acid sequence is generally between 18 and 50 bases. If the sequence is too long, the complementary binding time increases and the detection efficiency decreases; if the sequence is too short, the specificity of detection degenerates;
- No complementary interval. Nucleic acid sequences with a complementary interval can form a secondary structure by themselves and inhibit the complementary binding;
- The base composition ratio is constant. The base composition ratio of G and C must be within the range of 40–60%;
- Avoid multiple iterations of the same base. In order to reduce the false probability, the number of consecutive identical bases should be fewer than four.
2.1. Probe Immobilization
2.2. Complementary Hybridization
2.3. Signal Extraction and Processing
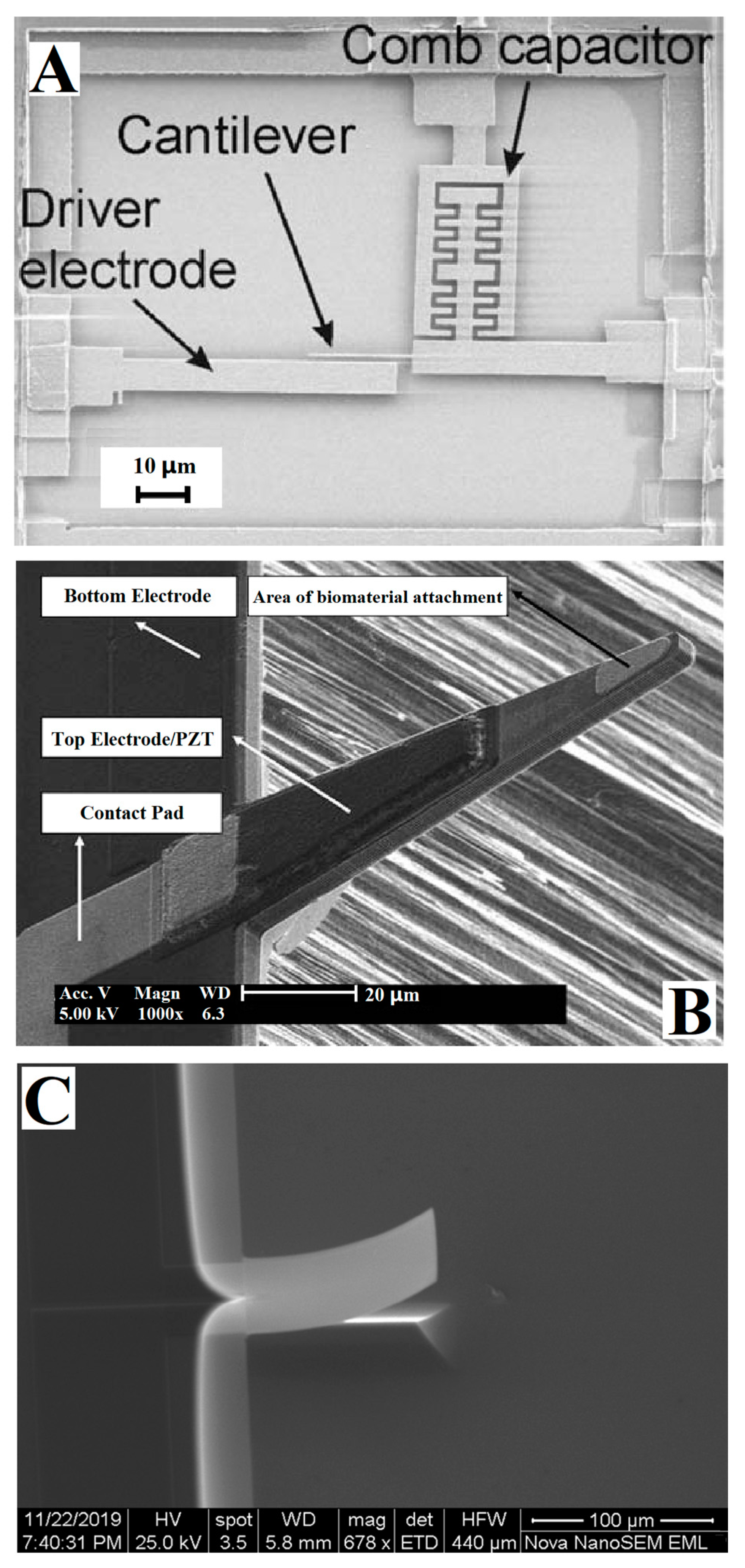
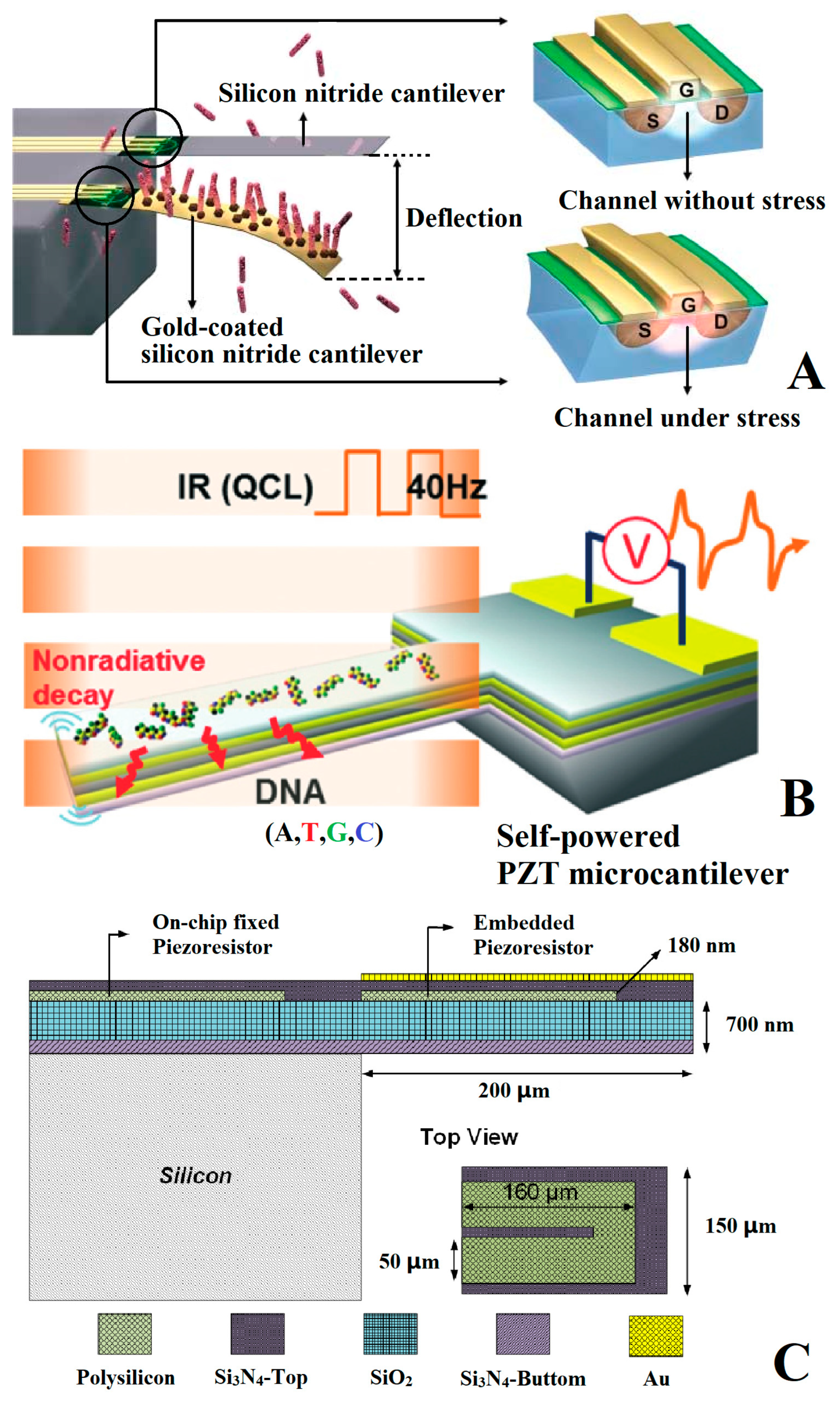
3. Detection Principle of Genetic-Probe-Modified Cantilever
3.1. Deflection Principle
3.2. Detection Model
3.3. Impact of Environmental Factors
3.3.1. Temperature
3.3.2. Salinity
3.3.3. Humidity
3.3.4. pH
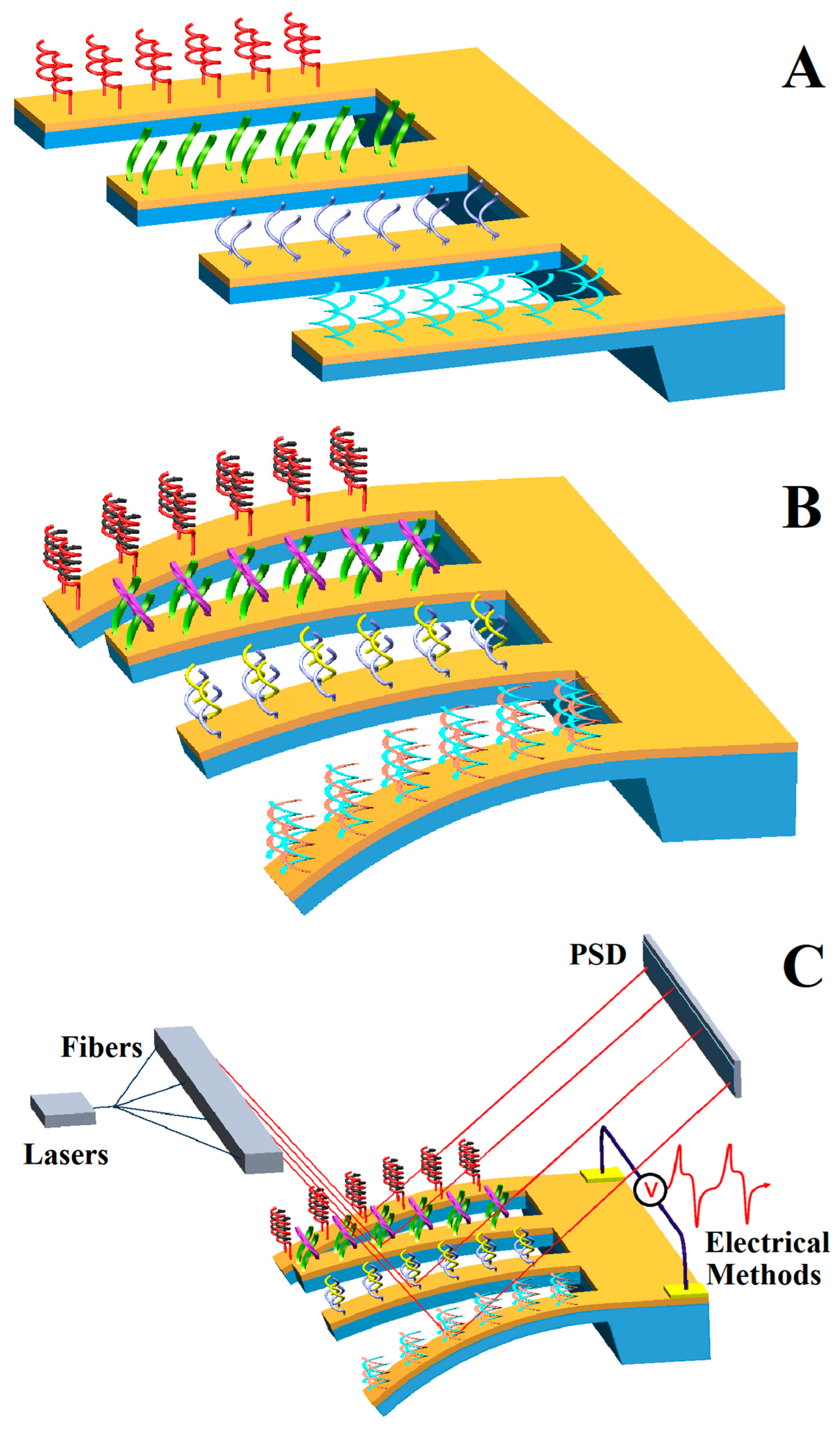
4. Application of Genetic-Probe-Modified Cantilever
4.1. Nucleic Acid
4.2. Viruses, Bacteria, and Cells
4.2.1. Viruses
4.2.2. Bacteria
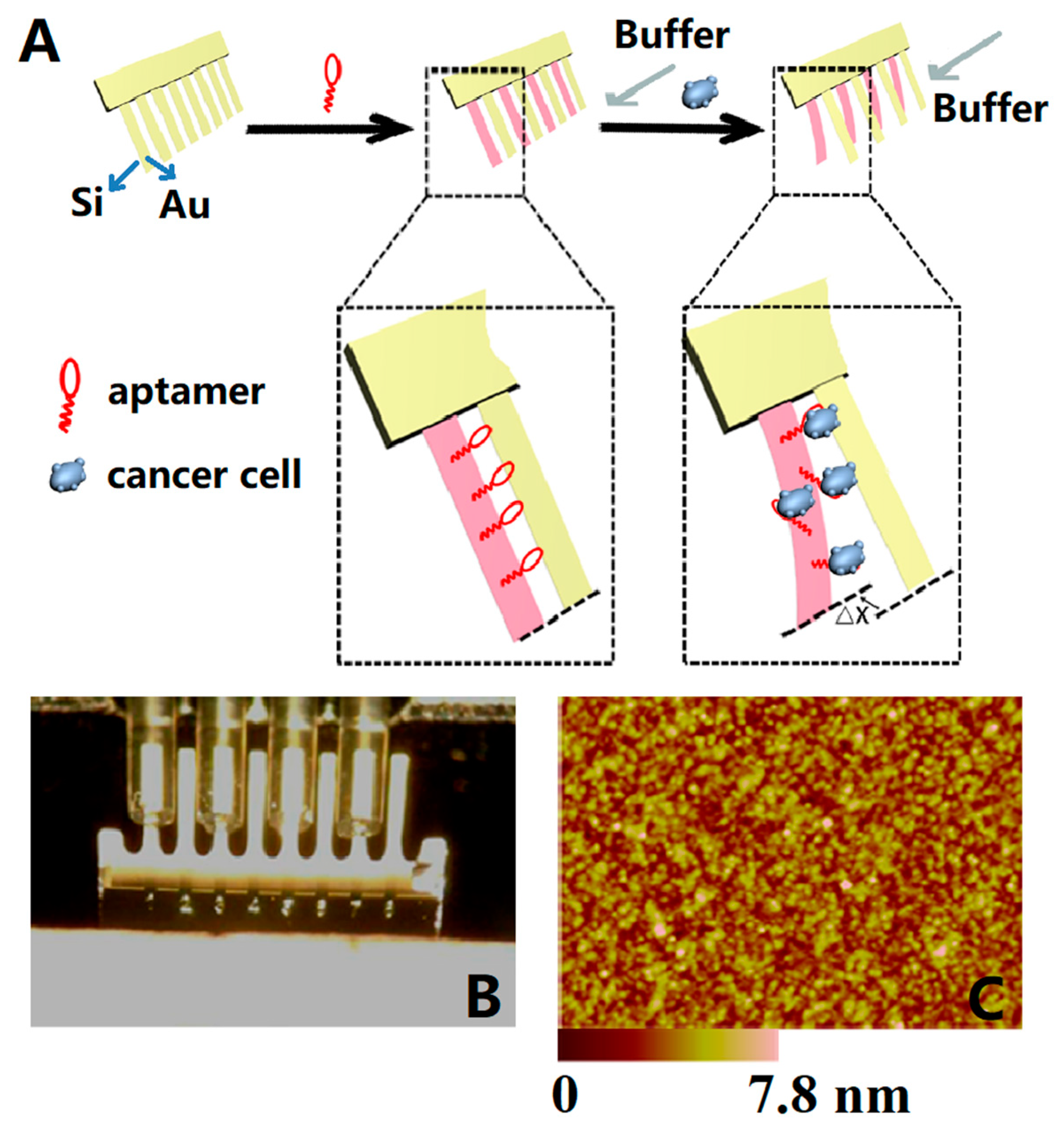
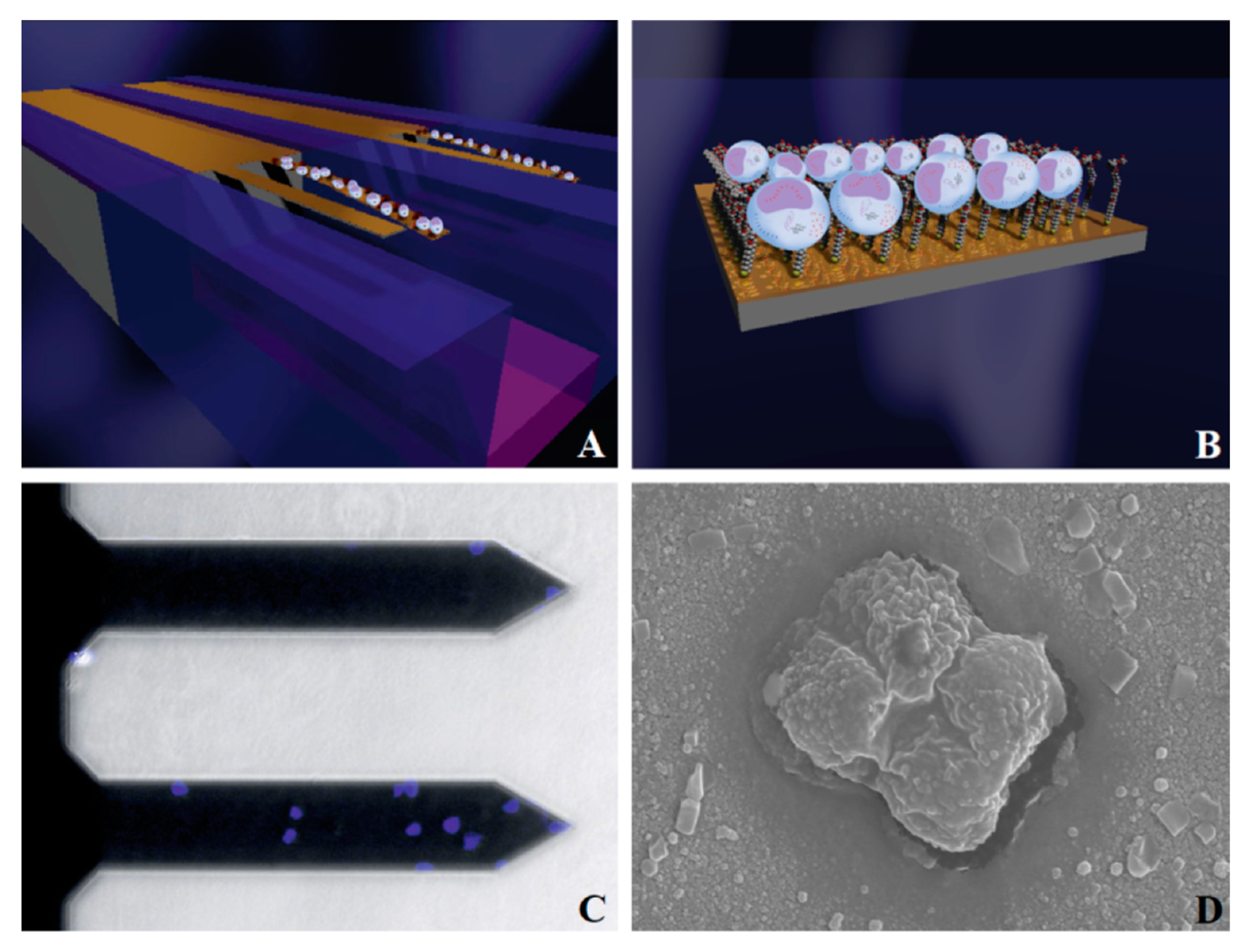
4.2.3. Cells
4.3. Other Substances
4.3.1. Proteins
4.3.2. Antibiotics
4.3.3. Heavy Metal Ions
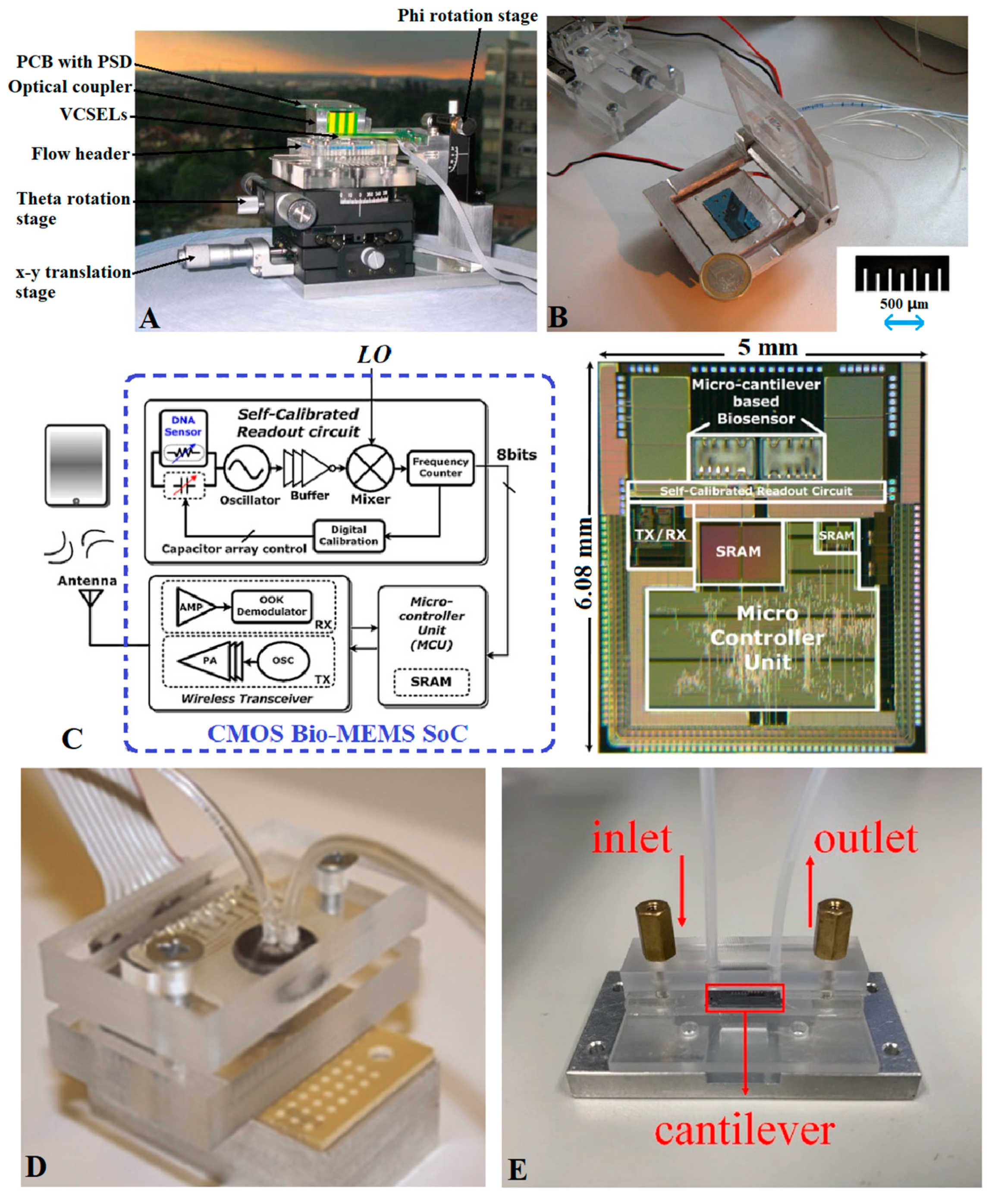
4.4. Integrated Detection
5. Conclusions and Prospects
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wilfinger, R.J.; Bardell, P.H.; Chhabra, D.S. The Resonistor: A Frequency Selective Device Utilizing the Mechanical Resonance of a Silicon Substrate. IBM J. Res. Dev. 1968, 12, 113–118. [Google Scholar] [CrossRef]
- Heng, T.M.S. Trimming of Microstrip Circuits Utilizing Microcantilever Air Gaps (Correspondence). IEEE Trans. Microw. Theory Tech. 1971, 19, 652–654. [Google Scholar] [CrossRef]
- Cleveland, J.P.; Manne, S.; Bocek, D.; Hansma, P.K. A nondestructive method for determining the spring constant of cantilevers for scanning force microscopy. Rev. Sci. Instrum. 1993, 64, 403–405. [Google Scholar] [CrossRef]
- Mahmoud, M.A. Validity and Accuracy of Resonance Shift Prediction Formulas for Microcantilevers: A Review and Comparative Study. Crit. Rev. Solid State Mater. Sci. 2016, 41, 386–429. [Google Scholar] [CrossRef]
- Peng, W.; Wei, Z.; Hai, W.; Yan, L.; Qin, H. Geometry and profile modification of microcantilevers for sensitivity enhancement in sensing applications. Sens. Mater. 2017, 29, 689–698. [Google Scholar]
- Goeders, K.M.; Colton, J.S.; Bottomley, L.A. Microcantilevers: Sensing Chemical Interactions via Mechanical Motion. Chem. Rev. 2008, 108, 522–542. [Google Scholar] [CrossRef]
- Thundat, T.; Wachter, E.A.; Sharp, S.L.; Warmack, R.J. Detection of mercury vapor using resonating microcantilevers. Appl. Phys. Lett. 1995, 66, 1695–1697. [Google Scholar] [CrossRef]
- Chen, G.Y.; Thundat, T.; Wachter, E.A.; Warmack, R.J. Adsorption-induced surface stress and its effects on resonance frequency of microcantilevers. J. Appl. Phys. 1995, 77, 3618–3622. [Google Scholar] [CrossRef]
- Buchapudi, K.R.; Huang, X.; Yang, X.; Ji, H.F.; Thundat, T. Microcantilever biosensors for chemicals and bioorganisms. Analyst 2011, 136, 1539–1556. [Google Scholar] [CrossRef]
- Ahmed, S.; Aslam, L.; Bilal, B.; Ali, S.; Kakkar, V. Investigation on Applicability and Suitability of Micro Cantilever Based Biosensors for DNA Detection. Adv. Biotech. Micro. 2017, 2, 555593. [Google Scholar] [CrossRef]
- Mouro, J.; Rui, P.; Paoletti, P.; Tiribilli, B. Microcantilever: Dynamical Response for Mass Sensing and Fluid Characterization. Sensors 2020, 21, 115. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Xu, B.; Shi, L.; Zhu, L.; Wei, X. Prospects and Challenges of AI and Neural Network Algorithms in MEMS Microcantilever Biosensors. Processes 2022, 10, 1658. [Google Scholar] [CrossRef]
- Britten, R.J.; Kohne, D.E. Repeated sequences in DNA: Hundreds of thousands of copies of DNA sequences have been incorporated into the genomes of higher organisms. Science 1968, 161, 529. [Google Scholar] [CrossRef] [PubMed]
- Wetmur, J.G. DNA probes: Applications of the principles of nucleic acid hybridization. CRC Crit. Rev. Biochem. 1991, 26, 227–259. [Google Scholar] [CrossRef] [PubMed]
- Pardue, M.L.; Gall, J.G. Molecular Hybridization of Radioactive DNA to the DNA of Cytological Preparations. Proc. Natl. Acad. Sci. USA 1969, 64, 600–604. [Google Scholar] [CrossRef]
- Friedman, R.L.; Stark, G.R. α-Interferon-induced transcription of HLA and metallothionein genes containing homologous upstream sequences. Nature 1985, 314, 637–639. [Google Scholar] [CrossRef]
- Bird, A.P.; Southern, E.M. Use of restriction enzymes to study eukaryotic DNA methylation: I. The methylation pattern in ribosomal DNA from Xenopus laevis. J. Mol. Biol. 1978, 118, 27. [Google Scholar] [CrossRef]
- Ellington, A.D.; Szostak, J.W. In vitro selection of RNA molecules that bind specific ligands. Nature 1990, 346, 818–822. [Google Scholar] [CrossRef]
- Whitesides, G.M.; Kriebel, J.K.; Love, J.C. Molecular engineering of surfaces using self-assembled monolayers. Sci. Prog. 2005, 88, 17–48. [Google Scholar] [CrossRef]
- Arroyohernández, M.; Tamayo, J.; Costakrämer, J.L. Stress and DNA Assembly Differences on Cantilevers Gold Coated by Resistive and E-Beam Evaporation Techniques. Langmuir ACS J. Surf. Colloids 2009, 25, 10633. [Google Scholar] [CrossRef]
- Fritz, J.; Baller, M.K.; Lang, H.P.; Rothuizen, H.; Vettiger, P.; Meyer, E.; Güntherodt, H.; Gerber, C.; Gimzewski, J.K. Translating biomolecular recognition into nanomechanics. Science 2000, 288, 316–318. [Google Scholar] [CrossRef] [PubMed]
- Marie, R.; Jensenius, H.; Thaysen, J.; Christensen, C.B.; Boisen, A. Adsorption kinetics and mechanical properties of thiol-modified DNA-oligos on gold investigated by microcantilever sensors. Ultramicroscopy 2002, 91, 29–36. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Cai, H.; Xu, Q.; He, P.; Fang, Y. Characterization of single-stranded DNA on chitosan-modified electrode and its application to the sequence-specific DNA detection. Fresenius J. Anal. Chem. 2001, 369, 428–432. [Google Scholar] [CrossRef] [PubMed]
- Dell’Atti, D.; Tombelli, S.; Minunni, M.; Mascini, M. Detection of clinically relevant point mutations by a novel piezoelectric biosensor. Biosens. Bioelectron. 2006, 21, 1876–1879. [Google Scholar] [CrossRef]
- Velanki, S.; Kelly, S.; Thundat, T.; Blake, D.A.; Ji, H.F. Detection of Cd(II) using antibody-modified microcantilever sensors. Ultramicroscopy 2007, 107, 1123–1128. [Google Scholar] [CrossRef]
- Zhang, D.; Chen, Y.; Chen, H.Y.; Xia, X.H. Silica-nanoparticle-based interface for the enhanced immobilization and sequence-specific detection of DNA. Anal. Bioanal. Chem. 2004, 379, 1025–1030. [Google Scholar] [CrossRef]
- Jiang, M.; Wang, J. Recognition and detection of oligonucleotides in the presence of chromosomal DNA based on entrapment within conducting-polymer networks. J. Electroanal. Chem. 2001, 500, 584–589. [Google Scholar] [CrossRef]
- Rupcich, N.; Nutiu, R.; Li, Y.; Brennan, J.D. Entrapment of fluorescent signaling DNA aptamers in sol-gel-derived silica. Anal. Chem. 2005, 77, 4300–4307. [Google Scholar] [CrossRef]
- Oliviero, G.; Bergese, P.; Canavese, G.; Chiari, M.; Colombi, P.; Cretich, M.; Damin, F.; Fiorilli, S.; Marasso, S.L.; Ricciardi, C.; et al. A biofunctional polymeric coating for microcantilever molecular recognition. Anal. Chim. Acta 2008, 655, 92. [Google Scholar] [CrossRef]
- Calleja, M.; Nordstrã, M.M.; Alvarez, M.; Mayo, J.; Lechuga, L.M.; Boisen, A. Highly sensitive polymer-based cantilever-sensors for DNA detection. Ultramicroscopy 2005, 105, 215–222. [Google Scholar] [CrossRef]
- Dugas, V.; Depret, G.; Chevalier, Y.; Nesme, X.; Souteyrand, É. Immobilization of single-stranded DNA fragments to solid surfaces and their repeatable specific hybridization: Covalent binding or adsorption. Sens. Actuators B 2004, 101, 112–121. [Google Scholar] [CrossRef]
- Vermeeren, V.; Wenmackers, S.; Daenen, M.; Haenen, K.; Williams, O.A.; Ameloot, M.; vandeVen, M.; Wagner, P.; Michiels, L. Topographical and Functional Characterization of the ssDNA Probe Layer Generated through EDC-Mediated Covalent Attachment to Nanocrystalline Diamond Using Fluorescence Microscopy. Langmuir 2008, 24, 9125–9134. [Google Scholar] [CrossRef] [PubMed]
- Mckenna, M.; Soberon, F.; Ricco, A.J.; Daniels, S.; Kelleher, S.M. Click chemistry as an immobilization method to improve oligonucleotide hybridization efficiency for nucleic acid assays. Sens. Actuators B Chem. 2016, 236, 286–293. [Google Scholar] [CrossRef]
- Tsekenis, G.; Chatzipetrou, M.; Tanner, J.; Chatzandroulis, S.; Thanos, D.; Tsoukalas, D.; Zergioti, I. Surface functionalization studies and direct laser printing of oligonucleotides toward the fabrication of a micromembrane DNA capacitive biosensor. Sens. Actuators 2012, 175, 123–131. [Google Scholar] [CrossRef]
- Beier, M.; Hoheisel, J.D. Versatile derivatisation of solid support media for covalent bonding on DNA-microchips. Nucleic Acids Res. 1999, 27, 1970–1977. [Google Scholar] [CrossRef]
- Alvarez, M.; Carrascosa, L.G.; Moreno, M.; Calle, A.; Zaballos, Á.; Lechuga, L.M.; Martínez-A, C.; Tamayo, J. Nanomechanics of the formation of DNA self-assembled monolayers and hybridization on microcantilevers. Langmuir 2004, 20, 9663–9668. [Google Scholar] [CrossRef]
- Mashhadizadeh, M.H.; Talemi, R.P. A new methodology for electrostatic immobilization of a non-labeled single strand DNA onto a self-assembled diazonium modified gold electrode and detection of its hybridization by differential pulse voltammetry. Talanta 2013, 103, 344–348. [Google Scholar] [CrossRef]
- Berger, R.; Delamarche, E.; Lang, H.P.; Gerber, C.; Gimzewski, J.K.; Meyer, E. Surface stress in the self-assembly of alkanethiols on Gold by a force microscopy technique. Science 1997, 276, 2021–2024. [Google Scholar] [CrossRef]
- Zhang, Z.; Chen, Q.; Knoll, W.; Foerch, R.; Holcomb, R.; Roitman, D. Plasma polymer film structure and DNA probe immobilization. Macromolecules 2003, 36, 7689–7694. [Google Scholar] [CrossRef]
- Demers, L.M.; Park, S.J.; Taton, T.A.; Li, Z.; Mirkin, C.A. Orthogonal assembly of nanoparticle building blocks on dip-pen nanolithographically generated templates of DNA. Angew. Chem. 2001, 40, 3071–3073. [Google Scholar] [CrossRef]
- Pépin, A.; Youinou, P.; Studer, V.; Lebib, A.; Chen, Y. Nanoimprint lithography for the fabrication of DNA electrophoresis chips. Microelectron. Eng. 2002, 61, 927–932. [Google Scholar] [CrossRef]
- Kim, K.H.; Sanedrin, R.G.; Ho, A.M.; Lee, S.W.; Moldovan, N.; Mirkin, C.; Espinosa, H.D. Direct Delivery and Submicrometer Patterning of DNA by a Nanofountain Probe. Adv. Mater. 2010, 20, 330–334. [Google Scholar] [CrossRef]
- Petersson, L.; Berthet, D.N.; Auger, A.; Borrebaeck, C.A.; Ikhlef, A.; Wingren, C. Generation of miniaturized planar ecombinant antibody arrays using a microcantilever-based printer. Nanotechnology 2014, 25, 275104. [Google Scholar] [CrossRef] [PubMed]
- Mishra, S.; Mukhopadhyay, R. Enhancing sensitivity in a piezoresistive cantilever-based label-free DNA detection assay using ssPNA sensor probes. J. Mater. Chem. B 2014, 2, 960. [Google Scholar]
- Chen, X.; Pan, Y.; Liu, H.; Bai, X.; Wang, N.; Zhang, B. Label-free detection of liver cancer cells by aptamer-based micro cantilever biosensor. Biosens. Bioelectron. 2016, 79, 353–358. [Google Scholar] [CrossRef]
- Cooper, O.; Phan, H.-P.; Fitzpatrick, T.; Dinh, T.; Huang, H.; Nguyen, N.-T.; Tiralongo, J. Picomolar detection of carbohydrate-lectin interactions on piezoelectrically printed microcantilever array. Biosens. Bioelectron. 2022, 205, 114088. [Google Scholar] [CrossRef]
- Southern, E.M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J. Mol. Biol. 1975, 98, 503–508 + IN3–IN5 + 509–517. [Google Scholar] [CrossRef]
- Alwine, J.C.; Kemp, D.J.; Stark, G.R. Method for Detection of Specific RNAs in Agarose Gels by Transfer to Diazobenzyloxymethyl-Paper and Hybridization with DNA Probes. Proc. Natl. Acad. Sci. USA 1977, 74, 5350–5354. [Google Scholar] [CrossRef]
- Mahmood, T.; Yang, P.C. Western blot: Technique, theory, and troubleshooting. N. Am. J. Med. Sci. 2012, 4, 429. [Google Scholar]
- Wu, G.; Datar, R.H.; Hansen, K.M.; Thundat, T.; Cote, R.J.; Majumdar, A. Bioassay of Prostate-specific Antigen (PSA) Using Microcantilevers. Nat. Biotechnol. 2001, 19, 856–860. [Google Scholar] [CrossRef]
- Zhang, H.Y.; Pan, H.Q.; Tang, J.L.; Zhang, B.L. Microcantilever Biosensor on the Biotin-avidin Interaction. Acta Chim. Sin. 2011, 69, 243–246. [Google Scholar]
- Hideo, T.; Jun’An, L.; Kazuhiro, O.; Kanesato, M.; Hiratani, K.; Baker, L.A. Efficient Biosensor Interfaces Based on Space-Controlled Self-Assembled Monolayers. Langmuir ACS J. Surf. Colloids 2009, 25, 1633–1637. [Google Scholar]
- Barker, J.M.; Mcinnes, J.L.; Murphy, P.J.; Symons, R.H. Dot-blot procedure with [32 P] DNA probes for the sensitive detection of avocado sunblotch and other viroids in plants. J. Virol. Methods 1985, 10, 87–98. [Google Scholar] [CrossRef] [PubMed]
- Nehls, P.; Adamkiewicz, J.; Rajewsky, M.F. Immuno-slot-blot: A highly sensitive immunoassay for the quantitation of carcinogen-modified nucleosides in DNA. J. Cancer Res. Clin. Oncol. 1984, 108, 23–29. [Google Scholar] [CrossRef]
- Pinkel, D.; Gray, J.W.; Trask, B.; Vand, E.G.; Fuscoe, J.; Van, D.H. Cytogenetic analysis by in situ hybridization with fluorescently labeled nucleic acid probes. Cold Spring Harb. Symp. Quant. Biol. 1986, 51, 151–157. [Google Scholar] [CrossRef] [PubMed]
- Dekken, H.V.; Pizzolo, J.G.; Kelsen, D.P.; Melamed, M.R. Targeted cytogenetic analysis of gastric tumors by in situ hybridization with a set of chromosome-specific DNA probes. Cancer 1990, 66, 491–497. [Google Scholar] [CrossRef]
- Wu, W.H.; Zhu, K.D. Proposition of a Silica Nanoparticle-Enhanced Hybrid Spin-Microcantilever Sensor Using Nonlinear Optics for Detection of DNA in Liquid. Sensors 2015, 15, 24848–24861. [Google Scholar] [CrossRef]
- Deng, Z.L.; Ge, Y.T.; Guan, W.G.; Nai, B.Z.; Qi, K.C. Nano gold Aggravating MEMS Cantilever Capacitance Detection Biochip. Adv. Mater. Res. 2010, 159, 429–433. [Google Scholar] [CrossRef]
- Ge, Y.; Yin, X.; Guo, C.; Yang, B. Study on gold nanoparticles aggravating MEMS cantilever gene detection biosensor. Appl. Mech. Mater. 2013, 336–338, 148–152. [Google Scholar] [CrossRef]
- Ge, Y.T.; Yin, X.T. Simulation of Gold Nanoparticles Aggravating MEMS Cantilever Optical Static Detection Biochip. Adv. Mater. Res. 2013, 694–697, 966–970. [Google Scholar] [CrossRef]
- Verd, J.; Uranga, A.; Abadal, G.; Teva, J.; Barniol, N. Monolithic mass sensor fabricated using a conventional technology with attogram resolution in air conditions. Appl. Phys. Lett. 2007, 91, 013501. [Google Scholar] [CrossRef]
- Li, Y.; Vancura, C.; Kirstein, K.U.; Lichtenberg, J.; Hierlemann, A. Monolithic Resonant-Cantilever-Based CMOS Microsystem for Biochemical Sensing. IEEE Trans. Circuits Syst. I Regul. Pap. 2008, 55, 2551–2560. [Google Scholar] [CrossRef]
- Mulhern, P.J.; Hubbard, T.; Arnold, C.S.; Blackford, B.L.; Jericho, M.H. A scanning force microscope with a fiber-optic-interferometer displacement sensor. Rev. Sci. Instrum. 1991, 62, 1280–1284. [Google Scholar] [CrossRef]
- Meyer, G.; Amer, N.M. Simultaneous measurement of lateral and normal forces with an optical-beam-deflection atomic force microscope. Appl. Phys. Lett. 1990, 57, 2089–2091. [Google Scholar] [CrossRef]
- Kang, K.; Sachan, A.; Nilsenhamilton, M.; Shrotriya, P. Aptamer functionalized microcantilever sensors for cocaine detection. Langmuir ACS J. Surf. Colloids 2011, 27, 14696–14702. [Google Scholar] [CrossRef]
- Yue, M.; Lin, H.; Dedrick, D.E.; Satyanarayana, S.; Majumdar, A.; Bedekar, A.S.; Jenkins, J.W.; Sundaram, S. A 2-D microcantilever array for multiplexed biomolecular analysis. J. Microelectromech. Syst. 2004, 13, 290–299. [Google Scholar] [CrossRef]
- Lechuga, L.; Tamayo, J.; Álvarez, M.; Carrascosa, L.; Yufera, A.; Rueda, A.; Plaza, J.; Zinoviev, K.; Domínguez, C.; Zaballos, A.; et al. A highly sensitive microsystem based on nanomechanical biosensors for genomics applications. Sens. Actuators B Chem. 2006, 118, 2–10. [Google Scholar] [CrossRef]
- Marvi, F.; Jafari, K. A Measurement Platform for Label-Free Detection of Biomolecules Based on a Novel Optical BioMEMS Sensor. IEEE Trans. Instrum. Meas. 2021, 70, 1–7. [Google Scholar] [CrossRef]
- Li, J.; Zhou, Y.X.; Guo, Y.X.; Wang, G.; Maier, R.R.J.; Hand, D.P.; MacPherson, W.N. Label-free ferrule-top optical fiber micro-cantilever biosensor. Sens. Actuators A Phys. 2018, 280, 505–512. [Google Scholar] [CrossRef]
- Forsen, E.; Abadal, G.; Ghatnekar-Nilsson, S.; Teva, J.; Verd, J.; Sandberg, R.; Svendsen, W.; Perez-Murano, F.; Esteve, J.; Figueras, E.; et al. Ultrasensitive mass sensor fully integrated with complementary metal-oxide-semiconductor circuitry. Appl. Phys. Lett. 2005, 87, 043507. [Google Scholar] [CrossRef]
- Algre, E. MEMS Ring Resonators for Laserless AFM with Sub-nano Newton Force Resolution. J. Microelectromech. Syst. 2012, 21, 385. [Google Scholar] [CrossRef]
- Lee, Y.; Lim, G.; Moon, W. A piezoelectric micro-cantilever bio-sensor using the mass-micro-balancing technique with self-excitation. Microsyst. Technol. 2007, 13, 563–567. [Google Scholar] [CrossRef]
- Loo, L.; Capobianco, J.A.; Wu, W.; Gao, X.T.; Shih, W.Y.; Wei-Heng Shih, W.H.; Pourrezaei, K.; Robinson, M.K.; Adams, G.P. Highly Sensitive Detection of HER2 Extracellular Domain in the Serum of Breast Cancer Patients by Piezoelectric Micro cantilevers. Anal. Chem. 2011, 83, 3392–3397. [Google Scholar] [CrossRef] [PubMed]
- Gunter, R.L.; Zhine, R.; Delinger, W.G.; Manygoats, K.; Kooser, A.; Porter, T.L. Investigation of DNA sensing using piezoresistive micro cantilever probes. IEEE Sens. J. 2004, 4, 430–433. [Google Scholar] [CrossRef]
- Tian, Y.; Liu, Y.; Wang, Y.; Xu, J.; Yu, X. A Flexible PI/Si/SiO2 Piezoresistive Microcantilever for Trace-Level Detection of Aflatoxin B1. Sensors 2021, 21, 1118. [Google Scholar] [CrossRef] [PubMed]
- Shekhawat, G.; Tark, S.-H.; Dravid, V.P. MOSFET-Embedded Microcantilevers for Measuring Deflection in Biomolecular Sensors. Science 2006, 311, 1592–1595. [Google Scholar] [CrossRef]
- Lee, D.; Hwang, K.S.; Kim, S.; Thundat, T. Rapid discrimination of DNA strands using an opto-calorimetric microcantilever sensor. Lab Chip 2014, 14, 4659–4664. [Google Scholar] [CrossRef]
- Ku, Y.F.; Huang, L.S.; Yen, Y.K. A Real-Time Thermal Self-Elimination Method for Static Mode Operated Freestanding Piezoresistive Micro cantilever-Based Biosensors. Biosensors 2018, 8, 18. [Google Scholar] [CrossRef]
- Rao, D.; Yan, T.; Qiao, Z.; Wang, Y.; Peng, Y.; Tu, H.; Wu, S.; Zhang, Q. Relay-type sensing mode: A strategy to push the limit on nanomechanical sensor sensitivity based on the magneto lever. Nano Res. 2022. [Google Scholar] [CrossRef]
- Mei, K.; Yan, T.; Wang, Y.; Rao, D.; Peng, Y.; Wu, W.; Chen, Y.; Ren, M.; Yang, J.; Wu, S.; et al. Magneto-Nanomechanical Array Biosensor for Ultrasensitive Detection of Oncogenic Exosomes for Early Diagnosis of Cancers. Small 2022, 2205445. [Google Scholar] [CrossRef]
- Strey, H.H.; Parsegian, V.A.; Podgornik, R. Equation of State for DNA Liquid Crystals: Fluctuation Enhanced Electrostatic Double Layer Repulsion. Phys. Rev. Lett. 1997, 78, 895–898. [Google Scholar] [CrossRef]
- Strey, H.H.; Podgornik, R.; Rau, D.C.; Parsegian, V.A. DNA—DNA interactions. Curr. Opin. Struct. Biol. 1998, 8, 309–313. [Google Scholar] [CrossRef] [PubMed]
- Strey, H.H.; Parsegian, V.A.; Podgornik, R. Equation of state for polymer liquid crystals: Theory and experiment. Phys. Rev. E 1999, 59, 999–1008. [Google Scholar] [CrossRef]
- Wu, G.; Ji, H.; Hansen, K.; Thundat, T.; Majumdar, A. Origin of nanomechanical cantilever motion generated from biomolecular interactions. Proc. Natl. Acad. Sci. USA 2001, 98, 1560–1564. [Google Scholar] [CrossRef]
- Hansen, K.M.; Ji, H.F.; Wu, G.; Datar, R.; Cote, R.; Majumdar, A.; Thundat, T. Cantilever-based optical deflection assay for discrimination of DNA single-nucleotide mismatches. Anal. Chem. 2001, 73, 1567–1571. [Google Scholar] [CrossRef] [PubMed]
- Mckendry, R.; Zhang, J.; Arntz, Y.; Strunz, T.; Hegner, M.; Lang, H.P.; Baller, M.K.; Certa, U.; Meyer, E.; Güntherodt, H.J.; et al. Multiple label-free biodetection and quantitative DNA-binding assays on a nanomechanical cantilever array. Proc. Natl. Acad. Sci. USA 2002, 99, 9783–9788. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Zhang, Y.; Ou-Yang, Z. Flexoelectric origin of nanomechanic deflection in DNA-microcantilever system. Biosens. Bioelectron. 2003, 18, 655–660. [Google Scholar] [CrossRef] [PubMed]
- Meyer, R.B. Piezoelectric Effects in Liquid Crystals. Phys. Rev. Lett. 1969, 22, 918–921. [Google Scholar] [CrossRef]
- Huber, F.; Hegner, M.; Gerber, C.; Güntherodt, H.-J.; Lang, H.P. Label free analysis of transcription factors using micro cantilever arrays. Biosens. Bioelectron. 2006, 21, 1599–1605. [Google Scholar] [CrossRef]
- Stachowiak, J.C.; Yue, M.; Castelino, K.; Chakraborty, A.; Majumdar, A. Chemomechanics of surface stresses induced by DNA hybridization. Langmuir 2006, 22, 263–268. [Google Scholar] [CrossRef]
- Godin, M.; Tabard-Cossa, V.; Miyahara, Y.; Monga, T.; Williams, P.J.; Beaulieu, L.Y.; Lennox, R.B.; Grutter, P. Cantilever-based sensing: The origin of surface stress and optimization strategies. Nanotechnology 2010, 21, 075501. [Google Scholar] [CrossRef]
- Hagan, M.F.; Majumdar, A.; Chakraborty, A.K. Nanomechanical Forces Generated by Surface Grafted DNA. J. Phys. Chem. B 2002, 106, 10163–10173. [Google Scholar] [CrossRef]
- Zhang, N.H.; Xing, J.J. An alternative model for elastic bending deformation of multilayered beams. J. Appl. Phys. 2006, 100, R53. [Google Scholar] [CrossRef]
- Zhang, N.H.; Chen, J.Z. An Alternative Two-Variable Model for Bending Problems of Multilayered Beams. J. Appl. Mech. 2008, 75, 044503. [Google Scholar] [CrossRef]
- Zhang, N.; Shan, J.; Xing, J. Piezoelectric properties of single-strand DNA molecular brush biolayers. Acta Mech. Solida Sin. 2007, 020, 206–210. [Google Scholar] [CrossRef]
- Zhang, N.H.; Shan, J.Y. An energy model for nanomechanical deflection of cantilever-DNA chip. J. Mech. Phys. Solids 2008, 56, 2328–2337. [Google Scholar] [CrossRef]
- Tan, Z.Q.; Li, J.J.; Zhang, N.H. An analytical model for nanomechanical behavior of microcantilever-DNA chip. In Proceedings of the International Conference on Smart Materials and Nanotechnology in Engineering, Weihai, China, 8–11 July 2009; p. 74933X. [Google Scholar]
- Zhang, N.H.; Chen, J.Z.; Wan, S.X. A Model for the Hybridization Exothermic Effect in Label-Free Biodetections by a Nanomechanical Cantilever-DNA Chip. Int. J. Thermophys. 2009, 30, 648–660. [Google Scholar] [CrossRef]
- Zhang, N.H.; Shan, J.Y. A model for nanomechanical behavior of microcantilever-ssDNA chip induced by electrostatic repulsion. In Piezoelectricity, Acoustic Waves, & Device Applications; IEEE: Piscataway, NJ, USA, 2010; p. 15. [Google Scholar]
- Zhang, N.H.; Shan, J.Y. Nanomechanical behaviors of microcantilever-based single-stranded DNA chips induced by counterion osmotic effects. Biomech. Model. Mechanobiol. 2011, 10, 229–234. [Google Scholar] [CrossRef]
- Zhang, N.H.; Meng, W.L.; Tan, Z.Q. A multi-scale model for the analysis of the inhomogeneity of elastic properties of DNA biofilm on microcantilevers. Biomaterials 2013, 34, 1833–1842. [Google Scholar] [CrossRef]
- Meng, W.L.; Zhang, N.H.; Tang, H.S.; Tan, Z.Q. Influence of disordered packing pattern on elastic modulus of single-stranded DNA film on substrate. Biomech. Model. Mechanobiol. 2015, 14, 1157–1165. [Google Scholar] [CrossRef]
- Junzheng, W.U.; Zhang, N. Clamped-end effect on static detection signals of DNA-microcantilever. Appl. Math. Mech. 2021, 42, 16. [Google Scholar]
- Merlo, M.; Negretto, F.; Soncini, M.; Montevecchi, F.M. Electrostatic Nanomechanics of Cantilever Biosensors. In Materials Science Forum; Trans Tech Publications, Ltd.: Bäch, Switzerland, 2007; pp. 595–601. [Google Scholar]
- Sushko, M.L.; Harding, J.H.; Shluger, A.L.; McKendry, R.A.; Watari, M. Physics of Nanomechanical Biosensing on Cantilever Arrays. Adv. Mater. 2008, 20, 3848–3853. [Google Scholar] [CrossRef]
- Sushko, M.L. Nanomechanics of organic/inorganic interfaces: A theoretical insight. Faraday Discuss. 2009, 143, 63–80. [Google Scholar] [CrossRef]
- Huang, L.; Seker, E.; Landers, J.P.; Begley, M.R.; Utz, M. Molecular interactions in surface-assembled monolayers of short double-stranded DNA. Langmuir ACS J. Surf. Colloids 2010, 26, 11574–11580. [Google Scholar] [CrossRef] [PubMed]
- Yue, M.; Dedrick, D.E.; Lin, H.; Satyanarayana, S.; Majumdar, A. Microcantilever Arrays for Multiplexed Biomolecular Analysis. In Proceedings of the ASME 2002 International Mechanical Engineering Congress and Exposition, New Orleans, LA, USA, 7–12 November 2002; pp. 585–589. [Google Scholar]
- Biswal, S.L.; Raorane, D.; Chaiken, A.; Birecki, H.; Majumdar, A. Nanomechanical detection of DNA melting on microcantilever surfaces. Anal. Chem. 2006, 78, 7104. [Google Scholar] [CrossRef]
- Biswal, S.L.; Raorane, D.; Chaiken, A.; Majumdar, A. Using a Microcantilever Array for Detecting Phase Transitions and Stability of DNA. Clin. Lab. Med. 2007, 27, 163–171. [Google Scholar] [CrossRef]
- Ng, J.D.; Dowell, J.J.; Kar, A.K.; Hansen, K.; George, M.A. Measurement of Temperature Induced Unfolding of DNA Hairpins by Microcantilever Sensors. Open J. Appl. Biosens. 2013, 2, 78–82. [Google Scholar] [CrossRef]
- Mertens, J.; Tamayo, J.; Kosaka, P.; Calleja, M. Observation of spermidine-induced attractive forces in self-assembled monolayers of single stranded DNA using a micro cantilever sensor. Appl. Phys. Lett. 2011, 98, 153704. [Google Scholar] [CrossRef]
- Wu, J.Z.; Zhou, M.H.; Zhang, N.H. The effect of microscopic attractive interactions on piezoelectric coefficients of nanoscale DNA films and its resultant mirocantilever-based biosensor signals. J. Phys. D Appl. Phys. 2017, 50, 415403. [Google Scholar] [CrossRef]
- Mertens, J.; Rogero, C.; Calleja, M.; Ramos, D.; Martín-Gago, J.A.; Briones, C.; Tamayo, J. Label-free detection of DNA hybridization based on hydration-induced tension in nucleic acid films. Nat. Nanotechnol. 2008, 3, 301–307. [Google Scholar] [CrossRef]
- Domínguez, C.M.; Kosaka, P.M.; Mokry, G.; Pini, V.; Malvar, O.; del Rey, M.; Ramos, D.; San Paulo, A.; Tamayo, J.; Calleja, M. Hydration Induced Stress on DNA Monolayers Grafted on Microcantilevers. Langmuir 2014, 30, 10962–10969. [Google Scholar] [CrossRef] [PubMed]
- Domínguez, C.M.; Ramos, D.; Mendietamoreno, J.I.; Fierro, J.L.G.; Mendieta, J.; Tamayo, J.; Calleja, M. Effect of water-DNA interactions on elastic properties of DNA self-ass embled monolayers. Sci. Rep. 2017, 7, 536. [Google Scholar] [CrossRef] [PubMed]
- Shu, W.; Liu, D.; Watari, M.; Riener, C.K.; Strunz, T.; Welland, M.E.; Balasubramanian, S.; McKendry, R.A. DNA molecular motor driven micromechanical cantilever arrays. J. Am. Chem. Soc. 2005, 127, 17054–17060. [Google Scholar] [CrossRef]
- Zhang, J.; Lang, H.P.; Yoshikawa, G.; Gerber, C. Optimization of DNA Hybridization Efficiency by pH-Driven Nanomechanical Bending. Langmuir 2012, 28, 6494–6501. [Google Scholar] [CrossRef] [PubMed]
- Zhang, N.-H.; Meng, W.-L.; Zhang, C.-Y.; Wu, J.-Z. The pH-dependent electrical potential and elastic modulus of a nanoscale DNA film and the resultant bending signal for a microcantilever biosensor. arXiv 2017, arXiv:1706.08196. [Google Scholar]
- Zhou, M.H.; Meng, W.L.; Zhang, C.Y.; Li, X.B.; Wu, J.Z.; Zhang, N.H. The pH-dependent elastic properties of nanoscale DNA films and the resultant bending signals for microcantilever biosensors. Soft Matter 2018, 14, 3028. [Google Scholar] [CrossRef] [PubMed]
- Su, M.; Li, S.; Dravid, V.P. Microcantilever resonance-based DNA detection with nanoparticle probes. Appl. Phys. Lett. 2003, 82, 3562–3564. [Google Scholar] [CrossRef]
- Ilic, B.; Yang, Y.; Aubin, K.; Reichenbach, R.; Krylov, S.; Craighead, H.G. Enumeration of DNA molecules bound to a nanomechanical oscillator. Nano Lett. 2005, 5, 925–929. [Google Scholar] [CrossRef]
- Zhang, J.; Lang, H.P.; Huber, F.; Bietsch, A.; Grange, W.; Certa, U.; McKendry, R.; Güntherodt, H.J.; Hegner, M.; Gerber, C. Rapid and label-free nanomechanical detection of biomarker transcripts in human RNA. Nat. Nanotechnol. 2006, 1, 214–220. [Google Scholar] [CrossRef]
- And, K.R.; Mutharasan, R. PEMC-based Method of Measuring DNA Hybridization at Femtomolar Concentration Directly in Human Serum and in the Presence of Copious Noncomplementary Strands. Anal. Chem. 2007, 79, 7392–7400. [Google Scholar]
- Miyachi, Y.; Shimizu, N.; Ogino, C.; Kondo, A. Selection of DNA aptamers using atomic force microscopy. Nucleic Acids Res. 2010, 38, e21. [Google Scholar] [CrossRef] [PubMed]
- Miyachi, Y.; Ogino, C.; Amino, T.; Kondo, A. Development of a novel aptamer-based sensing system using atomic force microscopy. J. Biosci. Bioeng. 2011, 112, 511–514. [Google Scholar] [CrossRef] [PubMed]
- Park, C.; Kang, J.; Baek, I.; You, J.; Jang, K.; Na, S. Highly sensitive and selective detection of single-nucleotide polymorphisms using gold nanoparticle MutS enzymes and a micro cantilever resonator. Talanta 2019, 205, 120154. [Google Scholar] [CrossRef] [PubMed]
- Stevenson, K.A.; Mehta, A.; Sachenko, P.; Hansen, K.M.; Thundat, T. Nanomechanical Effect of Enzymatic Manipulation of DNA on Microcantilever Surfaces. Langmuir 2002, 18, 8732–8736. [Google Scholar] [CrossRef]
- Muňoz, R.; Sandoval, F.A.; Wilson, C.; Melo, F. Pulling on super paramagnetic beads with micro cantilevers: Single molecule mechanical assay application. Phys. Biol. 2015, 12, 046011. [Google Scholar] [CrossRef]
- Velanki, S.; Ji, H.F. Detection of feline coronavirus using microcantilever sensors. Meas. Sci. Technol. 2006, 17, 2964–2968. [Google Scholar] [CrossRef]
- Hwang, K.; Lee, S.; Eom, K.; Lee, J.H.; Lee, Y.S.; Park, J.H.; Yoon, D.S.; Kim, T.S. Nanomechanical microcantilever operated in vibration modes with use of RNA aptamer as receptor molecules for label-free detection of HCV helicase. Biosens. Bioelectron. 2007, 23, 459–465. [Google Scholar] [CrossRef]
- Kim, T.S. In/Ex-situ Detection of HBV DNA Using Dynamic Micro cantilever. Adv. Mater. Res. 2009, 74, 337–340. [Google Scholar] [CrossRef]
- Cha, B.H.; Lee, S.M.; Park, J.C.; Hwang, K.S.; Kim, T.S. Detection of Hepatitis B Virus (HBV) DNA at femtomolar concentrations using a silica nanoparticle-enhanced micro cantilever sensor. Biosens. Bioelectron. 2010, 25, 130–135. [Google Scholar] [CrossRef]
- Kuan, S.; Chi, S.-C.; Cheng, Y.-J.; Chia, T.-J.; Huang, L.-S. Binding kinetics of grouper nervous necrosis viruses with functionalized antimicrobial peptides by nanomechanical detection. Biosens. Bioelectron. 2012, 31, 116–123. [Google Scholar] [CrossRef]
- Alodhayb, A.; Brown, N.; Rahman, S.M.S.; Harrigan, R.; Beaulieu, L.Y. Towards detecting the human immunodeficiency virus using microcantilever sensors. Appl. Phys. Lett. 2013, 102, 115–126. [Google Scholar] [CrossRef]
- Kim, H.H.; Jeon, H.J.; Cho, H.K.; Cheong, J.H.; Moon, H.S.; Go, J.S. Highly sensitive micro cantilever biosensors with enhanced sensitivity for detection of human papilloma virus infection. Sens. Actuators B Chem. 2015, 221, 1372–1383. [Google Scholar] [CrossRef]
- Wang, Y.; Yan, T.; Mei, K.; Rao, D.; Wu, W.; Chen, Y.; Peng, Y.; Wang, J.; Wu, S.; Zhang, Q. Nanomechanica1 assay for u1trasensitive and rapid detection of SARS-CoV-2 based on peptide nuc1eic acid. Nano Res. 2023, 16, 1183–1195. [Google Scholar] [CrossRef]
- Pablo, P.J.D. Atomic Force Microscopy of Viruses. Sub Cell. Biochem. 2013, 68, 247–271. [Google Scholar]
- Rijal, K.; Mutharasan, R. A method for DNA-based detection of E. coli O157: H7 in a proteinous background using piezoelectric-excited cantilever sensors. Analyst 2013, 138, 2943–2950. [Google Scholar] [CrossRef]
- Xu, T.; Yu, H.; Xu, P.; Xu, W.; Chen, W.; Chen, C.; Li, X. Real-time enzyme-digesting identification of double-strand DNA in a resonance-cantilever embedded micro-chamber. Lab A Chip 2014, 14, 1206–1214. [Google Scholar] [CrossRef]
- Zheng, F.; Wang, P.; Du, Q.; Chen, Y.; Liu, N. Simultaneous and Ultrasensitive Detection of Foodborne Bacteria by Gold Nanoparticles-Amplified Microcantilever Array Biosensor. Front. Chem. 2019, 7, 232. [Google Scholar] [CrossRef] [PubMed]
- Khemthongcharoen, N.; Wonglumsom, W.; Suppat, A.; Jaruwongrungsee, K.; Tuantranont, A.; Promptmas, C. Piezoresistive microcantilever-based DNA sensor for sensitive detection of pathogenic Vibrio cholerae, O1 in food sample. Biosens. Bioelectron. 2015, 63, 347–353. [Google Scholar] [CrossRef] [PubMed]
- Etayash, H.; Khan, M.F.; Kaur, K.; Thundat, T. Microfluidic cantilever detects bacteria and measures their susceptibility to antibiotics in small confined volumes. Nat. Commun. 2016, 7, 12947. [Google Scholar] [CrossRef]
- Wang, L.; Fu, D.; Liu, X.; Zhao, J.; Zhao, J.; Yuan, Q.; Zhu, Y.; Yang, J.; Yang, F. Highly sensitive biosensor based on a microcantilever and alternating current electrothermal Atechnology. J. Micromech. Microeng. 2021, 31, 015009. [Google Scholar] [CrossRef]
- Liu, X.; Wang, L.; Zhao, J.; Zhu, Y.; Yang, J.; Yang, F. Enhanced Binding Efficiency of Microcantilever Biosensor for the Detection of Yersinia. Sensors 2019, 19, 3326. [Google Scholar] [CrossRef] [PubMed]
- Lakshmi, S.; Almeida, A.; Bagchi, A.; Joseph, M. Detection of tuberculosis using MEMS. Microsyst. Technol. 2021, 27, 87–95. [Google Scholar] [CrossRef]
- Liu, Y.; Schweizer, L.M.; Wang, W.; Reuben, R.L.; Schweizer, M.; Shu, W. Label-free and real-time monitoring of yeast cell growth by the bending of polymer microcantilever biosensors. Sens. Actuators B Chem. 2013, 178, 621–626. [Google Scholar] [CrossRef]
- Ricciardi, C.; Fiorilli, S.; Bianco, S.; Canavese, G.; Castagna, R.; Ferrante, I.; Digregorio, G.; Marasso, S.L.; Napione, L.; Bussolino, F. Development of microcantilever-based biosensor array to detect Angiopoietin-1, a marker of tumor angiogenesis. Biosens. Bioelectron. 2010, 25, 1193–1198. [Google Scholar] [CrossRef]
- Le, T.T.T.; Pham, V.T.; Phan, T.N.K.; Pham, V.B.; Le, V.T.; Ton, D.H. Detecting the golgi protein 73 of liver cancer with micro cantilever. Adv. Nat. Sci. Nanosci. Nanotechnol. 2014, 5, 045016. [Google Scholar] [CrossRef][Green Version]
- Etayash, H.; Jiang, K.; Azmi, S.; Thundat, T.; Kaur, K. Real-time Detection of Breast Cancer Cells Using Peptide-functionalized Microcantilever Arrays. Sci. Rep. 2015, 5, 13967. [Google Scholar] [CrossRef]
- Patil, S.B.; Al-Jehani, R.M.; Etayash, H.; Turbe, V.; Jiang, K.; Bailey, J.; Al-Akkad, W.; Soudy, R.; Kaur, K.; McKendry, R.A.; et al. Modified cantilever arrays improve sensitivity and reproducibility of nano mechanical sensing in living cells. Commun. Biol. 2018, 1, 175. [Google Scholar] [CrossRef] [PubMed]
- Shim, S.; Kim, M.G.; Jo, K.; Kang, Y.S.; Lee, B.; Yang, S.; Shin, S.-M.; Lee, J.-H. Dynamic characterization of human breast cancer cells using a piezoresistive microcantilever. J. Biomech. Eng. 2010, 132, 104501. [Google Scholar] [CrossRef]
- Savran, C.A.; Knudsen, S.M.; Ellington, A.D.; Manalis, S.R. Micromechanical detection of proteins using aptamer-based receptor molecules. Anal. Chem. 2004, 76, 3194–3198. [Google Scholar] [CrossRef]
- Yoo, K.A.; Kim, J.H.; Nahm, B.H.; Kang, C.J.; Kim, Y.S. Fabrication and characteristics of microcantilever-based biosensor for detection of the protein-ligand binding. J. Phys. Conf. Ser. 2007, 61, 1308–1311. [Google Scholar] [CrossRef]
- Wang, B.; Huang, F.; Nguyen, T.H.; Lin, Q. Microcantilever based label free thermal characterization of bio-molecular affinity binding. In Proceedings of the IEEE International Conference on Micro Electro Mechanical Systems, Hong Kong, China, 24–28 January 2010; pp. 855–858. [Google Scholar]
- Bosco, F.G.; Bache, M.; Yang, J.; Chen, C.H.; Hwu, E.-T.; Lin, Q.; Boisen, A. Micromechanical PDGF recognition via lab-on-a-disc apt sensor arrays. Sens. Actuators A Phys. 2013, 195, 154–159. [Google Scholar] [CrossRef][Green Version]
- Zhai, L.; Wang, T.; Kang, K.; Zhao, Y.; Shrotriya, P.; Nilsen, H.M. An RNA aptamer-based microcantilever sensor to detect the inflammatory marker, mouse lipocalin-2. Anal. Chem. 2012, 84, 8763–8770. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.-W.; Tong, Z.-Y.; Liu, B.; Hao, L.-Q.; Mu, X.-H.; Zhang, J.-P.; Gao, C. Piezoresistive microcantilever aptasensor for ricin detection and kinetic analysis. Aip Adv. 2015, 5, 041324. [Google Scholar] [CrossRef]
- Agarwal, D.K.; Prasad, A.; Vinchurkar, M.; Gandhi, S.; Prabhakar, D.; Mukherji, S.; Rao, V.R. Detection of heart-type fatty acid-binding protein (h-FABP) using piezoresistive polymer microcantilevers functionalized by a dry method. Appl. Nanosci. 2018, 8, 1031–1042. [Google Scholar] [CrossRef]
- Agarwal, D.K.; Kushagra, A.; Ashwin, M.; Shukla, A.; Palaparthy, V. Sensitive detection of cardiac troponin-I protein using fabricated piezoresistive microcantilevers by a novel method of asymmetric biofunctionalization. Nanotechnology 2020, 31, 115503. [Google Scholar] [CrossRef] [PubMed]
- Ndieyira, J.W.; Watari, M.; Barrera, A.D.; Zhou, D.; Vögtli, M.; Batchelor, M.; Cooper, M.A.; Strunz, T.; Horton, M.A.; Abell, C.; et al. Nanomechanical detection of antibiotic-mucopeptide binding in a model for superbug drug resistance. Nat. Nanotechnol. 2008, 3, 691. [Google Scholar] [CrossRef] [PubMed]
- Hou, H.; Bai, X.; Xing, C.; Gu, N.Y.; Zhang, B.L.; Tang, J.L. Aptamer-based cantilever array sensors for oxytetracycline detection. Anal. Chem. 2013, 85, 2010–2014. [Google Scholar] [CrossRef]
- Bai, X.; Hou, H.; Zhang, B.; Tang, J. Label-free detection of kanamycin using aptamer-based cantilever array sensor. Biosens. Bioelectron. 2014, 56, 112–116. [Google Scholar] [CrossRef]
- Chen, X.; Bai, X.; Li, H.; Zhang, B. Aptamer-based microcantilever array biosensor for detection of fumonisin B-1. RSC Adv. 2015, 5, 35448–35452. [Google Scholar] [CrossRef]
- Li, H.; Bai, X.; Wang, N.; Chen, X.; Li, J.; Zhang, Z.; Tang, J. Aptamer-based microcantilever biosensor for ultrasensitive detection of tumor marker nucleolin. Talanta 2015, 146, 727. [Google Scholar] [CrossRef]
- Cherian, S.; Gupta, R.K.; Mullin, B.C.; Thundat, T. Detection of heavy metal ions using protein-functionalized microcantilever sensors. Biosens. Bioelectron. 2004, 19, 411–416. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.-M.; Pan, H.-Q.; Wu, S.-H.; Zhang, B.L. Interaction between tripeptide Gly-Gly-His and Cu2+ probed by microcantilevers. Chin. J. Anal. Chem. 2009, 37, 783–787. [Google Scholar] [CrossRef]
- Peng, R.P.; Chen, B.; Ji, H.F.; Wu, L.-Z.; Tung, C.-H. Highly sensitive and selective detection of beryllium ions using a microcantilever modified with benzo-9-crown-3 doped hydrogel. Analyst 2012, 137, 1220–1224. [Google Scholar] [CrossRef] [PubMed]
- Peng, R.P.; Xing, L.B.; Wang, X.J.; Wu, C.J.; Chen, B.; Ji, H.F.; Wu, L.Z.; Tung, C.H. Detection of Pb(2+) in Aqueous Solution by Using a DNA-modified Micro cantilever. Anal. Sci. Int. J. Jpn. Soc. Anal. Chem. 2016, 32, 1065–1069. [Google Scholar] [CrossRef]
- Peng, R.; Xing, L.B.; Wang, X.J.; Wu, C.-J.; Chen, B.; Ji, H.-F.; Wu, L.-Z.; Tung, C.-H. A beryllium-selective microcantilever sensor modified with benzo-9-crown-3 functionalized polymer brushes. Anal. Methods 2017, 9, 3356–3360. [Google Scholar] [CrossRef]
- You, J.; Song, Y.; Park, C.; Jang, K.; Na, S. A microcantilever-based silver ion sensor using DNA-functionalized gold nanoparticles as mass amplifier. Nanotechnology 2017, 28, 245501. [Google Scholar] [CrossRef]
- Wang, Y.; Rao, D.; Wu, X.; Zhang, Q.; Wu, S. Aptamer-based microcantilever-array biosensor for ultra-sensitive and rapid detection of okadaic acid. Microchem. J. 2020, 160, 105644. [Google Scholar] [CrossRef]
- Li, C.; Zhang, G.; Wu, S.; Zhang, Q. Aptamer-based microcantilever-array biosensor for profenofos detection. Anal. Chim. Acta 2018, 1020, 116–122. [Google Scholar] [CrossRef]
- Zhang, G.; Li, C.; Wu, S.; Zhang, Q. Label-free aptamer-based detection of microcystin-LR using a microcantilever array biosensor. Sens. Actuators B Chem. 2018, 260, 42–47. [Google Scholar] [CrossRef]
- Ricciardi, C.; Canavese, G.; Castagna, R.; Digregorio, G.; Ferrante, I.; Marasso, S.L.; Ricci, A.; Alessandria, V.; Rantsiou, K.; Cocolin, L.S. Online Portable Microcantilever Biosensors for Salmonella enterica Serotype Enteritidis Detection. Food Bioprocess Technol. 2010, 3, 956–960. [Google Scholar] [CrossRef]
- Huang, C.W.; Huang, Y.J.; Lin, T.H.; Lin, C.T.; Lee, J.K.; Chen, L.G.; Hsiao, P.Y.; Wu, B.R.; Hsueh, H.T.; Kuo, B.J.; et al. An Integrated Microcantilever-based Wireless DNA Chip for Hepatitis B Virus (HBV) DNA Detection. In Proceedings of the 15th International Conference on Miniaturized Systems for Chemistry and Life Science, Seattle, WA, USA, 2–6 October 2011. [Google Scholar]
- Huang, Y.-J.; Huang, C.-W.; Lin, T.-H.; Lin, C.-T.; Chen, L.-G.; Hsiao, P.-Y.; Wu, B.-R.; Hsueh, H.-T.; Kuo, B.-J. A CMOS Cantilever-Based Label-Free DNA SoC with Improved Sensitivity for Hepatitis B Virus Detection. IEEE Trans. Biomed. Circuits Syst. 2013, 7, 820–831. [Google Scholar] [CrossRef]
- Huang, C.W.; Hsueh, H.T.; Huang, Y.J.; Liao, H.H.; Tsai, H.H.; Juang, Y.Z.; Lin, T.H.; Lu, S.S.; Lin, C.T. A fully integrated wireless CMOS microcantilever lab chip for detection of DNA from Hepatitis B virus (HBV). Sens. Actuators B Chem. 2013, 181, 867–873. [Google Scholar] [CrossRef]
- Wang, S.; Wangm, J.; Zhu, Y.; Yang, J.; Yang, F. Cantilever with immobilized antibody for liver cancer biomarker detection. J. Semicond. 2014, 35, 60–65. [Google Scholar] [CrossRef]
- Wang, J.; Wang, S.; Wang, X.; Zhu, Y.; Yang, J.; Yang, F. Cantilever array sensor for multiple liver cancer biomarkers detection. In Proceedings of the Sensors, 2014 IEEE, Valencia, Spain, 2–5 November 2014; pp. 343–346. [Google Scholar]
- Wang, S.; Wang, J.; Zhu, Y.; Yang, J.; Yang, F. A new device for liver cancer biomarker detection with high accuracy. Sens. Bio-Sens. Res. 2015, 4, 40–45. [Google Scholar] [CrossRef]
- Wang, J.; Zhu, Y.; Wang, X.; Wang, S.; Yang, J.; Yang, F. A High-Throughput Cantilever Array Sensor for Multiple Liver Cancer Biomarkers Detection. IEEE Sens. J. 2016, 16, 4675–4682. [Google Scholar] [CrossRef]
- Liao, J.; Wang, J.; Li, N.; Zhu, Y.; Zhang, J.; Yang, J.; Yang, F. Resonance frequency tracking and measuring system for micro-electromechanical cantilever array sensors. Microsyst. Technol. 2017, 23, 2013–2021. [Google Scholar] [CrossRef]
- Zhao, J.; Wang, L.; Fu, D.; Zhao, D.; Wang, Y.; Yuan, Q.; Zhu, Y.; Yang, J.; Yang, F. Gold nanoparticles amplified microcantilever biosensor for detecting protein biomarkers with high sensitivity. Sens. Actuators A Phys. 2021, 321, 112563. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, H.; Yang, S.; Zeng, J.; Li, X.; Chuai, R. A Genosensor Based on the Modification of a Microcantilever: A Review. Micromachines 2023, 14, 427. https://doi.org/10.3390/mi14020427
Zhang H, Yang S, Zeng J, Li X, Chuai R. A Genosensor Based on the Modification of a Microcantilever: A Review. Micromachines. 2023; 14(2):427. https://doi.org/10.3390/mi14020427
Chicago/Turabian StyleZhang, He, Shuang Yang, Jian Zeng, Xin Li, and Rongyan Chuai. 2023. "A Genosensor Based on the Modification of a Microcantilever: A Review" Micromachines 14, no. 2: 427. https://doi.org/10.3390/mi14020427
APA StyleZhang, H., Yang, S., Zeng, J., Li, X., & Chuai, R. (2023). A Genosensor Based on the Modification of a Microcantilever: A Review. Micromachines, 14(2), 427. https://doi.org/10.3390/mi14020427







