Biosensing Systems Based on Graphene Oxide Fluorescence Quenching Effect
Abstract
:1. Introduction
2. The Role of GO in Fluorescence Detection
3. Fluorescence-Based GO Sensors for Diagnostics
3.1. Diagnosis of Viral Infections
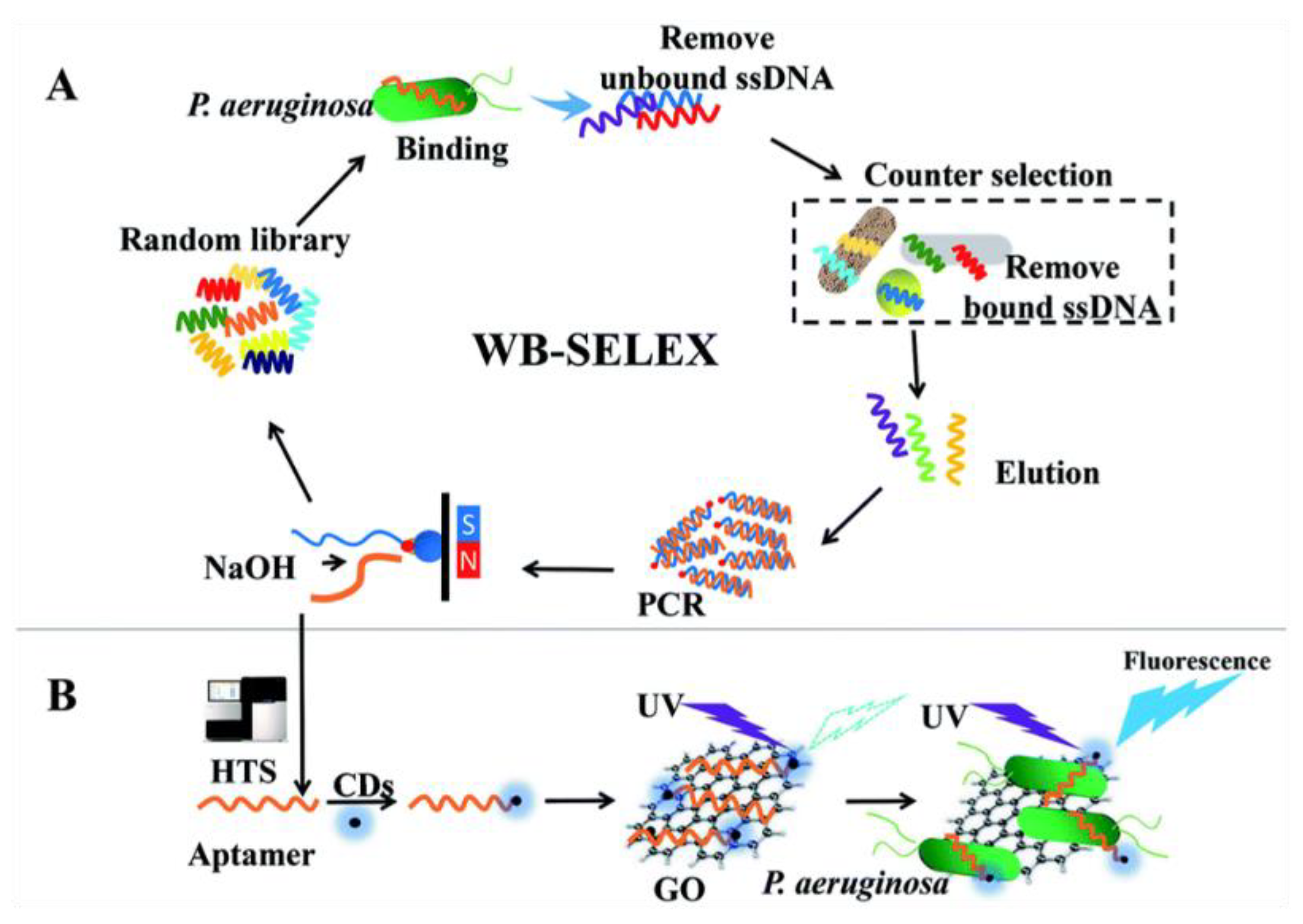
3.2. Detection of Bacteria
3.3. Detection of Cancer Cells and Biomarkers
4. Conclusions and Perspectives
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Speranza, G. Carbon Nanomaterials: Synthesis, Functionalization and Sensing Applications. Nanomaterials 2021, 11, 967. [Google Scholar] [CrossRef] [PubMed]
- Hummers, W.S.; Offeman, R.E. Preparation of Graphitic Oxide. J. Am. Chem. Soc. 1958, 80, 1339. [Google Scholar] [CrossRef]
- Feng, L.; Wu, L.; Qu, X.; Feng, L.; Wu, L.; Qu, X. New Horizons for Diagnostics and Therapeutic Applications of Graphene and Graphene Oxide. Adv. Mater. 2013, 25, 168–186. [Google Scholar] [CrossRef]
- Chung, C.; Kim, Y.K.; Shin, D.; Ryoo, S.R.; Hong, B.H.; Min, D.H. Biomedical Applications of Graphene and Graphene Oxide. Acc. Chem. Res. 2013, 46, 2211–2224. [Google Scholar] [CrossRef] [PubMed]
- Priyadarsini, S.; Mohanty, S.; Mukherjee, S.; Basu, S.; Mishra, M. Graphene and Graphene Oxide as Nanomaterials for Medicine and Biology Application. J. Nanostruct. Chem. 2018, 8, 123–137. [Google Scholar] [CrossRef] [Green Version]
- Dreyer, D.R.; Park, S.; Bielawski, C.W.; Ruoff, R.S. The Chemistry of Graphene Oxide. Chem. Soc. Rev. 2009, 39, 228–240. [Google Scholar] [CrossRef] [PubMed]
- Dideikin, A.T.; Vul’, A.Y. Graphene Oxide and Derivatives: The Place in Graphene Family. Front. Phys. 2019, 6, 149. [Google Scholar] [CrossRef]
- Aliyev, E.; Filiz, V.; Khan, M.M.; Lee, Y.J.; Abetz, C.; Abetz, V. Structural Characterization of Graphene Oxide: Surface Functional Groups and Fractionated Oxidative Debris. Nanomaterials 2019, 9, 1180. [Google Scholar] [CrossRef] [Green Version]
- Choi, Y.R.; Yoon, Y.G.; Choi, K.S.; Kang, J.H.; Shim, Y.S.; Kim, Y.H.; Chang, H.J.; Lee, J.H.; Park, C.R.; Kim, S.Y.; et al. Role of Oxygen Functional Groups in Graphene Oxide for Reversible Room-Temperature NO2 Sensing. Carbon 2015, 91, 178–187. [Google Scholar] [CrossRef]
- Sajjad, S.; Khan Leghari, S.A.; Iqbal, A. Study of Graphene Oxide Structural Features for Catalytic, Antibacterial, Gas Sensing, and Metals Decontamination Environmental Applications. ACS Appl. Mater. Interfaces 2017, 9, 43393–43414. [Google Scholar] [CrossRef]
- Lee, J.; Kim, J.; Kim, S.; Min, D.H. Biosensors Based on Graphene Oxide and Its Biomedical Application. Adv. Drug Deliv. Rev. 2016, 105, 275–287. [Google Scholar] [CrossRef]
- Samota, S.; Rani, R.; Chakraverty, S.; Kaushik, A. Biosensors for Simplistic Detection of Pathogenic Bacteria: A Review with Special Focus on Field-Effect Transistors. Mater. Sci. Semicond. Process 2022, 141, 106404. [Google Scholar] [CrossRef]
- Ivnitski, D.; Abdel-Hamid, I.; Atanasov, P.; Wilkins, E. Biosensors for Detection of Pathogenic Bacteria. Biosens. Bioelectron. 1999, 14, 599–624. [Google Scholar] [CrossRef]
- Kim, J.; Park, S.J.; Min, D.H. Emerging Approaches for Graphene Oxide Biosensor. Anal. Chem. 2017, 89, 232–248. [Google Scholar] [CrossRef]
- Nurrohman, D.T.; Chiu, N.-F. A Review of Graphene-Based Surface Plasmon Resonance and Surface-Enhanced Raman Scattering Biosensors: Current Status and Future Prospects. Nanomaterials 2021, 11, 216. [Google Scholar] [CrossRef]
- Roy, S.; Soin, N.; Bajpai, R.; Misra, D.S.; McLaughlin, J.A.; Roy, S.S. Graphene Oxide for Electrochemical Sensing Applications. J. Mater. Chem. 2011, 21, 14725–14731. [Google Scholar] [CrossRef]
- Liu, Y.Y.; Liu, C.Y.; Liu, Y.Y. Investigation on Fluorescence Quenching of Dyes by Graphite Oxide and Graphene. Appl. Surf. Sci. 2011, 257, 5513–5518. [Google Scholar] [CrossRef]
- Povedailo, V.A.; Ronishenko, B.V.; Stepuro, V.I.; Tsybulsky, D.A.; Shmanai, V.V.; Yakovlev, D.L. Fluorescence Quenching of Dyes by Graphene Oxide. J. Appl. Spectrosc. 2018, 85, 605–610. [Google Scholar] [CrossRef]
- Huang, S.T.; Shi, Y.; Li, N.B.; Luo, H.Q. Fast and Sensitive Dye-Sensor Based on Fluorescein/Reduced Graphene Oxide Complex. Analyst 2012, 137, 2593–2599. [Google Scholar] [CrossRef]
- Liu, D.; Wang, Q.; Chen, A.; Li, Q.; Sui, L.; Jin, M. Ultrafast Dynamics on Fluorescence Quenching of Rhodamine 6G by Graphene Oxide. Luminescence 2021, 36, 1300–1305. [Google Scholar] [CrossRef]
- Tran, L.H.; Lee, C.W.; Kang, T.J.; Jang, S.H. Graphene Oxide-Mediated Fluorescence Quenching of Green Fluorescent Protein for Biomedical Applications. Bull. Korean Chem. Soc. 2016, 37, 1265–1269. [Google Scholar] [CrossRef]
- Jang, H.; Lee, J.; Min, D.H. Graphene Oxide for Fluorescence-Mediated Enzymatic Activity Assays. J. Mater. Chem. B 2014, 2, 2452–2460. [Google Scholar] [CrossRef]
- Li, S.; Aphale, A.N.; Macwan, I.G.; Patra, P.K.; Gonzalez, W.G.; Miksovska, J.; Leblanc, R.M. Graphene Oxide as a Quencher for Fluorescent Assay of Amino Acids, Peptides, and Proteins. ACS Appl. Mater. Interfaces 2012, 4, 7069–7075. [Google Scholar] [CrossRef] [PubMed]
- Zheng, P.; Wu, N. Fluorescence and Sensing Applications of Graphene Oxide and Graphene Quantum Dots: A Review. Chem. Asian J. 2017, 12, 2343–2353. [Google Scholar] [CrossRef] [PubMed]
- Asha Jhonsi, M.; Nithya, C.; Kathiravan, A. Probing Electron Transfer Dynamics of Pyranine with Reduced Graphene Oxide. Phys. Chem. Chem. Phys. 2014, 16, 20878–20886. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Li, K.; He, Z.; Zhang, Y.; Zhao, Y.; Zhang, H.; Miao, Z. Investigation on Fluorescence Quenching Mechanism of Perylene Diimide Dyes by Graphene Oxide. Molecules 2016, 21, 1642. [Google Scholar] [CrossRef] [Green Version]
- Zhu, Y.; Cai, Y.; Xu, L.; Zheng, L.; Wang, L.; Qi, B.; Xu, C. Building An Aptamer/Graphene Oxide FRET Biosensor for One-Step Detection of Bisphenol A. ACS Appl. Mater. Interfaces 2015, 7, 7492–7496. [Google Scholar] [CrossRef]
- Swathi, R.S.; Sebastian, K.L. Long Range Resonance Energy Transfer from a Dye Molecule to Graphene Has (Distance)-4 Dependence. J. Chem. Phys. 2009, 130, 86101. [Google Scholar] [CrossRef]
- Wu, X.; Xing, Y.; Zeng, K.; Huber, K.; Zhao, J.X. Study of Fluorescence Quenching Ability of Graphene Oxide with a Layer of Rigid and Tunable Silica Spacer. Langmuir 2018, 34, 603–611. [Google Scholar] [CrossRef]
- Schaufele, F.; Demarco, I.; Day, R.N. FRET Imaging in the Wide-Field Microscope. In Molecular Imaging: FRET Microscopy and Spectroscopy; American Physiological Society: Rockville, MD, USA, 2005; pp. 72–94. ISBN 9780195177206. [Google Scholar]
- Islam, M.S.; Renner, F.; Azizighannad, S.; Mitra, S. Direct Incorporation of Nano Graphene Oxide (NGO) into Hydrophobic Drug Crystals for Enhanced Aqueous Dissolution. Colloids Surf. B Biointerfaces 2020, 189, 110827. [Google Scholar] [CrossRef]
- Pereira De Sousa, I.; Buttenhauser, K.; Suchaoin, W.; Partenhauser, A.; Perrone, M.; Matuszczak, B.; Bernkop-Schnürch, A. Thiolated Graphene Oxide as Promising Mucoadhesive Carrier for Hydrophobic Drugs. Int. J. Pharm. 2016, 509, 360–367. [Google Scholar] [CrossRef] [PubMed]
- Wei, L.; Lu, Z.; Ji, X.; Jiang, Y.; Ma, L. Self-Assembly of Hollow Graphene Oxide Microcapsules Directed by Cavitation for Loading Hydrophobic Drugs. ACS Appl. Mater. Interfaces 2021, 13, 2988–2996. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.-S.; Dong, X.-M.; Zhang, Z.-C.; Zhang, Z.-P.; Ban, C.-Y.; Zhou, Z.; Song, C.; Yan, S.-Q.; Xin, Q.; Liu, J.-Q.; et al. Co-Assembled Perylene/Graphene Oxide Photosensitive Heterobilayer for Efficient Neuromorphics. Nat. Commun. 2022, 13, 4996. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.; Yu, Y.; Hou, W.; Zhou, J.; Song, L. Effects of Particle Size and pH Value on the Hydrophilicity of Graphene Oxide. Appl. Surf. Sci. 2013, 273, 118–121. [Google Scholar] [CrossRef]
- Srisantitham, S.; Sukwattanasinitt, M.; Unarunotai, S. Effect of pH on Fluorescence Quenching of Organic Dyes by Graphene Oxide. Colloids Surf. A Physicochem. Eng. Asp. 2018, 550, 123–131. [Google Scholar] [CrossRef]
- Lakowicz, J.R. Principles of Fluorescence Spectroscopy; Springer: Berlin/Heidelberg, Germany, 2006; ISBN 0387312781. [Google Scholar]
- Ray, K.; Nakahara, H. Adsorption of Sulforhodamine Dyes in Cationic Langmuir-Blodgett Films: Spectroscopic and Structural Studies. J. Phys. Chem. B 2002, 106, 92–100. [Google Scholar] [CrossRef]
- Yan, X.; Niu, G.; Lin, J.; Jin, A.J.; Hu, H.; Tang, Y.; Zhang, Y.; Wu, A.; Lu, J.; Zhang, S.; et al. Enhanced Fluorescence Imaging Guided Photodynamic Therapy of Sinoporphyrin Sodium Loaded Graphene Oxide. Biomaterials 2015, 42, 94–102. [Google Scholar] [CrossRef] [Green Version]
- Qi, C.X.; Li, H.; Lam, J.W.Y.; Yuan, X.; Wei, J.; Tang, B.Z.; Zhang, H.; Qi, X.Y.; Li, H.; Yuan, X.T.; et al. Graphene Oxide as a Novel Nanoplatform for Enhancement of Aggregation-Induced Emission of Silole Fluorophores. Adv. Mater. 2012, 24, 4191–4195. [Google Scholar] [CrossRef]
- Geng, J.; Zhou, L.; Liu, B. Graphene Oxide Enhanced Fluorescence of Conjugated Polyelectrolytes with Intramolecular Charge Transfer Characteristics. Chem. Commun. 2013, 49, 4818–4820. [Google Scholar] [CrossRef]
- Bapli, A.; Kumar Gautam, R.; Seth, S.; Jana, R.; Pandit, S.; Seth, D.; Bapli, A.; Gautam, R.K.; Jana, R.; Pandit, S.; et al. Graphene Oxide as an Enhancer of Fluorescence. Chem. Asian J. 2020, 15, 1296–1300. [Google Scholar] [CrossRef]
- Toda, K.; Furue, R.; Hayami, S. Recent Progress in Applications of Graphene Oxide for Gas Sensing: A Review. Anal. Chim. Acta 2015, 878, 43–53. [Google Scholar] [CrossRef] [PubMed]
- Thakur, S.; Verma, A.; Alsanie, W.F.; Christie, G.; Thakur, V.K. On the Graphene and Its Derivative Based Polymer Nanocomposites for Glucose Sensing. Mater. Lett. 2022, 307, 130971. [Google Scholar] [CrossRef]
- Fu, L.; Mao, S.; Chen, F.; Zhao, S.; Su, W.; Lai, G.; Yu, A.; Lin, C. Te Graphene-Based Electrochemical Sensors for Antibiotic Detection in Water, Food and Soil: A Scientometric Analysis in CiteSpace (2011–2021). Chemosphere 2022, 297, 134127. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Wang, L.; Lu, H.; Dong, Q.; Li, H.; Liu, S. Graphene and Graphene-like Carbon Nanomaterials-Based Electrochemical Biosensors for Phytohormone Detection. Carbon Lett. 2022, 1, 1–16. [Google Scholar] [CrossRef]
- Zeng, L.; Cao, S.; Yin, H.; Xiong, J.; Lin, D.; Zeng, L.; Cao, S.; Yin, H.; Xiong, J.; Lin, D. Graphene Oxide-Based Biosensors; IntechOpen: London, UK, 2018; ISBN 978-1-78923-589-0. [Google Scholar]
- Peña-Bahamonde, J.; Nguyen, H.N.; Fanourakis, S.K.; Rodrigues, D.F. Recent Advances in Graphene-Based Biosensor Technology with Applications in Life Sciences. J. Nanobiotechnol. 2018, 16, 75. [Google Scholar] [CrossRef] [Green Version]
- Luo, Z.; Ang, M.J.Y.; Chan, S.Y.; Yi, Z.; Goh, Y.Y.; Yan, S.; Tao, J.; Liu, K.; Li, X.; Zhang, H.; et al. Combating the Coronavirus Pandemic: Early Detection, Medical Treatment, and a Concerted Effort by the Global Community. Research 2020, 2020, 6925296. [Google Scholar] [CrossRef]
- Vermisoglou, E.; Panáček, D.; Jayaramulu, K.; Pykal, M.; Frébort, I.; Kolář, M.; Hajdúch, M.; Zbořil, R.; Otyepka, M. Human Virus Detection with Graphene-Based Materials. Biosens. Bioelectron. 2020, 166, 112436. [Google Scholar] [CrossRef]
- Salama, A.M.; Yasin, G.; Zourob, M.; Lu, J. Fluorescent Biosensors for the Detection of Viruses Using Graphene and Two-Dimensional Carbon Nanomaterials. Biosensors 2022, 12, 460. [Google Scholar] [CrossRef]
- Huang, P.J.J.; Liu, J. Separation of Short Single- and Double-Stranded DNA Based on Their Adsorption Kinetics Difference on Graphene Oxide. Nanomaterials 2013, 3, 221–228. [Google Scholar] [CrossRef] [Green Version]
- Zeng, Y.; Zhou, L.; Yang, Z.; Yu, X.; Song, Z.; He, Y. High Sensitivity SARS-CoV-2 Detection Using Graphene Oxide-Multiplex QPCR. Anal. Chim. Acta 2022, 1234, 340533. [Google Scholar] [CrossRef]
- Zhang, Q.; Yin, B.; Hao, J.; Ma, L.; Huang, Y.; Shao, X.; Li, C.; Chu, Z.; Yi, C.; Hong, S.; et al. An AIEgen/Graphene Oxide Nanocomposite (AIEgen@GO)-Based Two-Stage “Turn-on” Nucleic Acid Biosensor for Rapid Detection of SARS-CoV-2 Viral Sequence. Aggregate 2022, 4, e195. [Google Scholar] [CrossRef]
- Alexaki, K.; Kyriazi, M.E.; Greening, J.; Taemaitree, L.; El-Sagheer, A.H.; Brown, T.; Zhang, X.; Muskens, O.L.; Kanaras, A.G. A SARS-Cov-2 Sensor Based on Upconversion Nanoparticles and Graphene Oxide. RSC Adv. 2022, 12, 18445–18449. [Google Scholar] [CrossRef] [PubMed]
- Choi, M.H.; Lee, J.; Seo, Y.J. Combined Recombinase Polymerase Amplification/RkDNA–Graphene Oxide Probing System for Detection of SARS-CoV-2. Anal. Chim. Acta 2021, 1158, 338390. [Google Scholar] [CrossRef]
- Jeong, S.; Kim, D.M.; An, S.Y.; Kim, D.H.; Kim, D.E. Fluorometric Detection of Influenza Viral RNA Using Graphene Oxide. Anal. Biochem. 2018, 561–562, 66–69. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Zhao, Q.; Huang, M.; Duan, Y.; Li, F.; Wang, T. Dual Detection of Hemagglutinin Proteins of H5N1 and H1N1 Influenza Viruses Based on FRET Combined With DNase I. Front. Microbiol. 2022, 13, 2302. [Google Scholar] [CrossRef] [PubMed]
- Wen, J.; Li, W.; Li, J.; Tao, B.; Xu, Y.; Li, H.; Lu, A.; Sun, S. Study on Rolling Circle Amplification of Ebola Virus and Fluorescence Detection Based on Graphene Oxide. Sens. Actuators B Chem. 2016, 227, 655–659. [Google Scholar] [CrossRef]
- Fu, M.Q.; Wang, X.C.; Dou, W.T.; Chen, G.R.; James, T.D.; Zhou, D.M.; He, X.P. Supramolecular Fluorogenic Peptide Sensor Array Based on Graphene Oxide for the Differential Sensing of Ebola Virus. Chem. Commun. 2020, 56, 5735–5738. [Google Scholar] [CrossRef] [Green Version]
- Wright, A.T.; Anslyn, E.V. Differential Receptor Arrays and Assays for Solution-Based Molecular Recognition. Chem. Soc. Rev. 2006, 35, 14–28. [Google Scholar] [CrossRef]
- Yeom, J. Current Status and Outlook of Mosquito-Borne Diseases in Korea. J. Korean Med. Assoc. 2017, 60, 468–474. [Google Scholar] [CrossRef] [Green Version]
- Park, G.; Park, H.; Park, S.-C.; Jang, M.; Yoon, J.; Ahn, J.-H.; Lee, T. Recent Developments in DNA-Nanotechnology-Powered Biosensors for Zika/Dengue Virus Molecular Diagnostics. Nanomaterials 2023, 13, 361. [Google Scholar] [CrossRef]
- Langerak, T.; Mumtaz, N.; Tolk, V.I.; Van Gorp, E.C.M.; Martina, B.E.; Rockx, B.; Koopmans, M.P.G. The Possible Role of Cross-Reactive Dengue Virus Antibodies in Zika Virus Pathogenesis. PLoS Pathog. 2019, 15, e1007640. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.S.; Kim, J.; Shin, H.; Min, D.H. Graphene Oxide-Based Molecular Diagnostic Biosensor for Simultaneous Detection of Zika and Dengue Viruses. 2D Mater. 2020, 7, 044001. [Google Scholar] [CrossRef]
- Guo, J.; Zhang, H.; Yang, J.; Zhang, Y.; Wang, J.; Yan, G. SsDNA-QDs/GO Multicolor Fluorescence System for Synchronous Screening of Hepatitis Virus DNA. Arab. J. Chem. 2023, 16, 104582. [Google Scholar] [CrossRef]
- Murray, C.J.; Ikuta, K.S.; Sharara, F.; Swetschinski, L.; Robles Aguilar, G.; Gray, A.; Han, C.; Bisignano, C.; Rao, P.; Wool, E.; et al. Global Burden of Bacterial Antimicrobial Resistance in 2019: A Systematic Analysis. Lancet 2022, 399, 629–655. [Google Scholar] [CrossRef]
- Limmathurotsakul, D.; Dunachie, S.; Fukuda, K.; Feasey, N.A.; Okeke, I.N.; Holmes, A.H.; Moore, C.E.; Dolecek, C.; van Doorn, H.R.; Shetty, N.; et al. Improving the Estimation of the Global Burden of Antimicrobial Resistant Infections. Lancet Infect. Dis. 2019, 19, e392–e398. [Google Scholar] [CrossRef] [PubMed]
- Davies, J.; Davies, D. Origins and Evolution of Antibiotic Resistance. Microbiol. Mol. Biol. Rev. 2010, 74, 417–433. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaper, J.B.; Nataro, J.P.; Mobley, H.L.T. Pathogenic Escherichia Coli. Nat. Rev. Microbiol. 2004, 2, 123–140. [Google Scholar] [CrossRef]
- Pang, Q.; Lou, D.; Li, S.; Wang, G.; Qiao, B.; Dong, S.; Ma, L.; Gao, C.; Wu, Z. Smart Flexible Electronics-Integrated Wound Dressing for Real-Time Monitoring and On-Demand Treatment of Infected Wounds. Adv. Sci. 2020, 7, 1902673. [Google Scholar] [CrossRef]
- Anand, T.; Narasa Raju, T.A.; Vishnu, C.; Venkateswar Rao, L.; Sharma, G. Development of Dot-ELISA for the Detection of Human Rotavirus Antigen and Comparison with RNA-PAGE. Lett. Appl. Microbiol. 2001, 32, 176–180. [Google Scholar] [CrossRef]
- Nair, S.; Kumar, V.; Kumar, R.; Jain, V.K.; Nagpal, S. Electrostatic Graphene Oxide-Based Biosensor for Rapid Direct Detection of E. coli. Int. J. Mater. Res. 2022, 113, 560–568. [Google Scholar] [CrossRef]
- Linklater, D.P.; Baulin, V.A.; Juodkazis, S.; Ivanova, E.P. Mechano-Bactericidal Mechanism of Graphene Nanomaterials. Interface Focus. 2018, 8, 20170060. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tu, Y.; Lv, M.; Xiu, P.; Huynh, T.; Zhang, M.; Castelli, M.; Liu, Z.; Huang, Q.; Fan, C.; Fang, H.; et al. Destructive Extraction of Phospholipids from Escherichia Coli Membranes by Graphene Nanosheets. Nat. Nanotechnol. 2013, 8, 594–601. [Google Scholar] [CrossRef] [PubMed]
- Chang, J.; Mao, S.; Zhang, Y.; Cui, S.; Zhou, G.; Wu, X.; Yang, C.H.; Chen, J. Ultrasonic-Assisted Self-Assembly of Monolayer Graphene Oxide for Rapid Detection of Escherichia Coli Bacteria. Nanoscale 2013, 5, 3620–3626. [Google Scholar] [CrossRef] [Green Version]
- Tong, S.Y.C.; Davis, J.S.; Eichenberger, E.; Holland, T.L.; Fowler, V.G. Staphylococcus aureus Infections: Epidemiology, Pathophysiology, Clinical Manifestations, and Management. Clin. Microbiol. Rev. 2015, 28, 603. [Google Scholar] [CrossRef] [Green Version]
- Lee, A.S.; De Lencastre, H.; Garau, J.; Kluytmans, J.; Malhotra-Kumar, S.; Peschel, A.; Harbarth, S. Methicillin-Resistant Staphylococcus aureus. Nat. Rev. Dis. Primers 2018, 4, 18033. [Google Scholar] [CrossRef] [Green Version]
- Ning, Y.; Gao, Q.; Zhang, X.; Wei, K.; Chen, L. A Graphene Oxide-Based Sensing Platform for the Determination of Methicillin-Resistant Staphylococcus aureus Based on Strand-Displacement Polymerization Recycling and Synchronous Fluorescent Signal Amplification. J. Biomol. Screen. 2016, 21, 851–857. [Google Scholar] [CrossRef] [Green Version]
- Hunsur Ravikumar, C.; Ira Gowda, M.; Balakrishna, R.G. An “OFF–ON” Quantum Dot–Graphene Oxide Bioprobe for Sensitive Detection of Micrococcal Nuclease of Staphylococcus aureus. Analyst 2019, 144, 3999–4005. [Google Scholar] [CrossRef]
- Pang, S.; Gao, Y.; Li, Y.; Liu, S.; Su, X. A Novel Sensing Strategy for the Detection of Staphylococcus aureus DNA by Using a Graphene Oxide-Based Fluorescent Probe. Analyst 2013, 138, 2749. [Google Scholar] [CrossRef]
- Chen, M.; Song, Y.; Han, L.; Zhou, D.; Wang, Y.; Pan, L.; Tu, K. An Ultrasensitive Upconversion Fluorescence Aptasensor Based on Graphene Oxide Release and Magnetic Separation for Staphylococcus aureus Detection. Food Anal. Methods 2022, 15, 2791–2800. [Google Scholar] [CrossRef]
- Griffin, A.J.; McSorley, S.J. Development of Protective Immunity to Salmonella, a Mucosal Pathogen with a Systemic Agenda. Mucosal Immunol. 2011, 4, 371–382. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Giannella, R.A. Salmonella. In Medical Microbiology, 4th ed.; University of Texas: Galveston, TX, USA, 1996; ISBN 0963117211. [Google Scholar]
- Cody, S.H.; Abbott, S.L.; Marfin, A.A.; Schulz, B.; Wagner, P.; Robbins, K.; Mohle-Boetani, J.C.; Vugia, D.J. Two Outbreaks of Multidrug-Resistant Salmonella Serotype Typhimurium DT104 Infections Linked to Raw-Milk Cheese in Northern California. JAMA 1999, 281, 1805–1810. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Duan, Y.F.; Ning, Y.; Song, Y.; Deng, L. Fluorescent Aptasensor for the Determination of Salmonella Typhimurium Based on a Graphene Oxide Platform. Microchim. Acta 2014, 181, 647–653. [Google Scholar] [CrossRef]
- Yu, S.; Xu, Q.; Huang, J.; Yi, B.; Aguilar, Z.P.; Xu, H. Rapid and Sensitive Detection of Salmonella in Milk Based on Hybridization Chain Reaction and Graphene Oxide Fluorescence Platform. J. Dairy Sci. 2021, 104, 12295–12302. [Google Scholar] [CrossRef] [PubMed]
- Das, K.; Abrol, S.; Verma, R.; Annapragada, H.; Katiyar, N.; Senthilkumar, M. Pseudomonas. Beneficial Microbes in Agro-Ecology: Bacteria and Fungi; Elsevier: Amsterdam, The Netherlands, 1996; pp. 133–148. ISBN 9780128234143. [Google Scholar] [CrossRef]
- Wang, H.; Chi, Z.; Cong, Y.; Wang, Z.; Jiang, F.; Geng, J.; Zhang, P.; Ju, P.; Dong, Q.; Liu, C. Development of a Fluorescence Assay for Highly Sensitive Detection of Pseudomonas Aeruginosa Based on an Aptamer-Carbon Dots/Graphene Oxide System. RSC Adv. 2018, 8, 32454–32460. [Google Scholar] [CrossRef]
- Parvin, N.; Jin, Q.; Wei, Y.; Yu, R.; Zheng, B.; Huang, L.; Zhang, Y.; Wang, L.; Zhang, H.; Gao, M.; et al. Few-Layer Graphdiyne Nanosheets Applied for Multiplexed Real-Time DNA Detection. Adv. Mater. 2017, 29, 1606755. [Google Scholar] [CrossRef]
- Sun, X.; Liu, Z.; Welsher, K.; Robinson, J.T.; Goodwin, A.; Zaric, S.; Dai, H. Nano-Graphene Oxide for Cellular Imaging and Drug Delivery. Nano Res. 2008, 1, 203–212. [Google Scholar] [CrossRef] [Green Version]
- Ock, K.; Jeon, W.I.; Ganbold, E.O.; Kim, M.; Park, J.; Seo, J.H.; Cho, K.; Joo, S.-W.; Lee, S.Y. Real-Time Monitoring of Glutathione-Triggered Thiopurine Anticancer Drug Release in Live Cells Investigated by Surface-Enhanced Raman Scattering. Anal. Chem. 2012, 84, 2172–2178. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Wei, L.; Wang, J.; Peng, F.; Luo, D.; Cui, R.; Niu, Y.; Qin, X.; Liu, Y.; Sun, H.; et al. Cell Imaging by Graphene Oxide Based on Surface Enhanced Raman Scattering. Nanoscale 2012, 4, 7084–7089. [Google Scholar] [CrossRef]
- Huang, J.; Zong, C.; Shen, H.; Liu, M.; Chen, B.; Ren, B.; Zhang, Z. Mechanism of Cellular Uptake of Graphene Oxide Studied by Surface-Enhanced Raman Spectroscopy. Small 2012, 8, 2577–2584. [Google Scholar] [CrossRef] [Green Version]
- Manikandan, M.; Nasser Abdelhamid, H.; Talib, A.; Wu, H.F. Facile Synthesis of Gold Nanohexagons on Graphene Templates in Raman Spectroscopy for Biosensing Cancer and Cancer Stem Cells. Biosens. Bioelectron. 2014, 55, 180–186. [Google Scholar] [CrossRef]
- Yim, D.B.; Kang, H.; Jeon, S.J.; Kim, H.I.; Yang, J.K.; Kang, T.W.; Lee, S.; Choo, J.; Lee, Y.S.; Kim, J.W.; et al. Graphene Oxide-Encoded Ag Nanoshells with Single-Particle Detection Sensitivity towards Cancer Cell Imaging Based on SERRS. Analyst 2015, 140, 3362–3367. [Google Scholar] [CrossRef]
- Hu, C.; Liu, Y.; Qin, J.; Nie, G.; Lei, B.; Xiao, Y.; Zheng, M.; Rong, J. Fabrication of Reduced Graphene Oxide and Sliver Nanoparticle Hybrids for Raman Detection of Absorbed Folic Acid: A Potential Cancer Diagnostic Probe. ACS Appl. Mater. Interfaces 2013, 5, 4760–4768. [Google Scholar] [CrossRef]
- Liu, Z.; Guo, Z.; Zhong, H.; Qin, X.; Wan, M.; Yang, B. Graphene Oxide Based Surface-Enhanced Raman Scattering Probes for Cancer Cell Imaging. Phys. Chem. Chem. Phys. 2013, 15, 2961–2966. [Google Scholar] [CrossRef]
- Sekhon, S.S.; Kaur, P.; Kim, Y.H.; Sekhon, S.S. 2D Graphene Oxide–Aptamer Conjugate Materials for Cancer Diagnosis. 2D Mater. Appl. 2021, 5, 21. [Google Scholar] [CrossRef]
- Tan, J.; Lai, Z.; Zhong, L.; Zhang, Z.; Zheng, R.; Su, J.; Huang, Y.; Huang, P.; Song, H.; Yang, N.; et al. A Graphene Oxide-Based Fluorescent Aptasensor for the Turn-on Detection of CCRF-CEM. Nanoscale Res. Lett. 2018, 13, 66. [Google Scholar] [CrossRef]
- Xiao, K.; Liu, J.; Chen, H.; Zhang, S.; Kong, J. A Label-Free and High-Efficient GO-Based Aptasensor for Cancer Cells Based on Cyclic Enzymatic Signal Amplification. Biosens. Bioelectron. 2017, 91, 76–81. [Google Scholar] [CrossRef]
- Wang, X.; Han, Q.; Yu, N.; Li, J.; Yang, L.; Yang, R.; Wang, C. Aptamer–Conjugated Graphene Oxide–Gold Nanocomposites for Targeted Chemo-Photothermal Therapy of Cancer Cells. J. Mater. Chem. B 2015, 3, 4036–4042. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.; Zhang, B.; Liu, S.; Tan, C.; Tan, Y.; Jiang, Y. A New Strategy Involving the Use of Peptides and Graphene Oxide for Fluorescence Turn-on Detection of Proteins. Sensors 2018, 18, 385. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ding, Y.; Ling, J.; Wang, H.; Zou, J.; Wang, K.; Xiao, X.; Yang, M. Fluorescent Detection of Mucin 1 Protein Based on Aptamer Functionalized Biocompatible Carbon Dots and Graphene Oxide. Anal. Methods 2015, 7, 7792–7798. [Google Scholar] [CrossRef]
- Wang, C.F.; Wang, Z.G.; Sun, X.Y.; Chen, M.J.; Lv, Y.K. An Ultrasensitive Fluorescent Aptasensor for Detection of Cancer Marker Proteins Based on Graphene Oxide–SsDNA. RSC Adv. 2018, 8, 41143–41149. [Google Scholar] [CrossRef]
- Viraka Nellore, B.P.; Kanchanapally, R.; Pramanik, A.; Sinha, S.S.; Chavva, S.R.; Hamme, A.; Ray, P.C. Aptamer-Conjugated Graphene Oxide Membranes for Highly Efficient Capture and Accurate Identification of Multiple Types of Circulating Tumor Cells. Bioconjug. Chem. 2015, 26, 235–242. [Google Scholar] [CrossRef] [Green Version]
- Chen, H.; Zhang, S.; Hsiao, Y.C.; Wang, Q.; Yu, J.S.; Li, W. Graphene Oxide and Fluorescent-Aptamer-Based Novel Aptasensors for Detection of Metastatic Colorectal Cancer Cells. Polymers 2022, 14, 40. [Google Scholar] [CrossRef]
- Ribeiro, B.V.; Cordeiro, T.A.R.; Oliveira e Freitas, G.R.; Ferreira, L.F.; Franco, D.L. Biosensors for the Detection of Respiratory Viruses: A Review. Talanta Open 2020, 2, 100007. [Google Scholar] [CrossRef]
- Ahmed, A.; Rushworth, J.V.; Hirst, N.A.; Millner, P.A. Biosensors for Whole-Cell Bacterial Detection. Clin. Microbiol. Rev. 2014, 27, 631. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xiao, X.; Zhang, Y.; Zhou, L.; Li, B.; Gu, L. Photoluminescence and Fluorescence Quenching of Graphene Oxide: A Review. Nanomaterials 2022, 12, 2444. [Google Scholar] [CrossRef] [PubMed]







| Target | LDR | LOD | Reference |
|---|---|---|---|
| S. aureus mecA gene | 1–40 nM | 0.5 nM | [79] |
| S. aureus | 103–107 CFU/mL | 3 × 102 CFU/mL | [79] |
| S. aureus MNase | 0.1–10 ng/mL | 0.3 ng/mL | [80] |
| S. aureus DNA | 0.0125–3.125 nM | 0.00625 nM | [81] |
| S. aureus | 86–8.6 × 107 CFU/mL | 13 CFU/mL | [82] |
| S. typhimurium | 1 × 103–1 × 108 CFU/mL | 102 CFU/mL | [86] |
| S. enteritidis | 4.2 × 101–4.2 × 108 CFU/mL | 4.2 × 102 CFU/mL | [87] |
| P. aeruginosa | 101–107 CFU/mL | 9 CFU/mL | [89] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Battisti, A.; Samal, S.K.; Puppi, D. Biosensing Systems Based on Graphene Oxide Fluorescence Quenching Effect. Micromachines 2023, 14, 1522. https://doi.org/10.3390/mi14081522
Battisti A, Samal SK, Puppi D. Biosensing Systems Based on Graphene Oxide Fluorescence Quenching Effect. Micromachines. 2023; 14(8):1522. https://doi.org/10.3390/mi14081522
Chicago/Turabian StyleBattisti, Antonella, Sangram Keshari Samal, and Dario Puppi. 2023. "Biosensing Systems Based on Graphene Oxide Fluorescence Quenching Effect" Micromachines 14, no. 8: 1522. https://doi.org/10.3390/mi14081522
APA StyleBattisti, A., Samal, S. K., & Puppi, D. (2023). Biosensing Systems Based on Graphene Oxide Fluorescence Quenching Effect. Micromachines, 14(8), 1522. https://doi.org/10.3390/mi14081522








