Continuous Flow Separation of Live and Dead Cells Using Gravity Sedimentation
Abstract
:1. Introduction
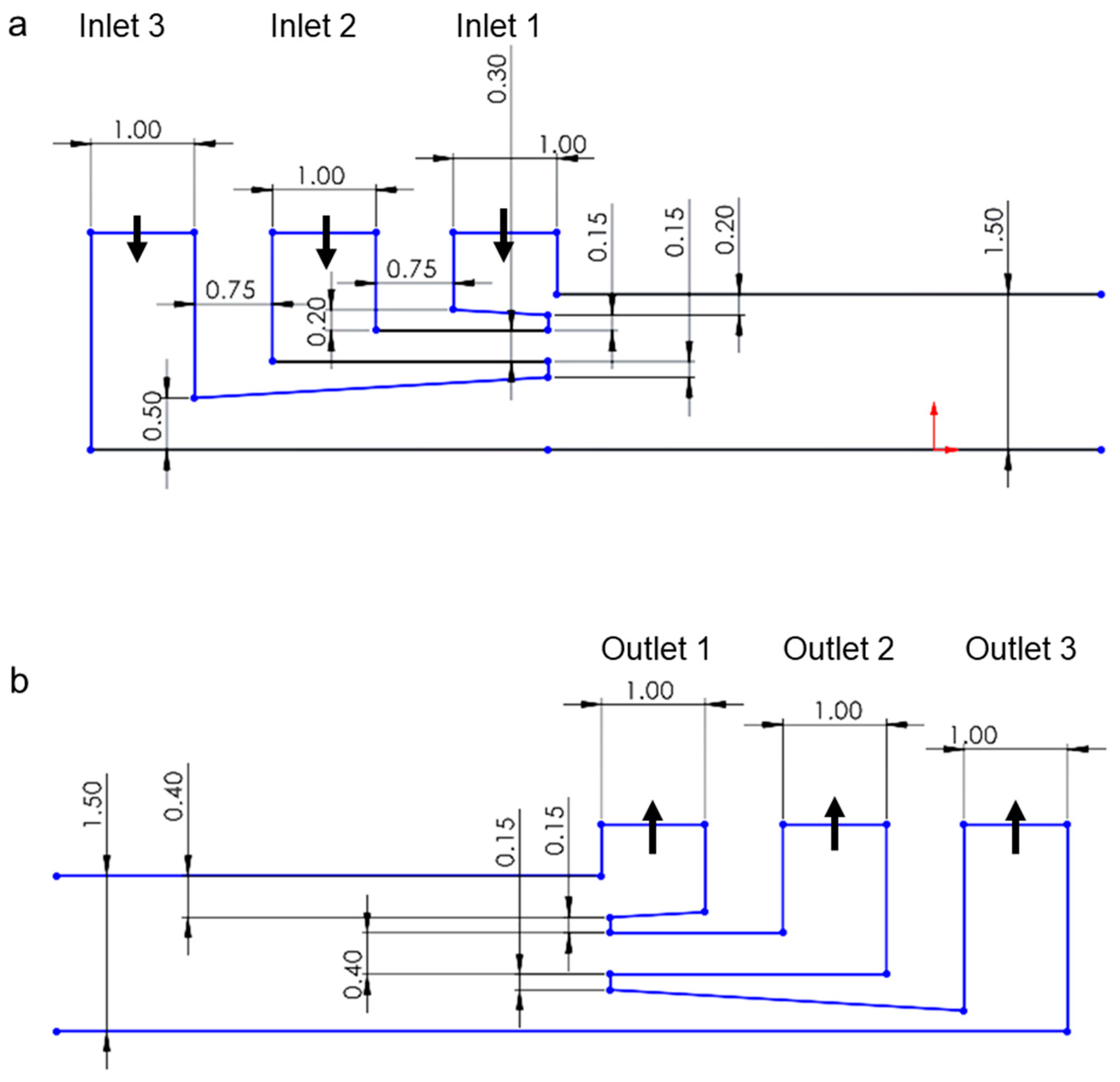
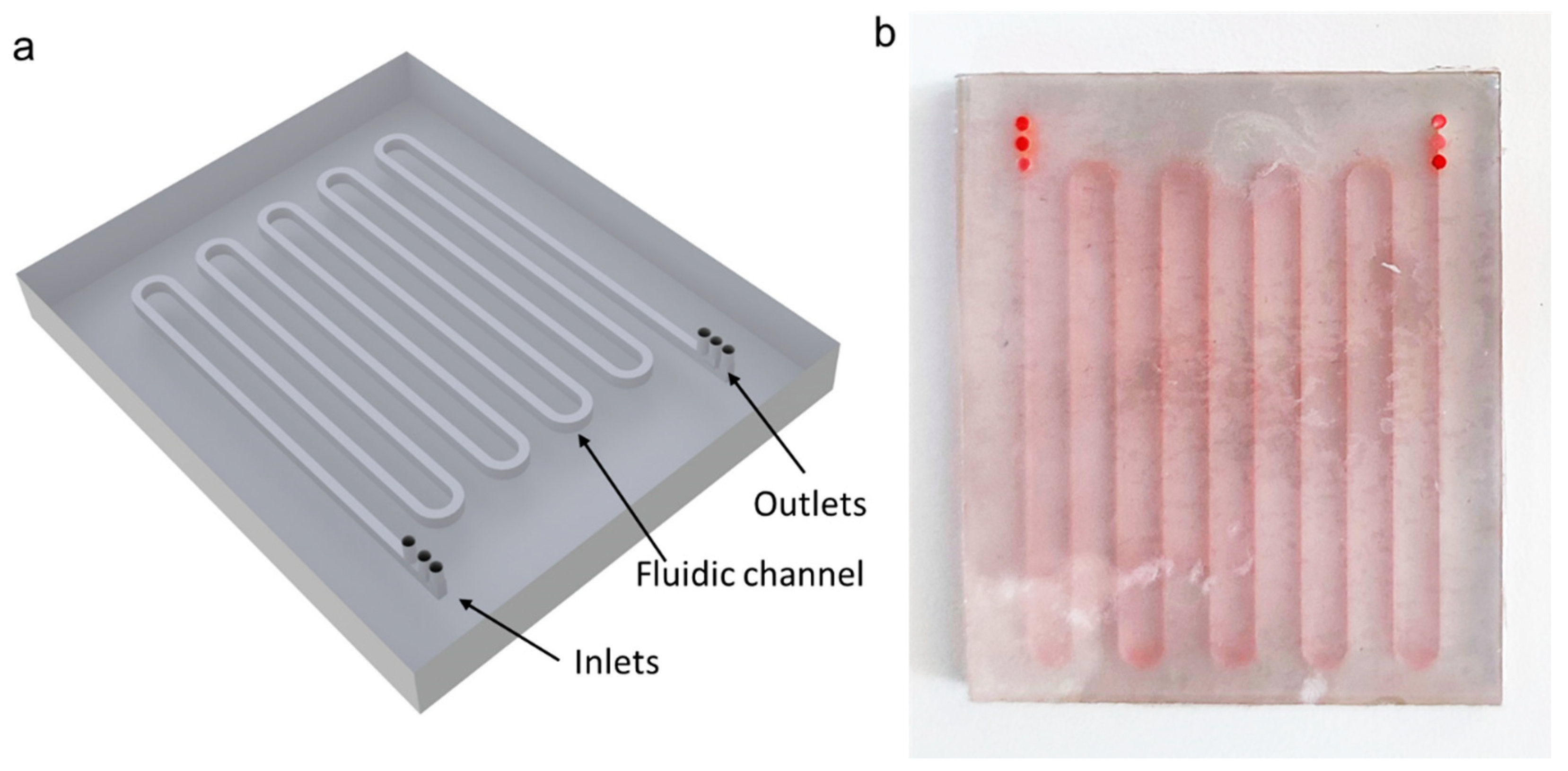
2. Materials and Methods
3. Results
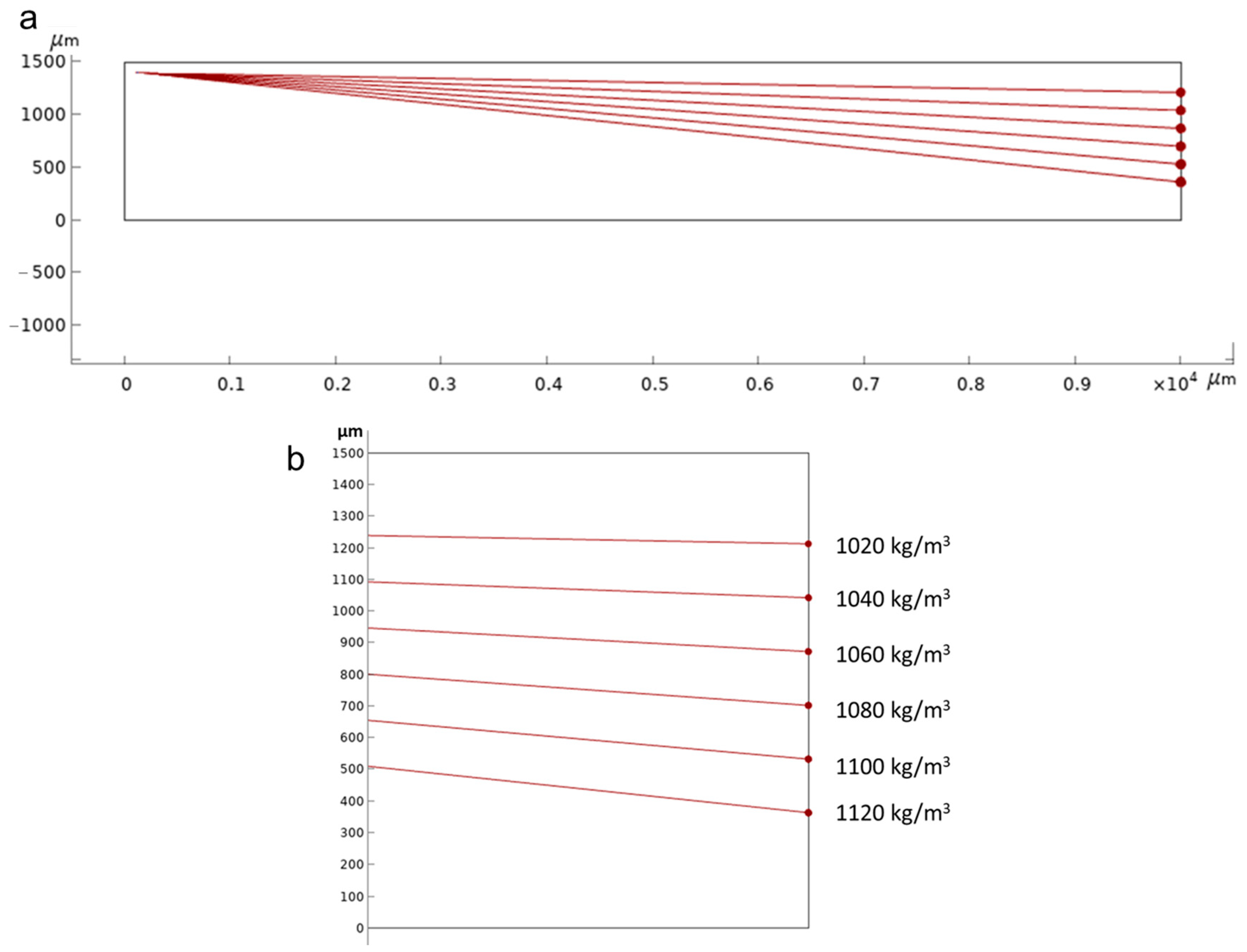
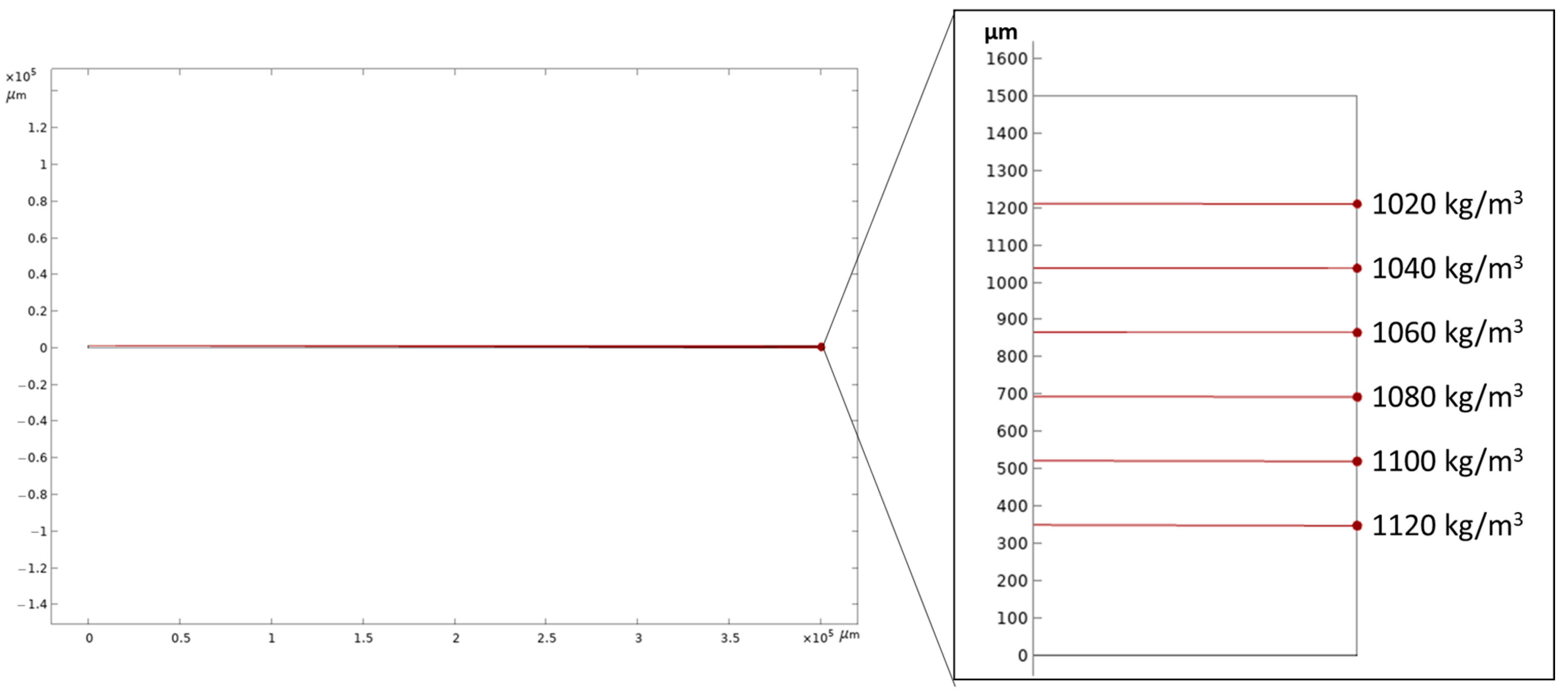
3.1. Numerical Simulation
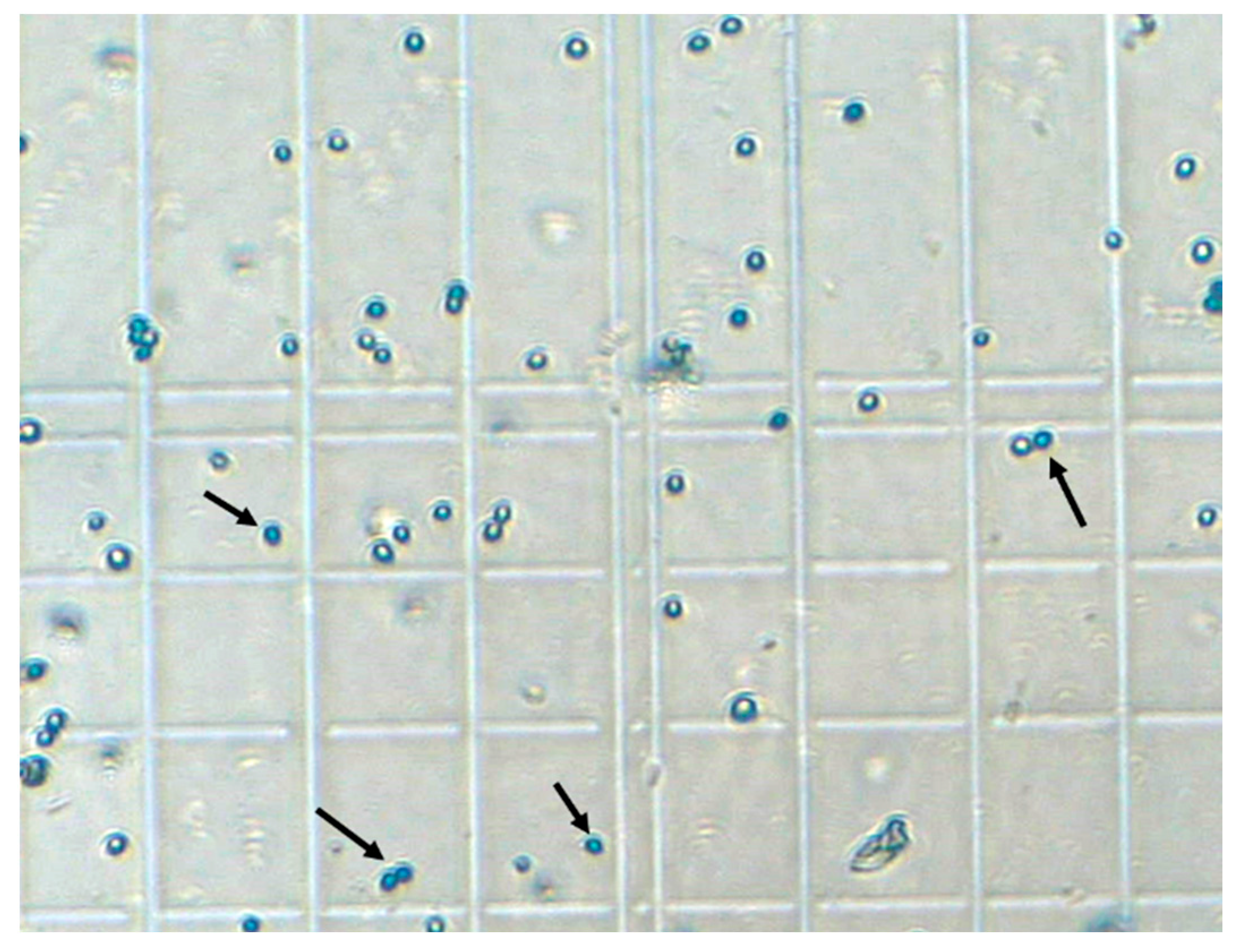
3.2. Experimental Characterization of Sedimentation
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bao, Q.; Zhao, Y.; Niess, H.; Conrad, C.; Schwarz, B.; Jauch, K.-W.; Huss, R.; Nelson, P.J.; Bruns, C.J. Mesenchymal Stem Cell-Based Tumor-Targeted Gene Therapy in Gastrointestinal Cancer. Stem Cells Dev. 2012, 21, 2355–2363. [Google Scholar] [CrossRef] [Green Version]
- Chiu, T.-K.; Chou, W.-P.; Huang, S.-B.; Wang, H.-M.; Lin, Y.-C.; Hsieh, C.-H.; Wu, M.-H. Application of Optically-Induced-Dielectrophoresis in Microfluidic System for Purification of Circulating Tumour Cells for Gene Expression Analysis-Cancer Cell Line Model. Sci. Rep. 2016, 6, 32851. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, J.; Liu, Y.; Zhao, Y.; Zhang, L.; Zhang, L.; Mao, H.; Huang, C. Nanotechnology-Assisted Isolation and Analysis of Circulating Tumor Cells on Microfluidic Devices. Micromachines 2020, 11, 774. [Google Scholar] [CrossRef] [PubMed]
- Phillips, M.A.; Gran, M.L.; Peppas, N. a Targeted Nanodelivery of Drugs and Diagnostics. Nano Today 2010, 5, 143–159. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meng, L.; Cai, F.; Jiang, P.; Deng, Z.; Li, F.; Niu, L.; Chen, Y.; Wu, J.; Zheng, H. On-Chip Targeted Single Cell Sonoporation with Microbubble Destruction Excited by Surface Acoustic Waves. Appl. Phys. Lett. 2014, 104, 073701. [Google Scholar] [CrossRef]
- Chan, J.K.Y.; Lam, P.Y.P. Human Mesenchymal Stem Cells and Their Paracrine Factors for the Treatment of Brain Tumors. Cancer Gene Ther. 2013, 20, 539–543. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Z.; Xu, H.; Yu, L.; Wang, J.; Meng, Q.; Mei, H.; Cai, Z.; Chen, W.; Huang, W. Patient-derived Renal Cell Carcinoma Organoids for Personalized Cancer Therapy. Clin. Transl. Med. 2022, 12, e970. [Google Scholar] [CrossRef]
- Wu, M.; Ozcelik, A.; Rufo, J.; Wang, Z.; Fang, R.; Jun Huang, T. Acoustofluidic Separation of Cells and Particles. Microsyst. Nanoeng. 2019, 5, 32. [Google Scholar] [CrossRef] [Green Version]
- Ozcelik, A.; Cevik, O. Microfluidic Methods Used in Exosome Isolation. Biocell 2023, 47, 959–964. [Google Scholar] [CrossRef]
- Gao, Y.; Wu, M.; Lin, Y.; Xu, J. Acoustic Microfluidic Separation Techniques and Bioapplications: A Review. Micromachines 2020, 11, 921. [Google Scholar] [CrossRef]
- Herzenberg, L.A.; Parks, D.; Sahaf, B.; Perez, O.; Roederer, M.; Herzenberg, L.A. The History and Future of the Fluorescence Activated Cell Sorter and Flow Cytometry: A View from Stanford. Clin. Chem. 2002, 48, 1819–1827. [Google Scholar] [CrossRef]
- Nawaz, A.A.; Chen, Y.; Nama, N.; Nissly, R.H.R.H.; Ren, L.; Ozcelik, A.; Wang, L.; McCoy, J.P.P.; Levine, S.J.S.J.; Huang, T.J.T.J. Acoustofluidic Fluorescence Activated Cell Sorter. Anal. Chem. 2015, 87, 12051–12058. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fu, A.Y.; Spence, C.; Scherer, A.; Arnold, F.H.; Quake, S.R. A Microfabricated Fluorescence-Activated Cell Sorter. Nat. Biotechnol. 1999, 17, 1109–1111. [Google Scholar] [CrossRef] [PubMed]
- Johansson, L.; Nikolajeff, F.; Johansson, S.; Thorslund, S. On-Chip Fluorescence-Activated Cell Sorting by an Integrated Miniaturized Ultrasonic Transducer. Anal. Chem. 2009, 81, 5188–5196. [Google Scholar] [CrossRef] [Green Version]
- Lin, Y.; Gao, C.; Gao, Y.; Wu, M.; Ahmadian Yazdi, A.; Xu, J. Acoustofluidic Micromixer on Lab-on-a-Foil Devices. Sens. Actuators B Chem. 2019, 287, 312–319. [Google Scholar] [CrossRef]
- Wu, M.; Gao, Y.; Ghaznavi, A.; Zhao, W.; Xu, J. AC Electroosmosis Micromixing on a Lab-on-a-Foil Electric Microfluidic Device. Sens. Actuators B Chem. 2022, 359, 131611. [Google Scholar] [CrossRef]
- Gao, Y.; Wu, M.; Lin, Y.; Zhao, W.; Xu, J. Acoustic Bubble-Based Bidirectional Micropump. Microfluid. Nanofluidics 2020, 24, 29. [Google Scholar] [CrossRef]
- Cai, H.; Ao, Z.; Wu, Z.; Song, S.; Mackie, K.; Guo, F. Intelligent Acoustofluidics Enabled Mini-Bioreactors for Human Brain Organoids. Lab Chip 2021, 21, 2194–2205. [Google Scholar] [CrossRef]
- Ao, Z.; Cai, H.; Wu, Z.; Ott, J.; Wang, H.; Mackie, K.; Guo, F. Controllable Fusion of Human Brain Organoids Using Acoustofluidics. Lab Chip 2021, 21, 688–699. [Google Scholar] [CrossRef]
- Ao, Z.; Cai, H.; Wu, Z.; Krzesniak, J.; Tian, C.; Lai, Y.Y.; Mackie, K.; Guo, F. Human Spinal Organoid-on-a-Chip to Model Nociceptive Circuitry for Pain Therapeutics Discovery. Anal. Chem. 2022, 94, 1365–1372. [Google Scholar] [CrossRef]
- Wu, Y.; Ao, Z.; Chen, B.; Muhsen, M.; Bondesson, M.; Lu, X.; Guo, F. Acoustic Assembly of Cell Spheroids in Disposable Capillaries. Nanotechnology 2018, 29, 504006. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ren, L.; Yang, S.; Zhang, P.; Qu, Z.; Mao, Z.; Huang, P.-H.; Chen, Y.; Wu, M.; Wang, L.; Li, P.; et al. Standing Surface Acoustic Wave (SSAW)-Based Fluorescence-Activated Cell Sorter. Small 2018, 14, 1801996. [Google Scholar] [CrossRef] [PubMed]
- Modak, N.; Datta, A.; Ganguly, R. Cell Separation in a Microfluidic Channel Using Magnetic Microspheres. Microfluid. Nanofluidics 2009, 6, 647–660. [Google Scholar] [CrossRef]
- Hejazian, M.; Li, W.; Nguyen, N.-T. Lab on a Chip for Continuous-Flow Magnetic Cell Separation. Lab Chip 2015, 15, 959–970. [Google Scholar] [CrossRef]
- Witek, M.A.; Freed, I.M.; Soper, S.A. Cell Separations and Sorting. Anal. Chem. 2020, 92, 105–131. [Google Scholar] [CrossRef]
- Bhagat, A.A.S.; Bow, H.; Hou, H.W.; Tan, S.J.; Han, J.; Lim, C.T. Microfluidics for Cell Separation. Med. Biol. Eng. Comput. 2010, 48, 999–1014. [Google Scholar] [CrossRef] [PubMed]
- Menachery, A.; Kumawat, N.; Qasaimeh, M. Label-Free Microfluidic Stem Cell Isolation Technologies. TrAC Trends Anal. Chem. 2017, 89, 1–12. [Google Scholar] [CrossRef]
- Song, J.; Song, M.; Kang, T.; Kim, D.; Lee, L.P. Label-Free Density Difference Amplification-Based Cell Sorting. Biomicrofluidics 2014, 8, 064108. [Google Scholar] [CrossRef] [Green Version]
- Wu, M.; Gao, Y.; Luan, Q.; Papautsky, I.; Chen, X.; Xu, J. Three-dimensional Lab-on-a-foil Device for Dielectrophoretic Separation of Cancer Cells. Electrophoresis 2023. [Google Scholar] [CrossRef]
- Ozcelik, A.; Huang, T.J. Acoustic tweezers for single-cell manipulation. In Handbook of Single Cell Technologies; Springer: Singapore, 2020; pp. 1–27. [Google Scholar]
- Akkoyun, F.; Gucluer, S.; Ozcelik, A. Potential of the Acoustic Micromanipulation Technologies for Biomedical Research. Biomicrofluidics 2021, 15, 061301. [Google Scholar] [CrossRef]
- Lee, M.G.; Shin, J.H.; Bae, C.Y.; Choi, S.; Park, J.-K. Label-Free Cancer Cell Separation from Human Whole Blood Using Inertial Microfluidics at Low Shear Stress. Anal. Chem. 2013, 85, 6213–6218. [Google Scholar] [CrossRef] [PubMed]
- Olofsson, K.; Hammarström, B.; Wiklund, M. Acoustic Separation of Living and Dead Cells Using High Density Medium. Lab Chip 2020, 20, 1981–1990. [Google Scholar] [CrossRef]
- Yildizhan, Y.; Erdem, N.; Islam, M.; Martinez-Duarte, R.; Elitas, M. Dielectrophoretic Separation of Live and Dead Monocytes Using 3D Carbon-Electrodes. Sensors 2017, 17, 2691. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zalis, M.C.; Reyes, J.F.; Augustsson, P.; Holmqvist, S.; Roybon, L.; Laurell, T.; Deierborg, T. Label-Free Concentration of Viable Neurons, HESCs and Cancer Cells by Means of Acoustophoresis. Integr. Biol. 2016, 8, 332–340. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Islam, M.; Brink, H.; Blanche, S.; DiPrete, C.; Bongiorno, T.; Stone, N.; Liu, A.; Philip, A.; Wang, G.; Lam, W.; et al. Microfluidic Sorting of Cells by Viability Based on Differences in Cell Stiffness. Sci. Rep. 2017, 7, 1997. [Google Scholar] [CrossRef]
- Norouzi, N.; Bhakta, H.C.; Grover, W.H. Sorting Cells by Their Density. PLoS ONE 2017, 12, e0180520. [Google Scholar] [CrossRef] [Green Version]
- Huh, D.; Bahng, J.H.; Ling, Y.; Wei, H.-H.; Kripfgans, O.D.; Fowlkes, J.B.; Grotberg, J.B.; Takayama, S. Gravity-Driven Microfluidic Particle Sorting Device with Hydrodynamic Separation Amplification. Anal. Chem. 2007, 79, 1369–1376. [Google Scholar] [CrossRef] [Green Version]
- Benincasa, M.-A.; Moore, L.R.; Williams, P.S.; Poptic, E.; Carpino, F.; Zborowski, M. Cell Sorting by One Gravity SPLITT Fractionation. Anal. Chem. 2005, 77, 5294–5301. [Google Scholar] [CrossRef]
- Herrmann, N.; Neubauer, P.; Birkholz, M. Spiral Microfluidic Devices for Cell Separation and Sorting in Bioprocesses. Biomicrofluidics 2019, 13, 061501. [Google Scholar] [CrossRef] [Green Version]
- Yoon, Y.; Lee, J.; Ra, M.; Gwon, H.; Lee, S.; Kim, M.Y.; Yoo, K.-C.; Sul, O.; Kim, C.G.; Kim, W.-Y.; et al. Continuous Separation of Circulating Tumor Cells from Whole Blood Using a Slanted Weir Microfluidic Device. Cancers 2019, 11, 200. [Google Scholar] [CrossRef] [Green Version]
- Catarino, S.O.; Rodrigues, R.O.; Pinho, D.; Miranda, J.M.; Minas, G.; Lima, R. Blood Cells Separation and Sorting Techniques of Passive Microfluidic Devices: From Fabrication to Applications. Micromachines 2019, 10, 593. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gucluer, S.; Guler, O. A Low-Cost Laser-Prototyped Microfluidic Device for Separating Cells and Bacteria. Appl. Sci. 2023, 13, 7938. [Google Scholar] [CrossRef]
- Javi, F.; Zaferani, M.; Lopez-Barbosa, N.; DeLisa, M.P.; Abbaspourrad, A. Sheathless Inertial Microfluidic Cell Separation via a Serpentine–Contraction–Expansion Device Coupled with a Combinatorial Extraction Regulator. Microfluid. Nanofluidics 2022, 26, 54. [Google Scholar] [CrossRef]
- Akkoyun, F.; Ozcelik, A. A Simple Approach for Controlling an Open-Source Syringe Pump. Eur. Mech. Sci. 2020, 4, 166–170. [Google Scholar] [CrossRef]
- Reuß, M.; Josić, D.; Popović, M.; Bronn, W.K. Viscosity of Yeast Suspensions. Eur. J. Appl. Microbiol. Biotechnol. 1979, 8, 167–175. [Google Scholar] [CrossRef]
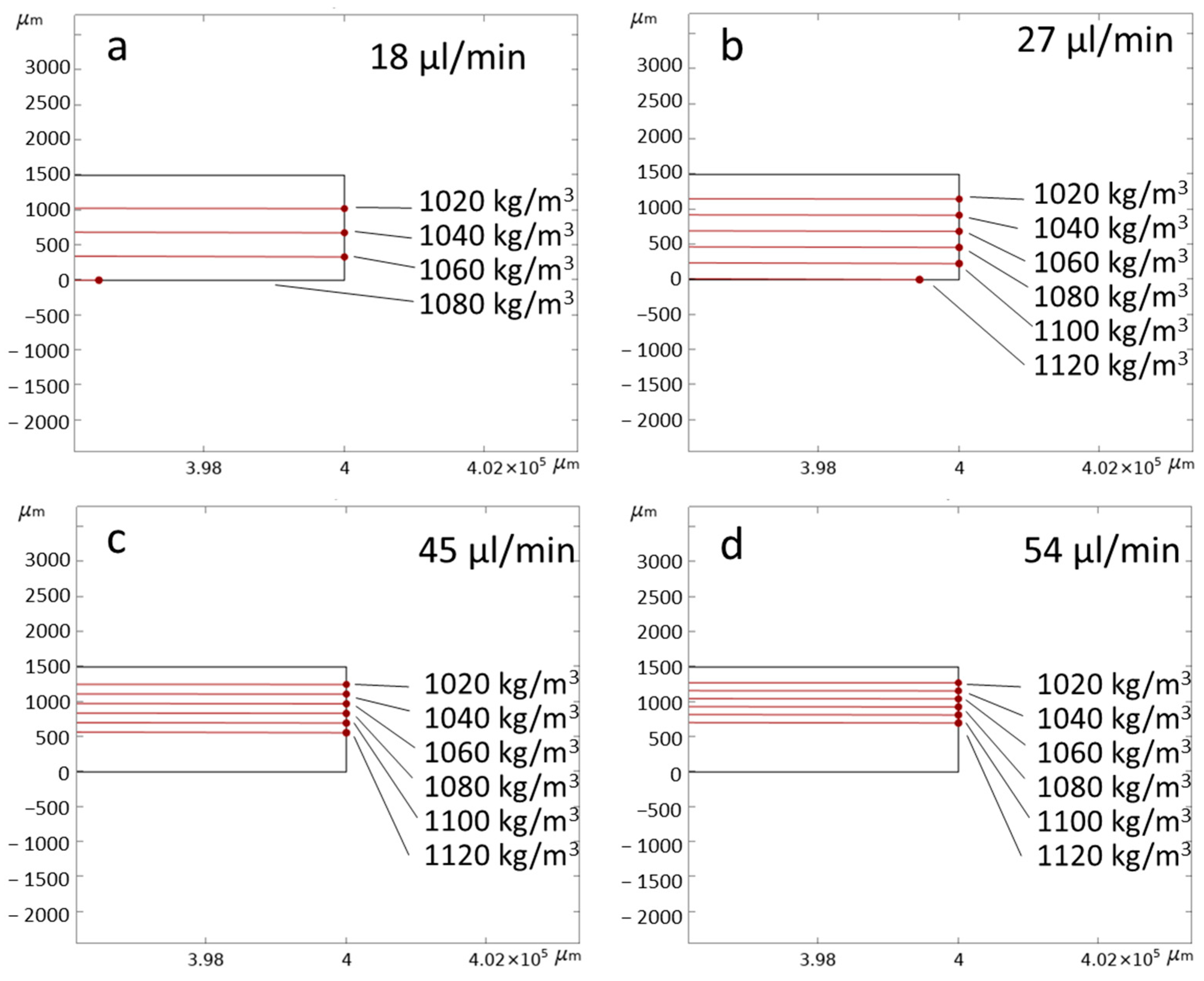

| Inlet 1 | Inlet 2 (Sample) | Inlet 3 | Total Flow Rate |
|---|---|---|---|
| 3 µL/min | 3 µL/min | 12 µL/min | 18 µL/min |
| 4.5 µL/min | 4.5 µL/min | 18 µL/min | 27 µL/min |
| 6 µL/min | 6 µL/min | 24 µL/min | 36 µL/min |
| 7.5 µL/min | 7.5 µL/min | 30 µL/min | 45 µL/min |
| 9 µL/min | 9 µL/min | 36 µL/min | 54 µL/min |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ozcelik, A.; Gucluer, S.; Keskin, T. Continuous Flow Separation of Live and Dead Cells Using Gravity Sedimentation. Micromachines 2023, 14, 1570. https://doi.org/10.3390/mi14081570
Ozcelik A, Gucluer S, Keskin T. Continuous Flow Separation of Live and Dead Cells Using Gravity Sedimentation. Micromachines. 2023; 14(8):1570. https://doi.org/10.3390/mi14081570
Chicago/Turabian StyleOzcelik, Adem, Sinan Gucluer, and Tugce Keskin. 2023. "Continuous Flow Separation of Live and Dead Cells Using Gravity Sedimentation" Micromachines 14, no. 8: 1570. https://doi.org/10.3390/mi14081570
APA StyleOzcelik, A., Gucluer, S., & Keskin, T. (2023). Continuous Flow Separation of Live and Dead Cells Using Gravity Sedimentation. Micromachines, 14(8), 1570. https://doi.org/10.3390/mi14081570






