Crystallization and Preliminary X-ray Diffraction Study of a Novel Bacterial Homologue of Mammalian Hormone-Sensitive Lipase (halip1) from Halocynthiibacter arcticus
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
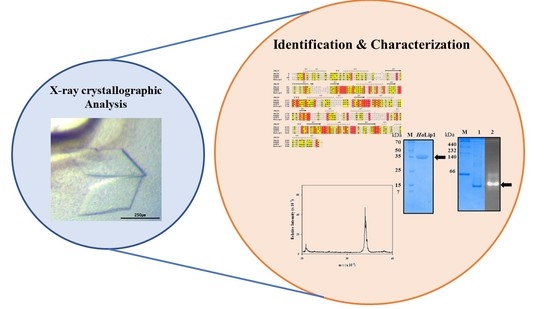
2.2. Cloning and Purification
2.3. Biochemical Characterization
2.4. Crystallization
2.5. X-ray Diffraction Data Collection and Data Processing
3. Results and Discussion
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Arner, P.; Langin, D. The role of neutral lipases in human adipose tissue lipolysis. Curr. Opin. Lipidol. 2007, 18, 246–250. [Google Scholar] [CrossRef] [PubMed]
- Trites, M.J.; Clugston, R.D. The role of adipose triglyceride lipase in lipid and glucose homeostasis: Lessons from transgenic mice. Lipids Health Dis. 2019, 18, 204. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lass, A.; Zimmermann, R.; Oberer, M.; Zechner, R. Lipolysis—A highly regulated multi-enzyme complex mediates the catabolism of cellular fat stores. Prog. Lipid Res. 2011, 50, 14–27. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krintel, C.; Klint, C.; Lindvall, H.; Mörgelin, M.; Holm, C. Quarternary structure and enzymological properties of the different hormone-sensitive lipase (HSL) isoforms. PLoS ONE 2010, 5, e11193. [Google Scholar] [CrossRef] [Green Version]
- Lampidonis, A.D.; Rogdakis, E.; Voutsinas, G.E.; Stravopodis, D.J. The resurgence of Hormone-Sensitive Lipase (HSL) in mammalian lipolysis. Gene 2011, 477, 1–11. [Google Scholar] [CrossRef]
- Kim, T.D. Bacterial Hormone-Sensitive Lipases (bHSLs): Emerging Enzymes for Biotechnological Applications. J. Microbiol. Biotechnol. 2017, 27, 1907–1915. [Google Scholar] [CrossRef] [Green Version]
- Osterlund, T.; Contreras, J.A.; Holm, C. Identification of essential aspartic acid and histidine residues of hormone-sensitive lipase: Apparent residues of the catalytic triad. FEBS Lett. 1997, 403, 259–262. [Google Scholar] [CrossRef] [Green Version]
- Ngo, T.D.; Ryu, B.H.; Ju, H.; Jang, E.; Park, K.; Kim, K.K.; Kim, T.D. Structural and functional analyses of a bacterial homologue of hormone-sensitive lipase from a metagenomic library. Acta Crystallogr. D Biol. Crystallogr. 2013, 69, 1726–1737. [Google Scholar] [CrossRef] [PubMed]
- Li, P.Y.; Ji, P.; Li, C.Y.; Zhang, Y.; Wang, G.L.; Zhang, X.Y.; Xie, B.B.; Qin, Q.L.; Chen, X.L.; Zhou, B.C.; et al. Structural basis for dimerization and catalysis of a novel esterase from the GTSAG motif subfamily of the bacterial hormone-sensitive lipase family. J. Biol. Chem. 2014, 289, 19031–19041. [Google Scholar] [CrossRef] [Green Version]
- Kim, B.Y.; Yoo, W.; Huong Luu Le, L.T.; Kim, K.K.; Kim, H.W.; Lee, J.H.; Kim, Y.O.; Kim, T.D. Characterization and mutation anaylsis of a cold-active bacterial hormone-sensitive lipase from Salinisphaera sp. P7–4. Arch. Biochem. Biophys. 2019, 663, 132–142. [Google Scholar] [CrossRef]
- Park, J.M.; Kang, C.H.; Won, S.M.; Oh, K.H.; Yoon, J.H. Characterization of a novel moderately thermophilic solvent-tolerant esterase isolated from a compost metagenome library. Front. Microbiol. 2020, 10, 3069. [Google Scholar] [CrossRef] [Green Version]
- Kryukova, M.V.; Petrovskaya, L.E.; Kryukova, E.A.; Lomakina, G.Y.; Yakimov, S.A.; Maksimov, E.G.; Boyko, K.M.; Popov, V.O.; Dolgikh, D.A.; Kirpichnikov, M.P. Thermal inactivation of a cold-active esterase PMGL3 isolated from the permafrost metagenomic library. Biomolecules 2019, 9, 880. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Le, L.T.H.L.; Yoo, W.; Lee, C.; Wang, Y.; Jeon, S.; Kim, K.K.; Lee, J.H.; Kim, T.D. Molecular characterization of a novel cold-active hormone-sensitive lipase (HaHSL) from Halocynthiibacter arcticus. Biomolecules 2019, 9, 704. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, J.; Huo, Y.-Y.; Ji, R.; Kuang, S.; Ji, C.; Xu, X.-W.; Li, J. Structural insights of a hormone sensitive lipase homologue Est22. Sci. Rep. 2016, 6, 28550. [Google Scholar] [CrossRef]
- Jayanath, G.; Mohandas, S.P.; Kachiprath, B.; Solomon, S.; Sajeevan, T.P.; Bright Singh, I.S.; Philip, R. A novel solvent tolerant esterase of GDSGG motif subfamily from solar saltern through metagenomic approach: Recombinant expression and characterization. Int. J. Biol. Macromol. 2018, 119, 393–401. [Google Scholar] [CrossRef] [PubMed]
- Baek, K.; Lee, Y.M.; Shin, S.C.; Hwang, K.; Hwang, C.Y.; Hong, S.G.; Lee, H.K. Halocynthiibacter arcticus sp. nov., isolated from Arctic marine sediment. Int. J. Syst. Evol. Microbiol. 2015, 65, 3861–3865. [Google Scholar] [CrossRef]
- Lee, Y.M.; Baek, K.; Lee, J.; Lee, H.K.; Park, H.; Shin, S.C. Complete genome sequence of Halocynthiibacter arcticus PAMC 20958(T) from an Arctic marine sediment sample. J. Biotechnol. 2016, 224, 12–13. [Google Scholar] [CrossRef] [PubMed]
- Sarmah, N.; Revathi, D.; Sheelu, G.; Yamuna Rani, K.; Sridhar, S.; Mehtab, V.; Sumana, C. Recent advances on sources and industrial applications of lipases. Biotechnol. Prog. 2018, 34, 5–28. [Google Scholar] [CrossRef] [PubMed]
- Oh, C.; Kim, T.D.; Kim, K.K. Carboxylic Ester Hydrolases in Bacteria: Active Site, Structure, Function and Application. Crystals 2019, 9, 597. [Google Scholar] [CrossRef] [Green Version]
- Bae, S.Y.; Ryu, B.H.; Jang, E.; Kim, S.; Kim, T.D. characterization and immobilization of a novel SGNH hydrolase (Est24) from Sinorhizobium meliloti. Appl. Microbiol. Biotechnol. 2013, 97, 1637–1647. [Google Scholar] [CrossRef]
- Hwang, H.; Kim, S.; Yoon, S.; Ryu, Y.; Lee, S.Y.; Kim, T.D. Characterization of a novel oligomeric SGNH-arylestersae from Sinorhizobium meliloti 1021. Int. J. Biol. Macromol. 2010, 46, 145–152. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.W.; Yoo, W.; Park, S.H.; Le, L.T.H.L.; Jeong, C.S.; Ryu, B.H.; Shin, S.C.; Kim, H.W.; Park, H.; Kim, K.K.; et al. Structural and functional characterization of a novel cold-active S-formylglutathione hydrolase (SfSFGH) homolog from Shewanella frigidimarina, a psychrophilic bacterium. Microb. Cell Fact. 2019, 18, 140. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ohara, K.; Unno, H.; Oshima, Y.; Hosoya, M.; Fujino, N.; Hirooka, K.; Takahashi, S.; Yamashita, S.; Kusunoki, M.; Nakayama, T. Structural insights into the low pH adaptation of a unique carboxylesterase from Ferroplasma: Altering the pH optima of two carboxylesterases. J. Biol. Chem. 2014, 289, 24499–24510. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, S.H.; Kang, P.A.; Han, K.; Lee, S.W.; Rhee, S. Crystal structure of chloramphenicol-metabolizing enzyme EstDL136 from a metagenome. PLoS ONE 2019, 14, e0210298. [Google Scholar] [CrossRef] [Green Version]
- Pereira, M.R.; Maester, T.C.; Mercaldi, G.F.; de Macedo Lemos, E.G.; Hyvönen, M.; Balan, A. From a metagenomic source to a high-resolution structure of a novel alkaline esterase. Appl. Microbiol. Biotechnol. 2017, 101, 4935–4949. [Google Scholar] [CrossRef]
- Nam, K.H.; Kim, M.Y.; Kim, S.J.; Priyadarshi, A.; Lee, W.H.; Hwang, K.Y. Structural and functional analysis of a novel EstE5 belonging to the subfamily of hormone-sensitive lipase. Biochem. Biophys. Res. Commun. 2009, 379, 553–556. [Google Scholar] [CrossRef]
- Li, P.Y.; Chen, X.L.; Ji, P.; Li, C.Y.; Wang, P.; Zhang, Y.; Xie, B.B.; Qin, Q.L.; Su, H.N.; Zhou, B.C.; et al. Interdomain hydrophobic interactions modulate the thermostability of microbial esterases from the hormone-sensitive lipase family. J. Biol. Chem. 2015, 290, 11188–11198. [Google Scholar] [CrossRef] [Green Version]
- Palm, G.J.; Fernández-Álvaro, E.; Bogdanović, X.; Bartsch, S.; Sczodrok, J.; Singh, R.K.; Böttcher, D.; Atomi, H.; Bornscheuer, U.T.; Hinrichs, W. The crystal structure of an esterase from the hyperthermophilic microorganism Pyrobaculum calidifontis VA1 explains its enantioselectivity. Appl. Microbiol. Biotechnol. 2011, 91, 1061–1072. [Google Scholar] [CrossRef]
- Santiago, M.; Ramírez-Sarmiento, C.A.; Zamora, R.A.; Parra, L.P. Discovery, molecular mechanisms, and industrial applications of cold-active eEnzymes. Front. Microbiol. 2016, 7, 1408. [Google Scholar] [CrossRef]
- Matthews, B.W. Solvent content of protein crystals. J. Mol. Biol. 1968, 33, 491–497. [Google Scholar] [CrossRef]



| Space Group | P212121 |
|---|---|
| Unit cell parameters (Å) | a = 54.6, b = 59.5, c = 82.9 |
| Wavelength (Å) | 0.9794 |
| Resolution (Å) | 50.00–1.30 (1.32–1.30) |
| Unique reflections | 67,202 (3311) |
| Completeness (%) | 99.4 (98.8) |
| Redundancy | 12.3 (12.2) |
| Rmeas † (%) | 8.7 (37.6) |
| Mean I/σ(I) | 57.0 (13.6) |
| CC1/2 (%) | 99.9 (95.1) |
| Wilson B (Å2) | 35.4 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jeon, S.; Hwang, J.; Yoo, W.; Do, H.; Kim, H.-W.; Kim, K.K.; Lee, J.H.; Kim, T.D. Crystallization and Preliminary X-ray Diffraction Study of a Novel Bacterial Homologue of Mammalian Hormone-Sensitive Lipase (halip1) from Halocynthiibacter arcticus. Crystals 2020, 10, 963. https://doi.org/10.3390/cryst10110963
Jeon S, Hwang J, Yoo W, Do H, Kim H-W, Kim KK, Lee JH, Kim TD. Crystallization and Preliminary X-ray Diffraction Study of a Novel Bacterial Homologue of Mammalian Hormone-Sensitive Lipase (halip1) from Halocynthiibacter arcticus. Crystals. 2020; 10(11):963. https://doi.org/10.3390/cryst10110963
Chicago/Turabian StyleJeon, Sangeun, Jisub Hwang, Wanki Yoo, Hackwon Do, Han-Woo Kim, Kyeong Kyu Kim, Jun Hyuck Lee, and T. Doohun Kim. 2020. "Crystallization and Preliminary X-ray Diffraction Study of a Novel Bacterial Homologue of Mammalian Hormone-Sensitive Lipase (halip1) from Halocynthiibacter arcticus" Crystals 10, no. 11: 963. https://doi.org/10.3390/cryst10110963
APA StyleJeon, S., Hwang, J., Yoo, W., Do, H., Kim, H.-W., Kim, K. K., Lee, J. H., & Kim, T. D. (2020). Crystallization and Preliminary X-ray Diffraction Study of a Novel Bacterial Homologue of Mammalian Hormone-Sensitive Lipase (halip1) from Halocynthiibacter arcticus. Crystals, 10(11), 963. https://doi.org/10.3390/cryst10110963