Deep Learning-Assisted Smartphone-Based Electrochemiluminescence Visual Monitoring Biosensor: A Fully Integrated Portable Platform
Abstract
1. Introduction
2. Enzymatic Reactions
3. Experimental Section
3.1. Chemicals, Materials, and Instrumentation
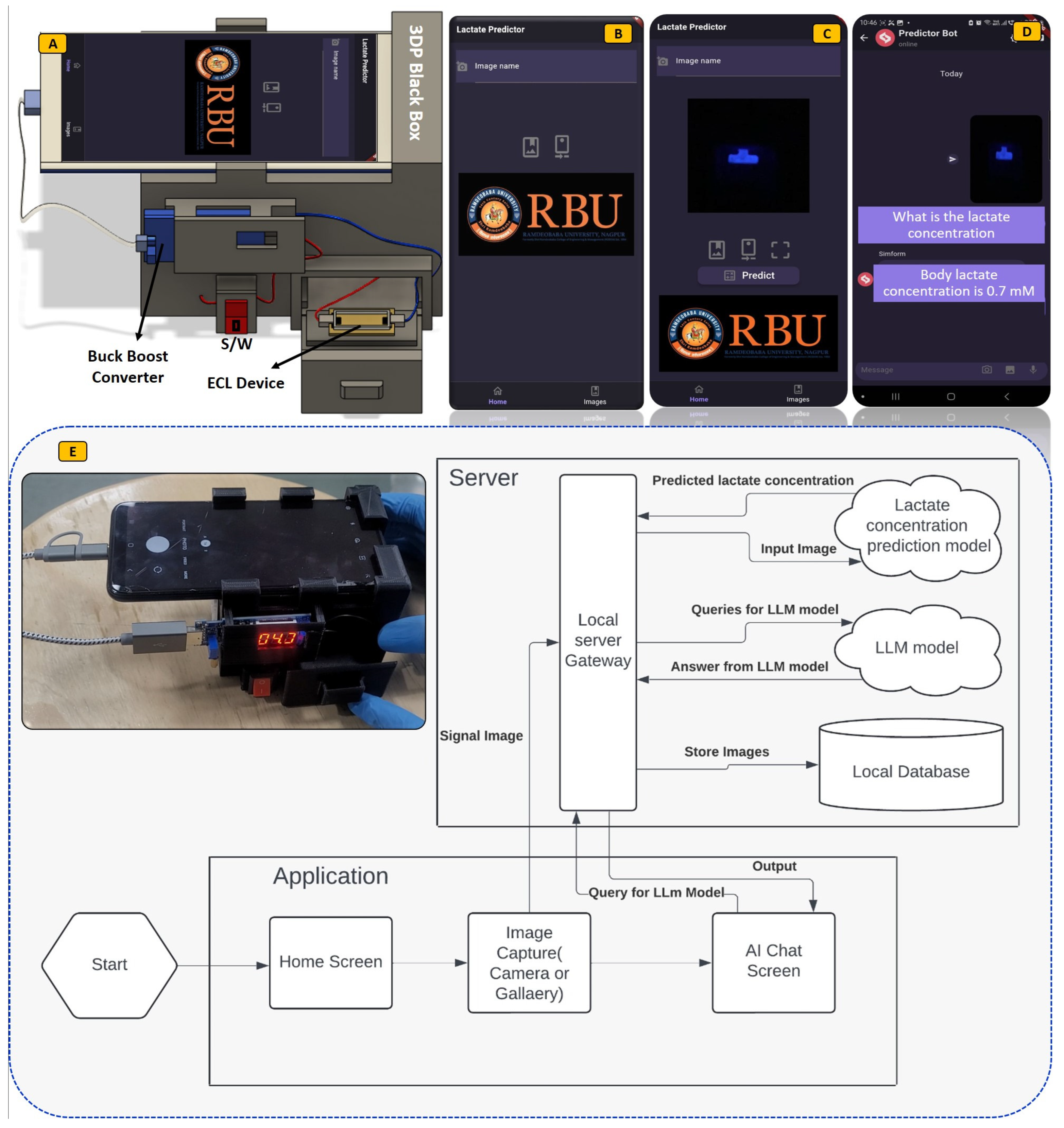
3.2. Fabrication Flow for ECL Biosensor
3.3. ECL Imaging and Analysis Mechanism
4. Results and Discussion
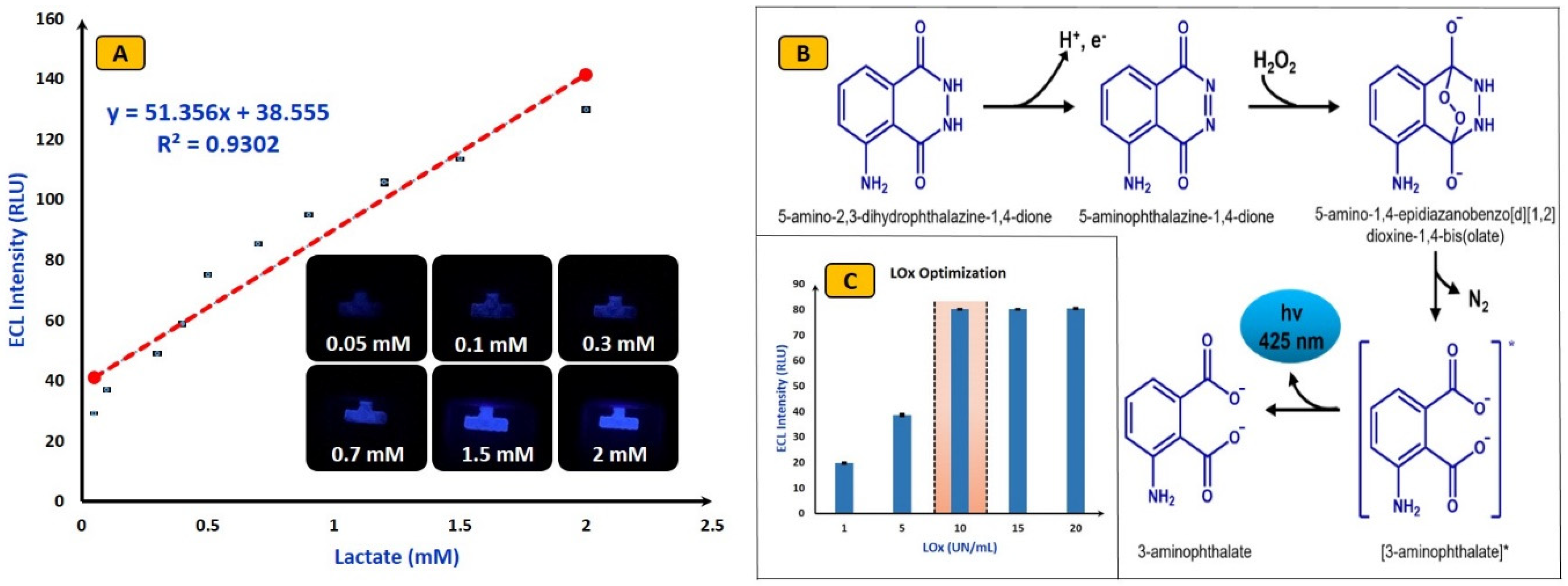
4.1. Parameter Optimization
4.2. Analytical Performance of ECL Biosensors
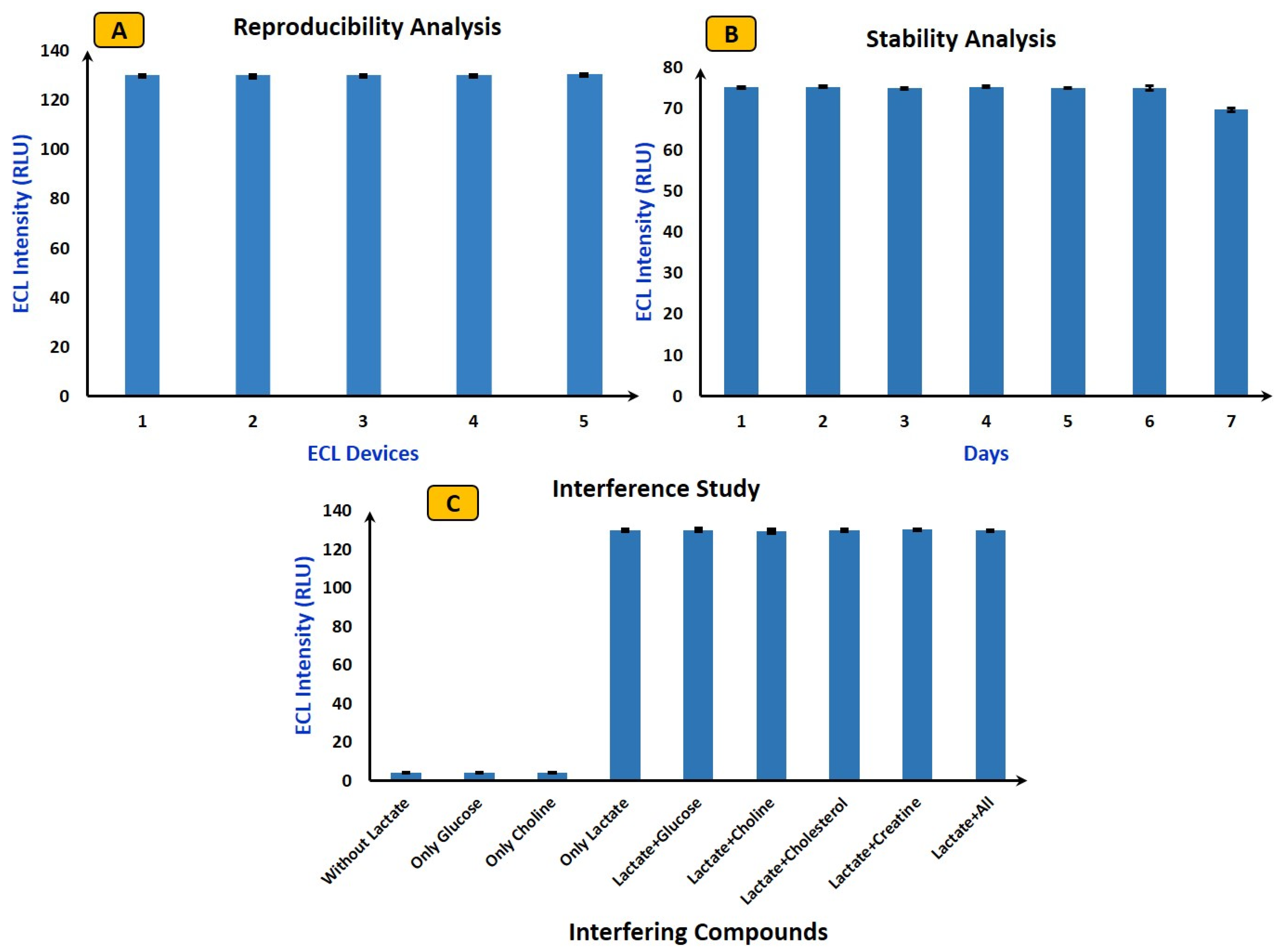
4.3. Reproducibility, Stability, and Interference Study Using ECL Biosensor
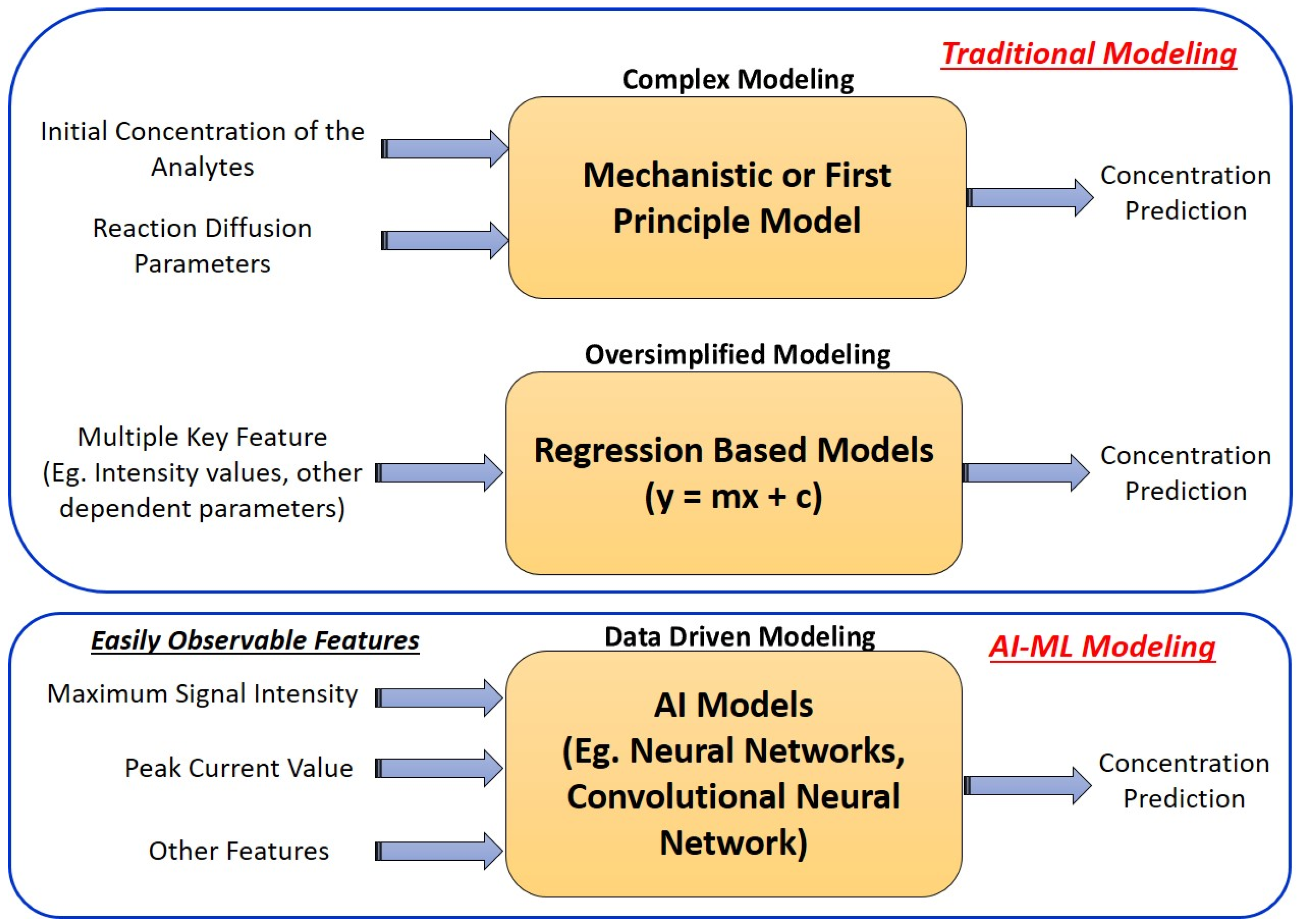
5. Deep Learning Modeling to Validate the Analytical Performance of ECL Biosensors
5.1. Dataset Statistics
5.2. Deep Learning Model Implementation
5.3. Comparative Analysis of Various Benchmarked Models
5.4. Performance Evaluation of Proposed Model through Mean Absolute Error (MAE)
6. Unknown Sample Analysis and Its Validation Using ML
7. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Luppa, P.B.; Mu, C.; Schlichtiger, A.; Schlebusch, H. Point-of-Care Testing (POCT): Current Techniques and Future Perspectives. Trends Anal. Chem. 2020, 30, 887–898. [Google Scholar] [CrossRef] [PubMed]
- Konwar, A.N.; Borse, V. Current Status of Point-of-Care Diagnostic Devices in the Indian Healthcare System with an Update on COVID-19 Pandemic. Sens. Int. 2020, 1, 100015. [Google Scholar] [CrossRef] [PubMed]
- Chan, H.N.; Tan, M.J.A.; Wu, H. Point-of-Care Testing: Applications of 3D Printing. Lab Chip 2017, 17, 2713–2739. [Google Scholar] [CrossRef]
- Zarei, M. Portable Biosensing Devices for Point-of-Care Diagnostics: Recent Developments and Applications. TrAC Trends Anal. Chem. 2017, 91, 26–41. [Google Scholar] [CrossRef]
- Gao, X.; Li, X.; Sun, X.; Zhang, J.; Zhao, Y.; Liu, X.; Li, F. DNA Tetrahedra-Cross-Linked Hydrogel Functionalized Paper for Onsite Analysis of DNA Methyltransferase Activity Using a Personal Glucose Meter. Anal. Chem. 2020, 92, 4592–4599. [Google Scholar] [CrossRef]
- Shahub, S.; Upasham, S.; Ganguly, A.; Prasad, S. Machine Learning Guided Electrochemical Sensor for Passive Sweat Cortisol Detection. Sens. Bio-Sens. Res. 2022, 38, 100527. [Google Scholar] [CrossRef]
- Xu, L.; Wu, R.; Zhu, X.; Wang, X.; Geng, X.; Xiong, Y.; Chen, T.; Wen, Y.; Ai, S. Intelligent Analysis of Maleic Hydrazide Using a Simple Electrochemical Sensor Coupled with Machine Learning. Anal. Methods 2021, 13, 4662–4673. [Google Scholar] [CrossRef]
- Kammarchedu, V.; Butler, D.; Ebrahimi, A. A Machine Learning-Based Multimodal Electrochemical Analytical Device Based on EMoSx-LIG for Multiplexed Detection of Tyrosine and Uric Acid in Sweat and Saliva. Anal. Chim. Acta 2022, 1232, 340447. [Google Scholar] [CrossRef]
- Hasti, V.R.; Navarkar, A.; Gore, J.P. A Data-Driven Approach Using Machine Learning for Early Detection of the Lean Blowout. Energy AI 2021, 5, 100099. [Google Scholar] [CrossRef]
- Rivera, E.C.; Swerdlow, J.J.; Summerscales, R.L.; Uppala, P.P.T.; Filho, R.M.; Neto, M.R.C.; Kwon, H.J. Data-Driven Modeling of Smartphone-Based Electrochemiluminescence Sensor Data Using Artificial Intelligence. Sensors 2020, 20, 625. [Google Scholar] [CrossRef]
- Bhaiyya, M.; Panigrahi, D.; Rewatkar, P.; Haick, H. Role of Machine Learning Assisted Biosensors in Point-of-Care-Testing For Clinical Decisions. ACS Sens. 2024. [Google Scholar] [CrossRef] [PubMed]
- Lu, Z.; Dai, S.; Liu, T.; Yang, J.; Sun, M.; Wu, C.; Su, G.H.; Wang, X.; Rao, H.; Yin, H.; et al. Machine Learning-Assisted Te–CdS@Mn3O4 Nano-Enzyme Induced Self-Enhanced Molecularly Imprinted Ratiometric Electrochemiluminescence Sensor with Smartphone for Portable and Visual Monitoring of 2,4-D. Biosens. Bioelectron. 2023, 222, 114996. [Google Scholar] [CrossRef]
- Kumar, A.; Jain, D.; Bahuguna, J.; Bhaiyya, M.; Dubey, S.K.; Javed, A.; Goel, S. Machine Learning Assisted and Smartphone Integrated Homogeneous Electrochemiluminescence Biosensor Platform for Sample to Answer Detection of Various Human Metabolites. Biosens. Bioelectron. 2023, 238, 115582. [Google Scholar] [CrossRef] [PubMed]
- Guan, W.; Liu, M.; Zhang, C. Electrochemiluminescence Detection in Microfluidic Cloth-Based Analytical Devices. Biosens. Bioelectron. 2016, 75, 247–253. [Google Scholar] [CrossRef]
- Calabria, D.; Lazzarini, E.; Pace, A.; Trozzi, I.; Zangheri, M.; Cinti, S.; Difonzo, M.; Valenti, G.; Guardigli, M.; Paolucci, F.; et al. Biosensors and Bioelectronics Smartphone-Based 3D-Printed Electrochemiluminescence Enzyme Biosensor for Reagentless Glucose Quantification in Real Matrices. Biosens. Bioelectron. 2023, 227, 115146. [Google Scholar] [CrossRef]
- Bhaiyya, M.; Pattnaik, P.K.; Goel, S. Multiplexed and Simultaneous Biosensing in a 3D-Printed Portable Six-Well Smartphone Operated Electrochemiluminescence Standalone Point-of-Care Platform. Microchim. Acta 2022, 189, 79. [Google Scholar] [CrossRef]
- Bhaiyya, M.L.; Srivastava, S.K.; Pattnaik, P.K.; Goel, S. Closed-Bipolar Mini Electrochemiluminescence Sensor to Detect Various Biomarkers: A Machine Learning Approach. IEEE Trans. Instrum. Meas. 2023, 72, 9510308. [Google Scholar] [CrossRef]
- Yang, X.Y.; Bai, Y.Y.; Huangfu, Y.Y.; Guo, W.J.; Yang, Y.J.; Pang, D.W.; Zhang, Z.L. Ultrasensitive Electrochemiluminescence Biosensor Based on Closed Bipolar Electrode for Alkaline Phosphatase Detection in Single Liver Cancer Cell. Anal. Chem. 2021, 93, 1757–1763. [Google Scholar] [CrossRef]
- Liu, M.; Liu, R.; Wang, D.; Liu, C.; Zhang, C. A Low-Cost, Ultraflexible Cloth-Based Microfluidic Device for Wireless Electrochemiluminescence Application. Lab Chip 2016, 16, 2860–2870. [Google Scholar] [CrossRef]
- Zhou, Z.; Xu, L.; Wu, S.; Su, B. A Novel Biosensor Array with a Wheel-like Pattern for Glucose, Lactate and Choline Based on Electrochemiluminescence Imaging. Analyst 2014, 139, 4934–4939. [Google Scholar] [CrossRef]
- Bhaiyya, M.; Kulkarni, M.B.; Pattnaik, P.K.; Goel, S. IoT Enabled PMT and Smartphone Based Electrochemiluminescence Platform to Detect Choline and Dopamine Using 3D-Printed Closed Bipolar Electrodes. Luminescence 2022, 37, 357–365. [Google Scholar] [CrossRef] [PubMed]
- Salve, M.; Mandal, A.; Amreen, K.; Rao, B.V.V.S.N.P.; Pattnaik, P.K.; Goel, S. A Portable 3-D Printed Electrochemiluminescence Platform with Pencil Graphite Electrodes for Point-of-Care Multiplexed Analysis with Smartphone-Based Read Out. IEEE Trans. Instrum. Meas. 2021, 70. [Google Scholar] [CrossRef]
- Rivera, E.C.; Taylor, J.W.; Summerscales, R.L.; Kwon, H.J. Quenching Behavior of the Electrochemiluminescence of Ru(Bpy)32+/TPrA System by Phenols on a Smartphone-Based Sensor. ChemistryOpen 2021, 10, 842–847. [Google Scholar] [CrossRef] [PubMed]
- Kerr, E.; Knezevic, S.; Francis, P.S.; Hogan, C.F.; Valenti, G.; Paolucci, F.; Kanoufi, F.; Sojic, N. Electrochemiluminescence Amplification in Bead-Based Assays Induced by a Freely Diffusing Iridium(III) Complex. ACS Sens. 2023, 8, 933–939. [Google Scholar] [CrossRef]
- Sakanoue, K.; Fiorani, A.; Santo, C.I.; Irkham; Valenti, G.; Paolucci, F.; Einaga, Y. Boron-Doped Diamond Electrode Outperforms the State-of-the-Art Electrochemiluminescence from Microbeads Immunoassay. ACS Sens. 2022, 7, 1145–1155. [Google Scholar] [CrossRef]
- Richter, M.M. Electrochemiluminescence (ECL). Chem. Rev. 2004, 104, 3003–3036. [Google Scholar] [CrossRef]
- Marquette, C.A.; Blum, L.J. Electro-Chemiluminescent Biosensing. Anal. Bioanal. Chem. 2008, 390, 155–168. [Google Scholar] [CrossRef]
- Liu, R.; Zhang, C.; Liu, M. Open Bipolar Electrode-Electrochemiluminescence Imaging Sensing Using Paper-Based Microfluidics. Sens. Actuators B Chem. 2015, 216, 255–262. [Google Scholar] [CrossRef]
- Zhang, F.; He, Y.; Fu, K.; Fu, L.; Zhang, B.; Wang, H.; Zou, G. Dual-Wavebands-Resolved Electrochemiluminescence Multiplexing Immunoassay with Dichroic Mirror Assistant Photomultiplier-Tubes as Detectors. Biosens. Bioelectron. 2018, 115, 77–82. [Google Scholar] [CrossRef]
- D’Alton, L.; Carrara, S.; Barbante, G.J.; Hoxley, D.; Hayne, D.J.; Francis, P.S.; Hogan, C.F. A Simple, Low-Cost Instrument for Electrochemiluminescence Immunoassays Based on a Raspberry Pi and Screen-Printed Electrodes. Bioelectrochemistry 2022, 146, 108107. [Google Scholar] [CrossRef] [PubMed]
- Bhatia, D.; Paul, S.; Acharjee, T.; Ramachairy, S.S. Biosensors and Their Widespread Impact on Human Health. Sens. Int. 2024, 5, 100257. [Google Scholar] [CrossRef]
- Mehrotra, P. Biosensors and Their Applications—A Review. J. Oral Biol. Craniofacial Res. 2016, 6, 153–159. [Google Scholar] [CrossRef]
- Yang, X.; Pan, P.; Tu, L.; Liao, Z.; Niu, H.; Zang, C.; Li, M.; Liu, J.; Yang, Z.; Qi, Y.; et al. Novel Detection of Acrylamide by Electrochemiluminescence Sensor and Optical Imaging Analysis. Int. J. Electrochem. Sci. 2019, 14, 7380–7388. [Google Scholar] [CrossRef]
- Srivastava, S.K.; Bhaiyya, M.; Dudala, S.; Hota, C.; Goel, S. A Machine Learning Approach for Electrochemiluminescence Based Point of Care Testing Device to Detect Multiple Biomarkers. Sens. Actuators A Phys. 2023, 350, 114135. [Google Scholar] [CrossRef]
- Zhang, Y.; Cui, Y.; Sun, M.; Wang, T.; Liu, T.; Dai, X.; Zou, P.; Zhao, Y.; Wang, X.; Wang, Y.; et al. Deep Learning-Assisted Smartphone-Based Molecularly Imprinted Electrochemiluminescence Detection Sensing Platform: Protable Device and Visual Monitoring Furosemide. Biosens. Bioelectron. 2022, 209, 114262. [Google Scholar] [CrossRef] [PubMed]
- Badidi, E. Edge AI for Early Detection of Chronic Diseases and the Spread of Infectious Diseases: Opportunities, Challenges, and Future Directions. Future Internet 2023, 15, 370. [Google Scholar] [CrossRef]
- Xiao, Y.; Xu, L.; Qi, L.W. Electrochemiluminescence Bipolar Electrode Array for the Multiplexed Detection of Glucose, Lactate and Choline Based on a Versatile Enzymatic Approach. Talanta 2017, 165, 577–583. [Google Scholar] [CrossRef] [PubMed]
- Bhaiyya, M.; Kumar, P.S.; Pattnaik, P.K.; Shankar, K.; Goel, S. Stereolithography 3D Printed Electrochemiluminescence Platform with Random Grade Graphite Electrode: Detection of H2O2 and Cholesterol Using a Smartphone. IEEE Sens. J. 2022, 23, 750–757. [Google Scholar] [CrossRef]
- Du, F.; Dong, Z.; Guan, Y.; Zeid, A.M.; Ma, D.; Feng, J.; Yang, D.; Xu, G. Single-Electrode Electrochemical System for the Visual and High-Throughput Electrochemiluminescence Immunoassay. Anal. Chem. 2022, 94, 2189–2194. [Google Scholar] [CrossRef]
- Roda, A.; Guardigli, M.; Calabria, D.; Maddalena Calabretta, M.; Cevenini, L.; Michelini, E. A 3D-Printed Device for a Smartphone-Based Chemiluminescence Biosensor for Lactate in Oral Fluid and Sweat. Analyst 2014, 139, 6494–6501. [Google Scholar] [CrossRef]
- Taylor, J.; Ccopa-Rivera, E.; Kim, S.; Campbell, R.; Summerscales, R.; Kwon, H. Machine Learning Analysis for Phenolic Compound Monitoring Using a Mobile Phone-Based Ecl Sensor. Sensors 2021, 21, 6004. [Google Scholar] [CrossRef] [PubMed]



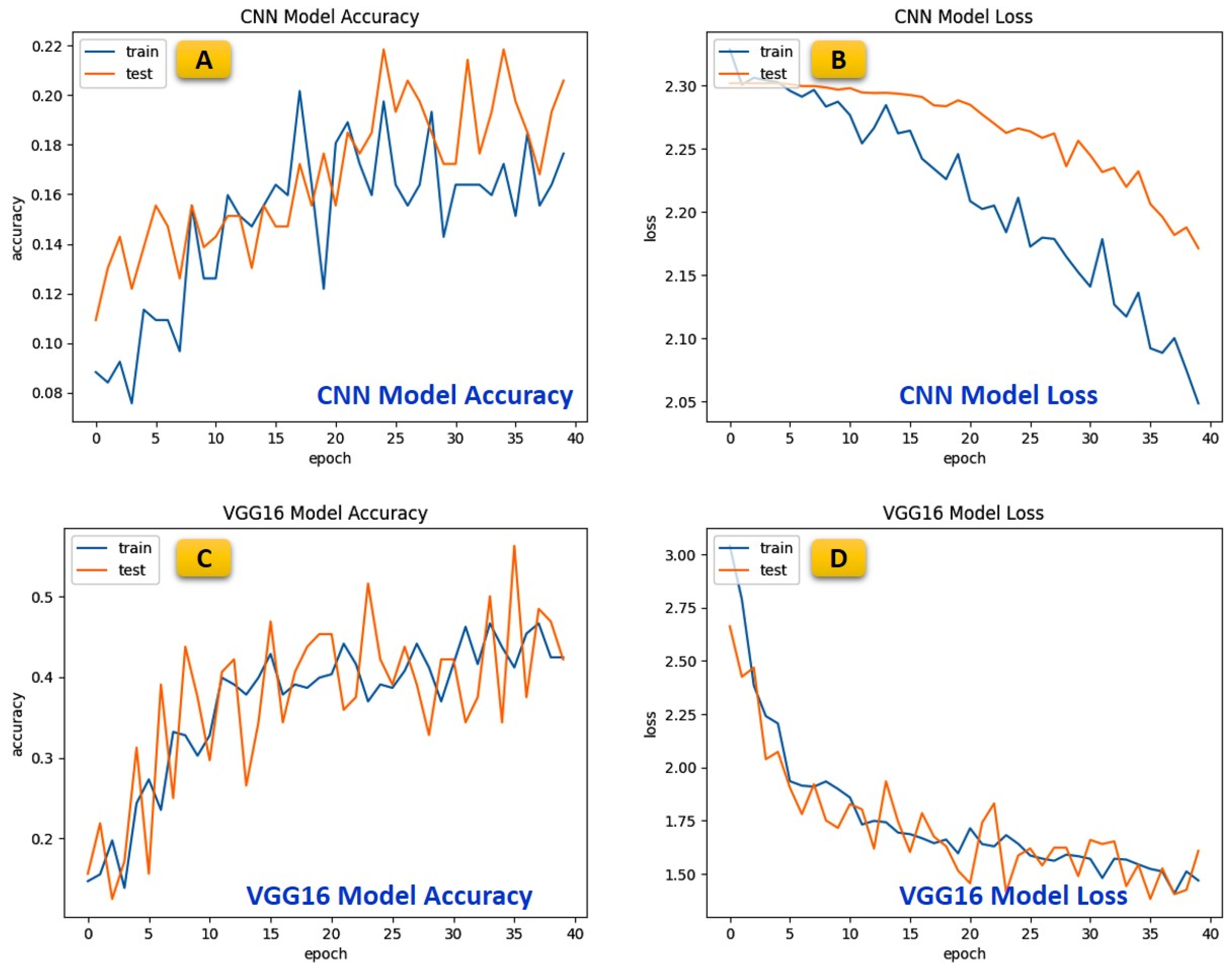

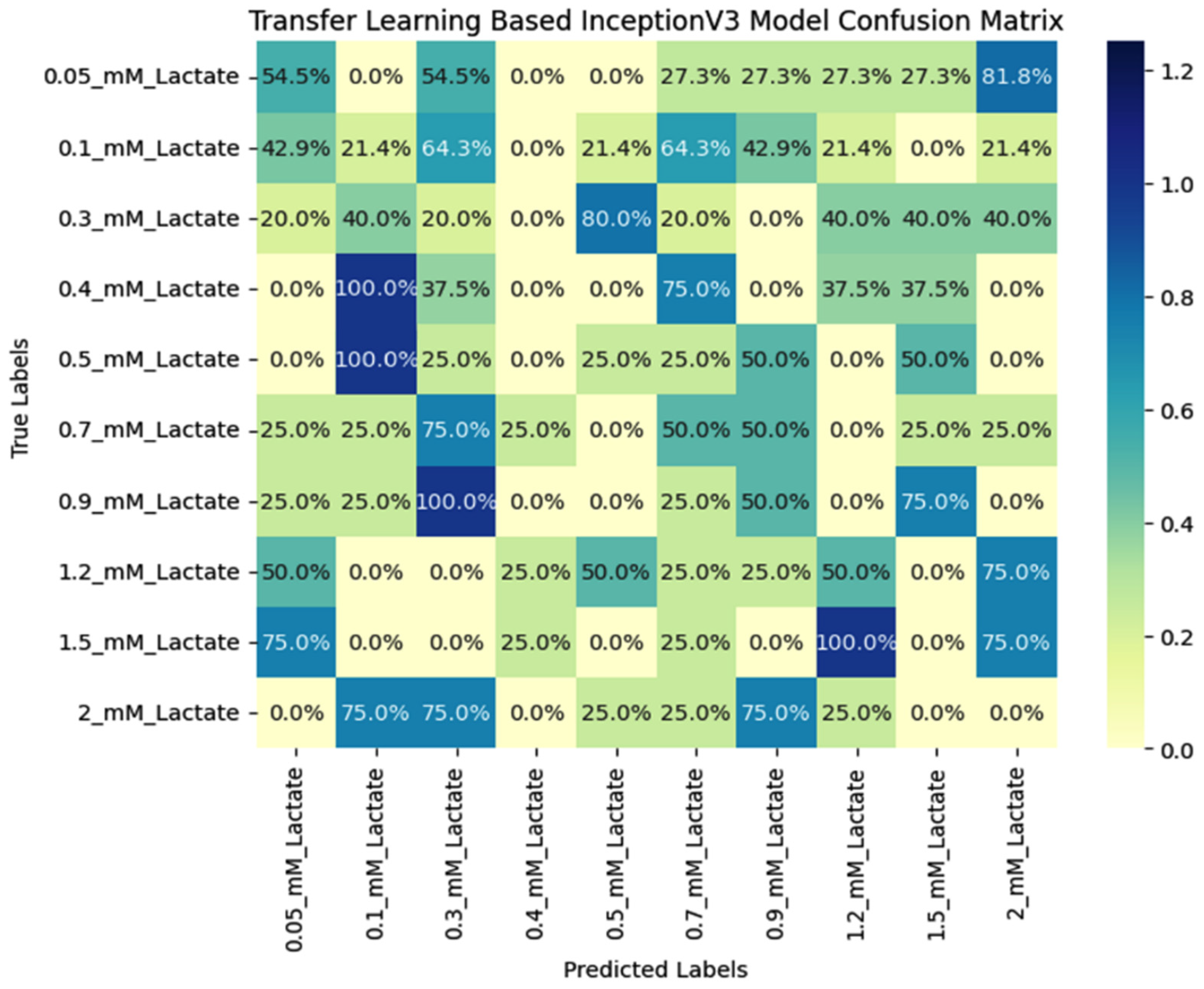
| Class | Before Data Augmentation | After Data Augmentation | ||||
|---|---|---|---|---|---|---|
| Test | Train | Total | Test | Train | Total | |
| 0.1_mM_Lactate | 9 | 36 | 45 | 90 | 360 | 450 |
| 0.3_mM_Lactate | 9 | 36 | 45 | 90 | 360 | 450 |
| 0.4_mM_Lactate | 9 | 36 | 45 | 90 | 360 | 450 |
| 0.05_mM_Lactate | 9 | 36 | 45 | 90 | 360 | 450 |
| 0.5_mM_Lactate | 9 | 36 | 45 | 90 | 360 | 450 |
| 0.7_mM_Lactate | 9 | 36 | 45 | 90 | 360 | 450 |
| 0.9_mM_Lactate | 9 | 36 | 45 | 90 | 360 | 450 |
| 1.2_mM_Lactate | 9 | 36 | 45 | 90 | 360 | 450 |
| 1.5_mM_Lactate | 9 | 36 | 45 | 90 | 360 | 450 |
| 2.0_mM_Lactate | 9 | 36 | 45 | 90 | 360 | 450 |
| Total | 90 | 360 | 450 | 900 | 3600 | 4500 |
| Parameter | Value |
|---|---|
| input shape | 224 × 224 |
| optimizer | SGD, Adam |
| learning rate | 0.001 |
| pooling | GlobalAveragePooling2D |
| activation function | ReLU |
| dropout | 0.5 |
| padding | ZeroPadding2D |
| epochs | 40 |
| verbose | 1, 2 |
| validation steps | 8 |
| include top | False |
| layer trainable | False |
| output layers | Dense, 10 |
| shuffle | True |
| loss | categorical_crossentropy |
| metrics | Accuracy |
| Epoch/Model | CNN | VGG16 | VGG19 | DenseNet121 | AlexNet | InceptionV3 |
|---|---|---|---|---|---|---|
| 10 | 12.61 | 30.25 | 28.99 | 10.50 | 35.29 | 53.33 |
| 15 | 15.55 | 39.92 | 34.45 | 11.76 | 49.16 | 75.83 |
| 20 | 12.18 | 39.92 | 39.50 | 10.50 | 52.52 | 90.00 |
| 25 | 19.75 | 39.08 | 38.66 | 14.71 | 57.98 | 93.33 |
| 30 | 14.29 | 36.97 | 36.13 | 16.81 | 58.82 | 97.50 |
| 35 | 17.23 | 43.70 | 46.22 | 20.17 | 59.66 | 97.50 |
| 40 | 17.65 | 42.44 | 37.39 | 15.55 | 70.59 | 97.73 |
| Peak Accuracy | 19.75 | 43.70 | 46.22 | 20.17 | 70.59 | 97.73 |
| Test Instance | Actual Values | Predicted Value | Mean Absolute Error Value |
|---|---|---|---|
| 1 | 104.857 | 102.89 | 1.967 |
| 2 | 106.65 | 110.14 | 3.49 |
| 3 | 107.499 | 110.36 | 2.861 |
| 4 | 105.799 | 101.97 | 3.829 |
| 5 | 110.573 | 105.23 | 5.343 |
| 6 | 109.688 | 106.55 | 3.138 |
| 7 | 108.303 | 104.31 | 3.993 |
| 8 | 106.65 | 110.14 | 3.49 |
| 9 | 110.137 | 107.4 | 2.737 |
| 10 | 105.22 | 105.58 | 0.36 |
| 11 | 36.552 | 37.246 | 0.694 |
| 12 | 37.882 | 40.606 | 2.724 |
| 13 | 40.497 | 40.953 | 0.456 |
| 14 | 38.685 | 38.787 | 0.102 |
| 15 | 39.5 | 41.141 | 1.641 |
| 16 | 124 | 128.482 | 4.482 |
| 17 | 125.05 | 128.713 | 3.663 |
| 18 | 126.53 | 119.174 | 7.356 |
| 19 | 129.8 | 127.125 | 2.675 |
| 20 | 122.9325 | 127.442 | 4.5095 |
| Average Mean Absolute Error Value | 2.975525 | ||
| Analyte | Actual Value (mM) | LAB Testing (mM) | Testing Using ECL Device | Corresponding ECL Image | ML Prediction |
|---|---|---|---|---|---|
| Lactate | 0.2 | 0.22 | 0.24 |  | 0.2 |
| 0.35 | 0.34 | 0.42 |  | 0.4 | |
| 1.1 | 1.18 | 1.2 |  | 1.2 | |
| 1.35 | 1.42 | 1.4 |  | 1.3 | |
| 1.8 | 1.9 | 1.85 |  | 1.8 |
| Sr. No. | Device Fabrication Method | Chemistry Used | LoD | AI-ML Models Used | Application | Real Sample Analysis | Ref. No |
|---|---|---|---|---|---|---|---|
| 1. | Screen-printed electrodes | Ru(bpy)2+3/TPrA | - | Random forest and Feedforward neural network | Ru(bpy)2+3 | - | [10] |
| 2. | 3D Printing | Luminol/H2O2 | 0.04 mM for glucose and 0.1 mM for lactate | Regression-based ML models | Glucose, Lactate | Blood serum | [13] |
| 3. | 3D Printing | Luminol/H2O2 | 0.49, 0.01, 0.09, and 0.3 mM | Regression-based analyses were carried out for the prediction | cholesterol, choline, lactate, and glucose | Blood serum | [17] |
| 4. | ITO glass | Luminol/H2O2 | 14 mM for glucose, 40 mM for lactate, and 97 mM for choline | - | Glucose, lactate, choline | Blood serum | [20] |
| 5. | Conventional three-electrode system | Molecularly imprinted polymer | 0.25 µM | Deep Learning | Furosemide | Human urine | [35] |
| 6. | 3D Printing | Luminol/H2O2 | 0.5 mM | - | Lactate | Sweat | [40] |
| 7. | Screen-printed electrodes | Ru(bpy)2+3/TPrA | - | Single or Multilayer Neural Net | phenolic compounds | - | [41] |
| 8. | Screen-printed electrodes | Luminol/H2O2 | 5.14 µM | Deep Learning | Lactate | Commercial Lactate | This work |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bhaiyya, M.; Rewatkar, P.; Pimpalkar, A.; Jain, D.; Srivastava, S.K.; Zalke, J.; Kalambe, J.; Balpande, S.; Kale, P.; Kalantri, Y.; et al. Deep Learning-Assisted Smartphone-Based Electrochemiluminescence Visual Monitoring Biosensor: A Fully Integrated Portable Platform. Micromachines 2024, 15, 1059. https://doi.org/10.3390/mi15081059
Bhaiyya M, Rewatkar P, Pimpalkar A, Jain D, Srivastava SK, Zalke J, Kalambe J, Balpande S, Kale P, Kalantri Y, et al. Deep Learning-Assisted Smartphone-Based Electrochemiluminescence Visual Monitoring Biosensor: A Fully Integrated Portable Platform. Micromachines. 2024; 15(8):1059. https://doi.org/10.3390/mi15081059
Chicago/Turabian StyleBhaiyya, Manish, Prakash Rewatkar, Amit Pimpalkar, Dravyansh Jain, Sanjeet Kumar Srivastava, Jitendra Zalke, Jayu Kalambe, Suresh Balpande, Pawan Kale, Yogesh Kalantri, and et al. 2024. "Deep Learning-Assisted Smartphone-Based Electrochemiluminescence Visual Monitoring Biosensor: A Fully Integrated Portable Platform" Micromachines 15, no. 8: 1059. https://doi.org/10.3390/mi15081059
APA StyleBhaiyya, M., Rewatkar, P., Pimpalkar, A., Jain, D., Srivastava, S. K., Zalke, J., Kalambe, J., Balpande, S., Kale, P., Kalantri, Y., & Kulkarni, M. B. (2024). Deep Learning-Assisted Smartphone-Based Electrochemiluminescence Visual Monitoring Biosensor: A Fully Integrated Portable Platform. Micromachines, 15(8), 1059. https://doi.org/10.3390/mi15081059







