High-Efficiency Single-Cell Containment Microdevices Based on Fluid Control
Abstract
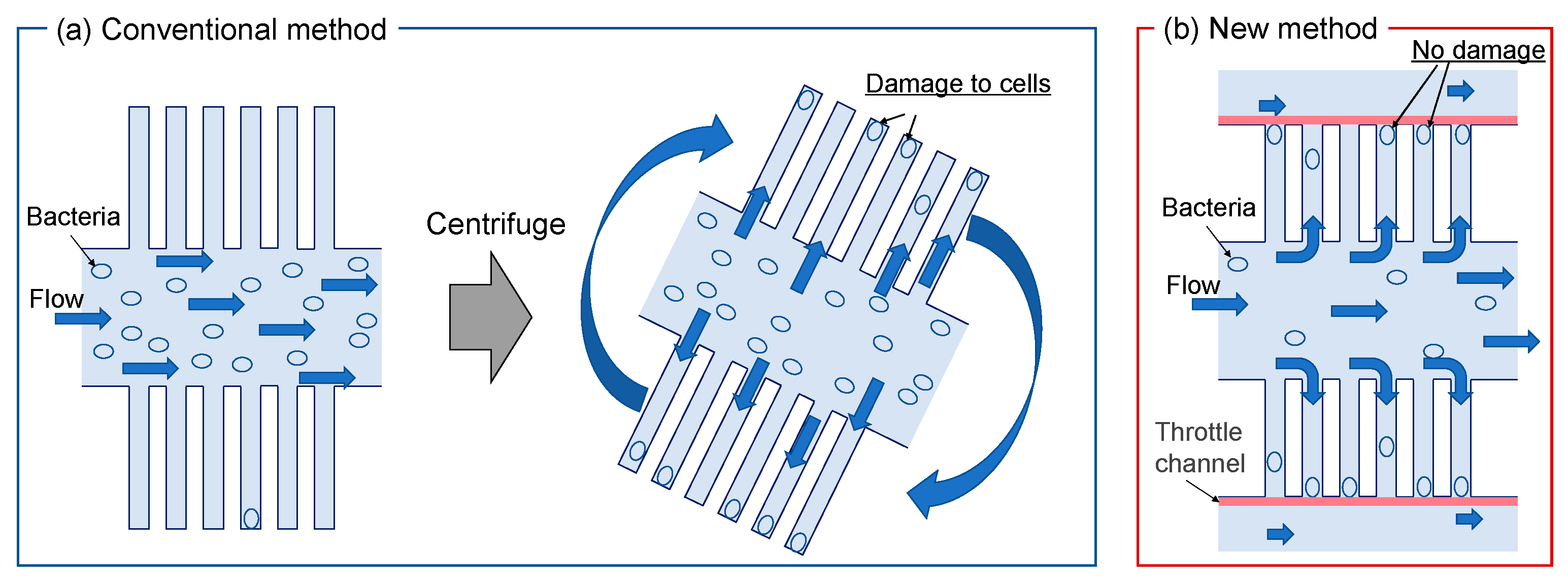
1. Introduction
2. Materials and Methods
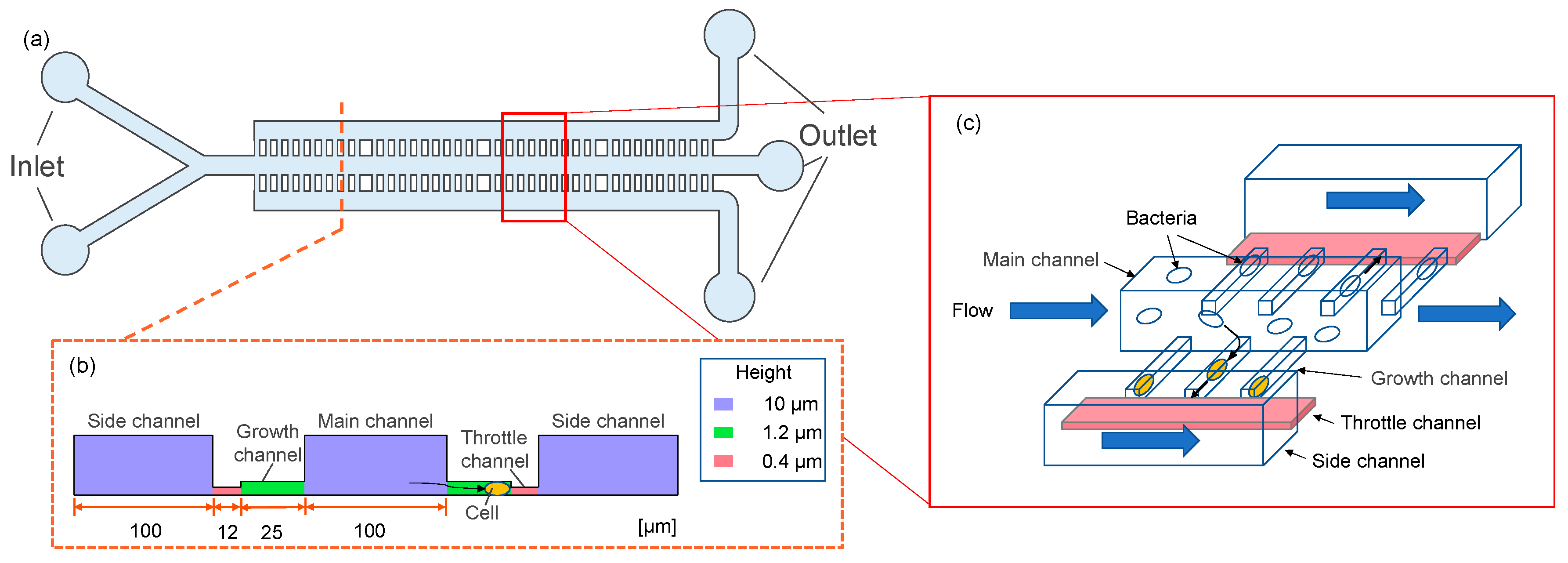
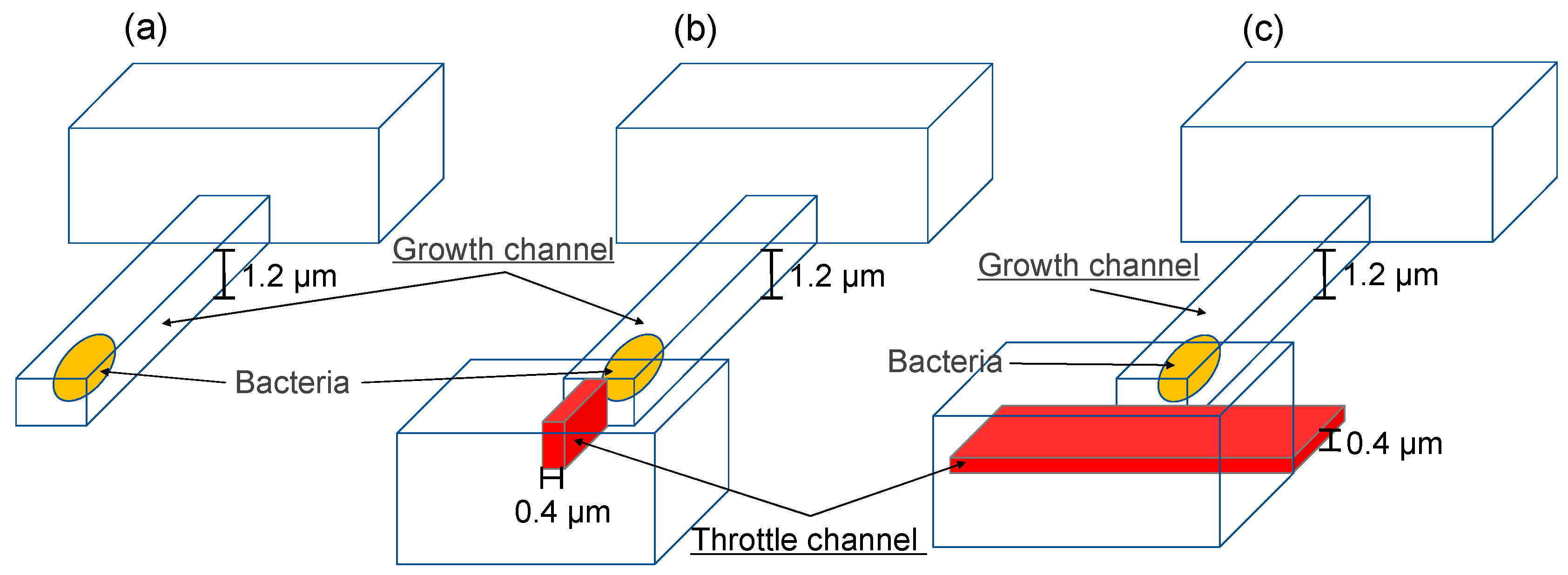
2.1. Device Design
2.2. Device Fabrication
2.3. Fluid Simulation
2.4. Bacterial Culture and Fluid Experiments
2.4.1. Bacterial Strains and Culture Conditions
2.4.2. Procedure for Microfluidic Experiments
3. Results and Discussion
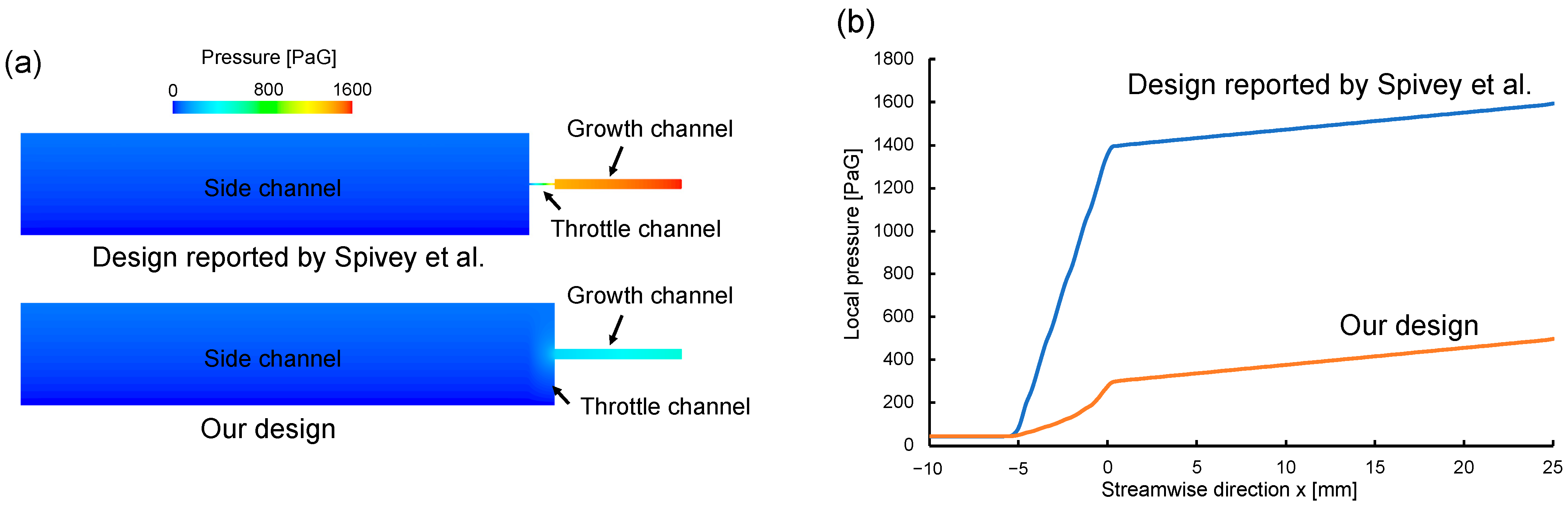
3.1. Computational Fluid Dynamics Model and Simulations
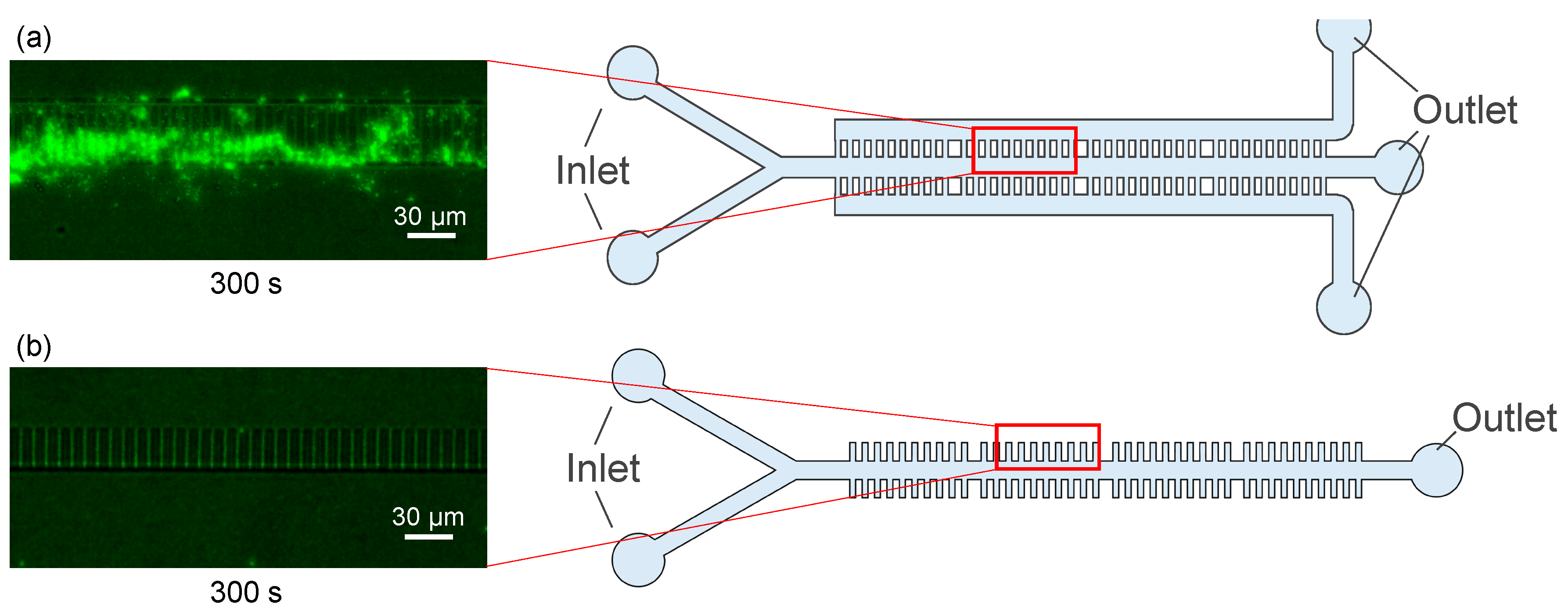
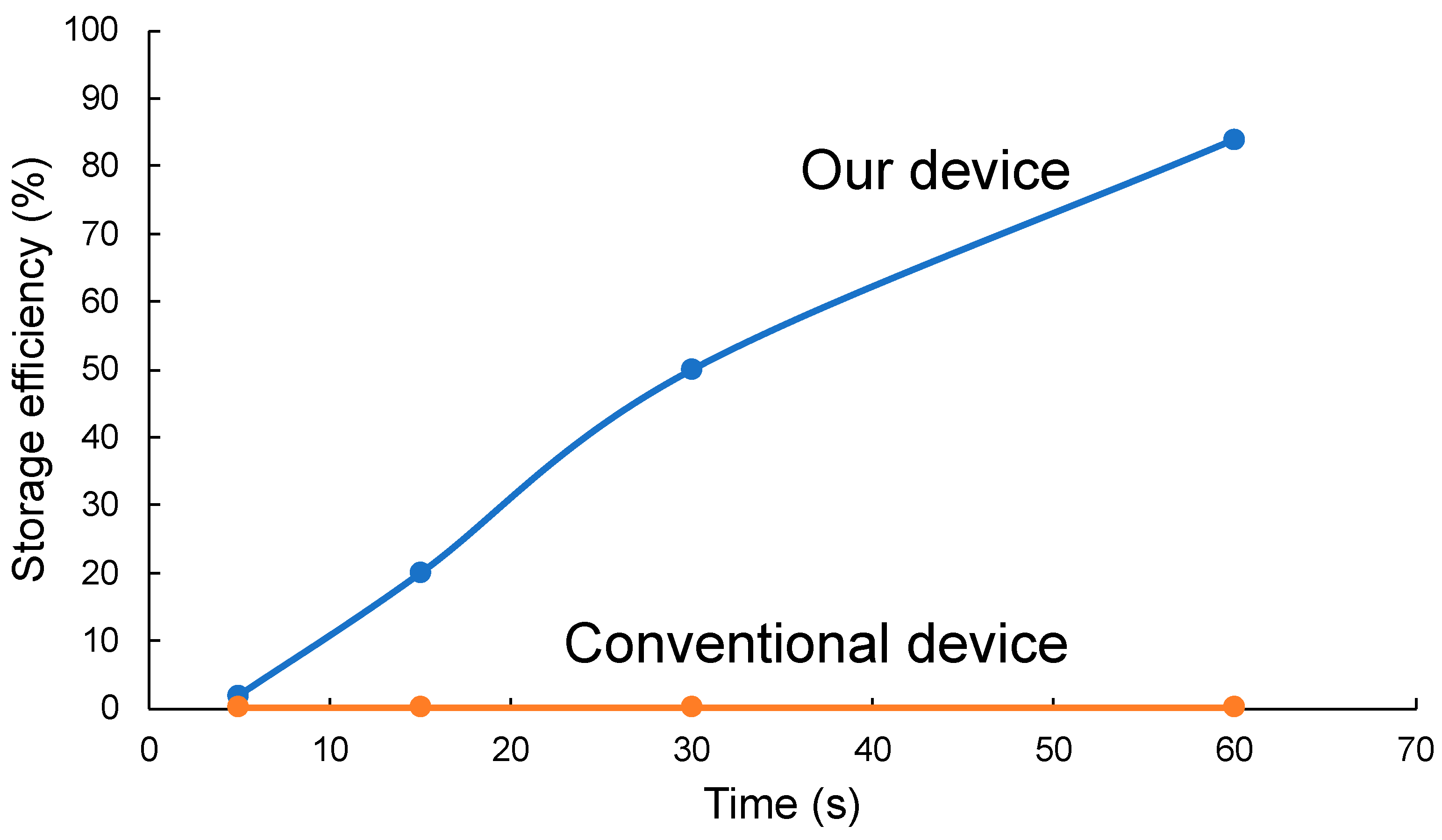
3.2. Fluid Experiments (with Microbeads)
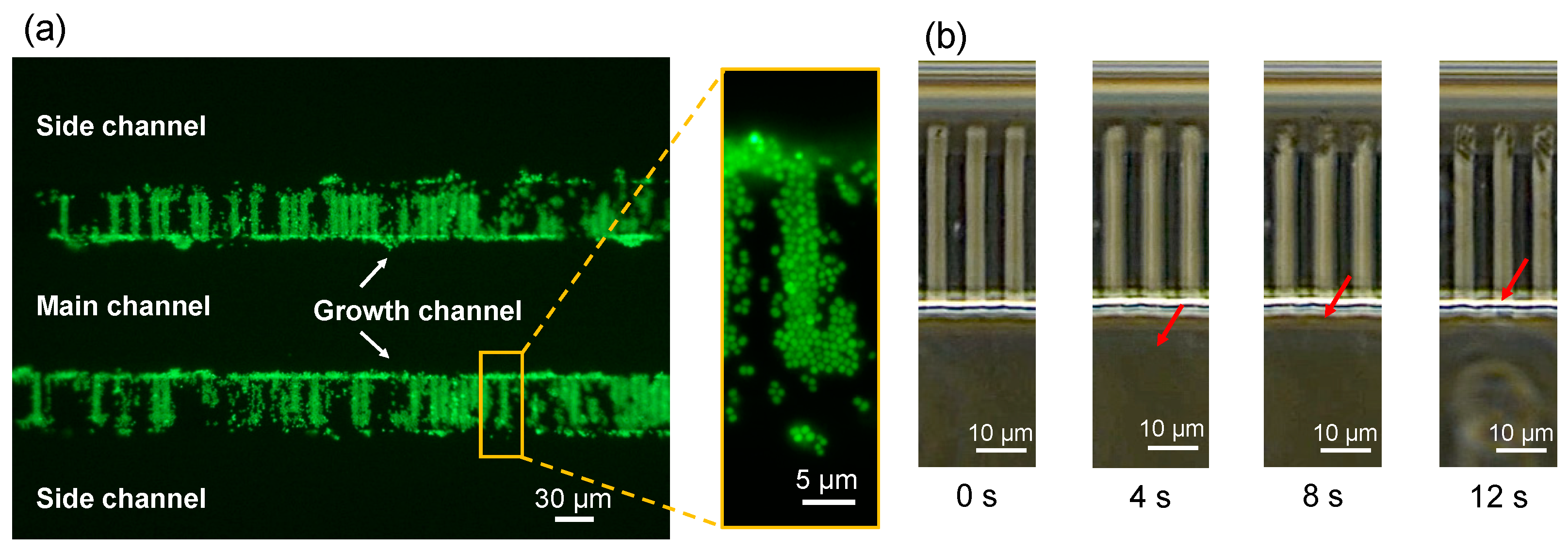
3.3. Fluid Experiments (with Bacteria)
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Grünberger, A.; Wiechert, W.; Kohlheyer, D. Single-cell microfluidics: Opportunity for bioprocess development. Curr. Opin. Biotechnol. 2014, 29, 15–23. [Google Scholar] [CrossRef] [PubMed]
- Bai, Y.; Gao, M.; Wen, L.; He, C.; Chen, Y.; Liu, C.; Fu, X.; Huang, S. Applications of Microfluidics in Quantitative Biology. Biotechnol. J. 2018, 13, 1700170. [Google Scholar] [CrossRef] [PubMed]
- Potvin-Trottier, L.; Luro, S.; Paulsson, J. Microfluidics and single-cell microscopy to study stochastic processes in bacteria. Curr. Opin. Microbiol. 2018, 43, 186–192. [Google Scholar] [CrossRef]
- Probst, C.; Grünberger, A.; Wiechert, W.; Kohlheyer, D. Microfluidic growth chambers with optical tweezers for full spatial single-cell control and analysis of evolving microbes. J. Microbiol. Methods 2013, 95, 470–476. [Google Scholar] [CrossRef] [PubMed]
- Grünberger, A.; Probst, C.; Helfrich, S.; Nanda, A.; Stute, B.; Wiechert, W.; von Lieres, E.; Nöh, K.; Frunzke, J.; Kohlheyer, D. Spatiotemporal Microbial Single-Cell Analysis Using a High-Throughput Microfluidics Cultivation Platform. Cytometry A 2015, 87, 1101. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.; Kim, S.H.; Lee, S.K.; Kim, T. Microfluidic device for analyzing preferential chemotaxis and chemoreceptor sensitivity of bacterial cells toward carbon sources. Analyst 2011, 136, 3238–3243. [Google Scholar] [CrossRef]
- Bell, L.; Seshia, A.; Lando, D.; Laue, E.; Palayret, M.; Lee, S.F.; Klenerman, D. A microfluidic device for the hydrodynamic immobilisation of living fission yeast cells for super-resolution imaging. Sens. Actuators B Chem. 2014, 192, 36–41. [Google Scholar] [CrossRef]
- Leea, P.J.; Hunga, P.J. Microfluidic application-specific integrated device for monitoring direct cell-cell communication via gap junctions between individual cell pairs. Appl. Phys. Lett. 2005, 86, 223902. [Google Scholar] [CrossRef]
- JMoffitt, R.; Lee, J.B.; Cluzel, P. Applications of Microfluidics in Quantitative Biology. Lab. Chip 2012, 12, 1487. [Google Scholar]
- Guo, P.; Hall, E.W.; Schirhagl, R.; Mukaibo, H.; Martin, C.R.; Zare, R.N. Microfluidic capture and release of bacteria in a conical nanopore array. Lab. Chip 2012, 12, 558–561. [Google Scholar] [CrossRef]
- Probst, C.; Grünberger, A.; Wiechert, W.; Kohlheyer, D. Polydimethylsiloxane (PDMS) Sub-Micron Traps for Single-Cell Analysis of Bacteria. Micromachines 2013, 4, 357–369. [Google Scholar] [CrossRef]
- Priest, D.G.; Tanaka, N.; Tanaka, Y.; Taniguchi, Y. Micro-patterned agarose gel devices for single-cell high-throughput microscopy of E. coli cells. Sci. Rep. 2017, 7, 17750. [Google Scholar] [CrossRef] [PubMed]
- Wu, F.; Dekker, C. Nanofabricated structures and microfluidic devices for bacteria: From techniques to biology. Chem. Soc. Rev. 2016, 45, 268–280. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Wu, X.; Lin, F. Recent developments in microfluidics-based chemotaxis studies. Lab. Chip 2013, 13, 2484–2499. [Google Scholar] [CrossRef] [PubMed]
- Moolman, M.C.; Huang, Z.; Krishnan, S.T.; Kerssemakers, J.W.J.; Dekker, N.H. Electron beam fabrication of a microfluidic device for studying submicron-scale bacteria. J. Nanobiotechnol. 2013, 11, 12. [Google Scholar] [CrossRef]
- Yang, D.; Jennings, A.D.; Borrego, E.; Retterer, S.T.; Männik, J. Analysis of Factors Limiting Bacterial Growth in PDMS Mother Machine Devices. Front. Microbiol. 2018, 9, 871. [Google Scholar] [CrossRef]
- Wang, P.; Robert, L.; Pelletier, J.; Dang, W.L.; Taddei, F.; Wright, A.; Jun, S. Robust Growth of Escherichia coli. Curr. Biol. 2010, 20, 1099–1103. [Google Scholar] [CrossRef]
- Bamford, R.A.; Smith, A.; Metz, J.; Glover, G.; Titball, R.W.; Pagliara, S. Investigating the physiology of viable but non-culturable bacteria by microfluidics and time-lapse microscopy. BMC Biol. 2017, 15, 121. [Google Scholar] [CrossRef]
- Nakaoka, H.; Wakamoto, Y. Aging, mortality, and the fast growth trade-off of Schizosaccharomyces pombe. PLoS Biol. 2017, 15, e2001109. [Google Scholar] [CrossRef]
- Cama, J.; Voliotis, M.; Metz, J.; Smith, A.; Iannucci, J.; Keyser, U.F.; Tsaneva-Atanasova, K.; Pagliara, S. Single-cell microfluidics facilitates the rapid quantification of antibiotic accumulation in Gram- negative bacteria. Lab Chip 2020, 20, 2765–2775. [Google Scholar] [CrossRef]
- Kaiser, M.; Jug, F.; Julou, T.; Deshpande, S.; Pfohl, T.; Silander, O.K.; Myers, G.; van Nimwegen, E. Monitoring single-cell gene regulation under dynamically controllable conditions with integrated microfluidics and software. Nat. Commun. 2018, 9, 212. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, N.; Isshiki, R.; Kawai, Y.; Tanaka, D.; Sekiguchi, T.; Matsumoto, S.; Tsuneda, S. Stochastic expression of lactate dehydrogenase A induces Escherichia coli persister formation. J. Biosci. Bioeng. 2018, 126, 30–37. [Google Scholar] [CrossRef] [PubMed]
- Spivey, E.C.; Xhemalce, B.; Shear, J.B.; Finkelstein, I.J. 3D-Printed Microfluidic Microdissector for High-Throughput Studies of Cellular Aging. Anal. Chem. 2014, 86, 7406–7412. [Google Scholar] [CrossRef]
- Rowata, A.C.; Birda, J.C.; Agrestia, J.J.; Randob, O.J.; Weitza, D.A. Tracking lineages of single cells in lines using a microfluidic device. Proc. Natl. Acad. Sci. USA 2009, 106, 18153. [Google Scholar] [CrossRef]
- Kato, F.; Nakamura, M.; Sugai, M. The development of fluorescent protein tracing vectors for multicolor imaging of clinically isolated Staphylococcus aureus. Sci. Rep. 2017, 7, 2865. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, Y.; Oscherwitz, J.; Cease, K.B.; Chan, S.M.; Muñoz-Planillo, R.; Hasegawa, M.; Villaruz, A.E.; Cheung, G.Y.; McGavin, M.J.; Travers, J.B.; et al. Staphylococcus δ-toxin induces allergic skin disease by activating mast cells. Nature 2013, 503, 397–401. [Google Scholar] [CrossRef] [PubMed]
- Monk, I.R.; Foster, T.J. Genetic manipulation of Staphylococci—Breaking through the barrier. Front. Cell. Infect. Microbiol. 2012, 2, 49. [Google Scholar] [CrossRef] [PubMed]
- Gulig, P.A.; Curtiss, R., 3rd. Plasmid-associated virulence of Salmonella typhimurium. Plasmid-associated virulence of Salmonella typhimurium. Infect. Immun. 1987, 55, 2891–2901. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tanaka, D.; Ishihara, J.; Takahashi, H.; Kobayashi, M.; Miyazaki, A.; Kajiya, S.; Fujita, R.; Maekawa, N.; Yamazaki, Y.; Takaya, A.; et al. High-Efficiency Single-Cell Containment Microdevices Based on Fluid Control. Micromachines 2023, 14, 1027. https://doi.org/10.3390/mi14051027
Tanaka D, Ishihara J, Takahashi H, Kobayashi M, Miyazaki A, Kajiya S, Fujita R, Maekawa N, Yamazaki Y, Takaya A, et al. High-Efficiency Single-Cell Containment Microdevices Based on Fluid Control. Micromachines. 2023; 14(5):1027. https://doi.org/10.3390/mi14051027
Chicago/Turabian StyleTanaka, Daiki, Junichi Ishihara, Hiroki Takahashi, Masashi Kobayashi, Aya Miyazaki, Satsuki Kajiya, Risa Fujita, Naoki Maekawa, Yuriko Yamazaki, Akiko Takaya, and et al. 2023. "High-Efficiency Single-Cell Containment Microdevices Based on Fluid Control" Micromachines 14, no. 5: 1027. https://doi.org/10.3390/mi14051027
APA StyleTanaka, D., Ishihara, J., Takahashi, H., Kobayashi, M., Miyazaki, A., Kajiya, S., Fujita, R., Maekawa, N., Yamazaki, Y., Takaya, A., Nakamura, Y., Furuya, M., Sekiguchi, T., & Shoji, S. (2023). High-Efficiency Single-Cell Containment Microdevices Based on Fluid Control. Micromachines, 14(5), 1027. https://doi.org/10.3390/mi14051027





