Effects of Proline Substitutions on the Thermostable LOV Domain from Chloroflexus aggregans
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sequence Analysis
2.2. Cloning, Protein Expression and Purification
2.3. Spectroscopic Characterization
2.4. Crystallization
2.5. Acquisition and Treatment of Diffraction Data
2.6. Structure Determination and Refinement
2.7. Molecular Dynamics (MD) Simulations
3. Results
3.1. Identification of Positions for Proline Substitutions
3.2. Characterization of Ala→Pro CagFbFP Mutants
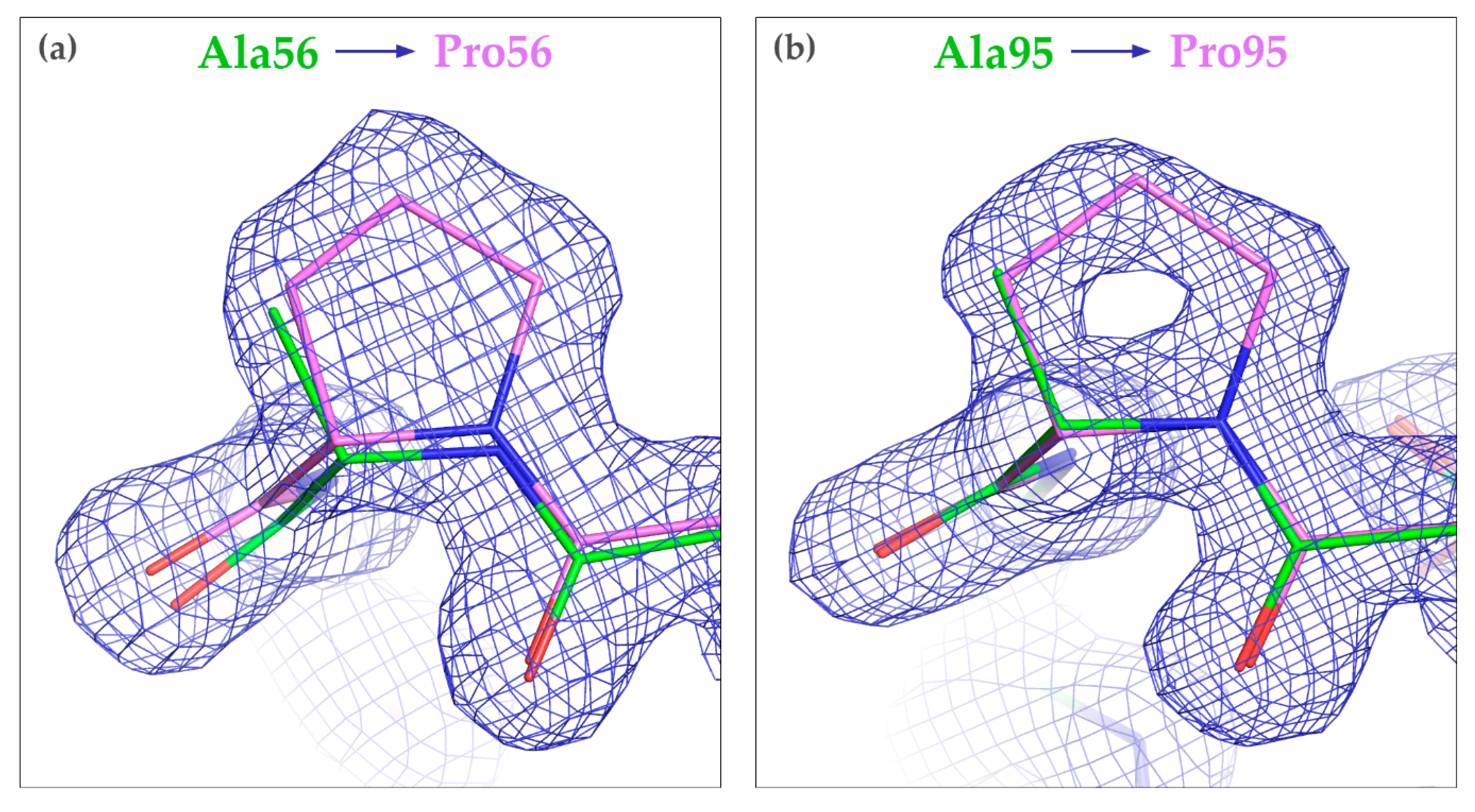
3.3. Crystallization of the Mutated CagFbFP Variants
3.4. MD Simulations of the A58P Variant
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| FbFP | Flavin-based Fluorescent Protein |
| LOV | Light-Oxygen-Voltage |
| WT | Wild Type |
References
- Losi, A.; Gärtner, W. Solving Blue Light Riddles: New Lessons from Flavin-binding LOV Photoreceptors. Photochem. Photobiol. 2017, 93, 141–158. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Glantz, S.T.; Carpenter, E.J.; Melkonian, M.; Gardner, K.H.; Boyden, E.S.; Wong, G.K.-S.; Chow, B.Y. Functional and topological diversity of LOV domain photoreceptors. Proc. Natl. Acad. Sci. USA 2016, 113, E1442–E1451. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Möglich, A. Signal transduction in photoreceptor histidine kinases. Protein Sci. 2019, 28, 1923–1946. [Google Scholar] [CrossRef] [PubMed]
- Conrad, K.S.; Manahan, C.C.; Crane, B.R. Photochemistry of flavoprotein light sensors. Nat. Chem. Biol. 2014, 10, 801–809. [Google Scholar] [CrossRef]
- Losi, A.; Gardner, K.H.; Möglich, A. Blue-Light Receptors for Optogenetics. Chem. Rev. 2018, 118, 10659–10709. [Google Scholar] [CrossRef]
- Glantz, S.T.; Berlew, E.E.; Jaber, Z.; Schuster, B.S.; Gardner, K.H.; Chow, B.Y. Directly light-regulated binding of RGS-LOV photoreceptors to anionic membrane phospholipids. Proc. Natl. Acad. Sci. USA 2018, 115, E7720–E7727. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Buckley, A.M.; Petersen, J.; Roe, A.J.; Douce, G.R.; Christie, J.M. LOV-based reporters for fluorescence imaging. Curr. Opin. Chem. Biol. 2015, 27, 39–45. [Google Scholar] [CrossRef] [Green Version]
- Mukherjee, A.; Schroeder, C.M. Flavin-based fluorescent proteins: Emerging paradigms in biological imaging. Curr. Opin. Biotechnol. 2015, 31, 16–23. [Google Scholar] [CrossRef]
- Drepper, T.; Eggert, T.; Circolone, F.; Heck, A.; Krauß, U.; Guterl, J.-K.; Wendorff, M.; Losi, A.; Gärtner, W.; Jaeger, K.-E. Reporter proteins for in vivo fluorescence without oxygen. Nat. Biotechnol. 2007, 25, 443–445. [Google Scholar] [CrossRef]
- Kim, N.M.; Sinnott, R.W.; Sandoval, N.R. Transcription factor-based biosensors and inducible systems in non-model bacteria: Current progress and future directions. Curr. Opin. Biotechnol. 2020, 64, 39–46. [Google Scholar] [CrossRef]
- Chia, H.E.; Marsh, E.N.G.; Biteen, J.S. Extending fluorescence microscopy into anaerobic environments. Curr. Opin. Chem. Biol. 2019, 51, 98–104. [Google Scholar] [CrossRef] [PubMed]
- Ozbakir, H.F.; Anderson, N.T.; Fan, K.-C.; Mukherjee, A. Beyond the Green Fluorescent Protein: Biomolecular Reporters for Anaerobic and Deep-Tissue Imaging. Bioconjug. Chem. 2020, 31, 293–302. [Google Scholar] [CrossRef] [PubMed]
- Shu, X.; Lev-Ram, V.; Deerinck, T.J.; Qi, Y.; Ramko, E.B.; Davidson, M.W.; Jin, Y.; Ellisman, M.H.; Tsien, R.Y. A genetically encoded tag for correlated light and electron microscopy of intact cells, tissues, and organisms. PLoS Biol. 2011, 9, e1001041. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Westberg, M.; Etzerodt, M.; Ogilby, P.R. Rational design of genetically encoded singlet oxygen photosensitizing proteins. Curr. Opin. Struct. Biol. 2019, 57, 56–62. [Google Scholar] [CrossRef]
- Endres, S.; Wingen, M.; Torra, J.; Ruiz-González, R.; Polen, T.; Bosio, G.; Bitzenhofer, N.L.; Hilgers, F.; Gensch, T.; Nonell, S.; et al. An optogenetic toolbox of LOV-based photosensitizers for light-driven killing of bacteria. Sci. Rep. 2018, 8, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Gushchin, I.; Gordeliy, V. Microbial Rhodopsins. Subcell. Biochem. 2018, 87, 19–56. [Google Scholar]
- Pudasaini, A.; El-Arab, K.K.; Zoltowski, B.D. LOV-based optogenetic devices: Light-driven modules to impart photoregulated control of cellular signaling. Front. Mol. Biosci. 2015, 2, 18. [Google Scholar] [CrossRef] [Green Version]
- Weber, A.M.; Kaiser, J.; Ziegler, T.; Pilsl, S.; Renzl, C.; Sixt, L.; Pietruschka, G.; Moniot, S.; Kakoti, A.; Juraschitz, M.; et al. A blue light receptor that mediates RNA binding and translational regulation. Nat. Chem. Biol. 2019, 15, 1085–1092. [Google Scholar] [CrossRef]
- Chapman, S.; Faulkner, C.; Kaiserli, E.; Garcia-Mata, C.; Savenkov, E.I.; Roberts, A.G.; Oparka, K.J.; Christie, J.M. The photoreversible fluorescent protein iLOV outperforms GFP as a reporter of plant virus infection. Proc. Natl. Acad. Sci. USA 2008, 105, 20038–20043. [Google Scholar] [CrossRef] [Green Version]
- Christie, J.M.; Hitomi, K.; Arvai, A.S.; Hartfield, K.A.; Mettlen, M.; Pratt, A.J.; Tainer, J.A.; Getzoff, E.D. Structural tuning of the fluorescent protein iLOV for improved photostability. J. Biol. Chem. 2012, 287, 22295–22304. [Google Scholar] [CrossRef] [Green Version]
- Song, X.; Wang, Y.; Shu, Z.; Hong, J.; Li, T.; Yao, L. Engineering a More Thermostable Blue Light Photo Receptor Bacillus subtilis YtvA LOV Domain by a Computer Aided Rational Design Method. PLOS Comput. Biol. 2013, 9, e1003129. [Google Scholar] [CrossRef] [PubMed]
- Higgins, S.A.; Ouonkap, S.V.Y.; Savage, D.F. Rapid and Programmable Protein Mutagenesis Using Plasmid Recombineering. ACS Synth. Biol. 2017, 6, 1825–1833. [Google Scholar] [CrossRef] [PubMed]
- Ko, S.; Hwang, B.; Na, J.-H.; Lee, J.; Jung, S.T. Engineered Arabidopsis Blue Light Receptor LOV Domain Variants with Improved Quantum Yield, Brightness, and Thermostability. J. Agric. Food Chem. 2019, 67, 12037–12043. [Google Scholar] [CrossRef] [PubMed]
- Wingen, M.; Jaeger, K.-E.; Gensch, T.; Drepper, T. Novel Thermostable Flavin-binding Fluorescent Proteins from Thermophilic Organisms. Photochem. Photobiol. 2017, 93, 849–856. [Google Scholar] [CrossRef]
- Nazarenko, V.V.; Remeeva, A.; Yudenko, A.; Kovalev, K.; Dubenko, A.; Goncharov, I.M.; Kuzmichev, P.; Rogachev, A.V.; Buslaev, P.; Borshchevskiy, V.; et al. A thermostable flavin-based fluorescent protein from Chloroflexus aggregans: A framework for ultra-high resolution structural studies. Photochem. Photobiol. Sci. Off. J. Eur. Photochem. Assoc. Eur. Soc. Photobiol. 2019, 18, 1793–1805. [Google Scholar] [CrossRef]
- Porebski, B.T.; Buckle, A.M. Consensus protein design. Protein Eng. Des. Sel. 2016, 29, 245–251. [Google Scholar] [CrossRef] [Green Version]
- Pezeshgi Modarres, H.; Mofrad, M.R.; Sanati-Nezhad, A. Protein thermostability engineering. RSC Adv. 2016, 6, 115252–115270. [Google Scholar] [CrossRef]
- Kazlauskas, R. Engineering more stable proteins. Chem. Soc. Rev. 2018, 47, 9026–9045. [Google Scholar] [CrossRef]
- Goldenzweig, A.; Fleishman, S.J. Principles of Protein Stability and Their Application in Computational Design. Annu. Rev. Biochem. 2018, 87, 105–129. [Google Scholar] [CrossRef]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X version 2.0. Bioinforma Oxf. Engl. 2007, 23, 2947–2948. [Google Scholar] [CrossRef] [Green Version]
- Waterhouse, A.M.; Procter, J.B.; Martin, D.M.A.; Clamp, M.; Barton, G.J. Jalview Version 2—A multiple sequence alignment editor and analysis workbench. Bioinforma. Oxf. Engl. 2009, 25, 1189–1191. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Johnson, M.; Zaretskaya, I.; Raytselis, Y.; Merezhuk, Y.; McGinnis, S.; Madden, T.L. NCBI BLAST: A better web interface. Nucleic Acids Res. 2008, 36, W5-9. [Google Scholar] [CrossRef] [PubMed]
- Studier, F.W. Protein production by auto-induction in high-density shaking cultures. Protein Expr. Purif. 2005, 41, 207–234. [Google Scholar] [CrossRef] [PubMed]
- Kabsch, W. XDS. Acta Crystallogr. D Biol. Crystallogr. 2010, 66, 125–132. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Evans, P. Scaling and assessment of data quality. Acta Crystallogr. D Biol. Crystallogr. 2005, 62, 72–82. [Google Scholar] [CrossRef]
- Vagin, A.; Teplyakov, A. Molecular replacement with MOLREP. Acta Crystallogr. D Biol. Crystallogr. 2009, 66, 22–25. [Google Scholar] [CrossRef]
- Emsley, P.; Lohkamp, B.; Scott, W.G.; Cowtan, K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 2010, 66, 486–501. [Google Scholar] [CrossRef] [Green Version]
- Murshudov, G.N.; Skubák, P.; Lebedev, A.A.; Pannu, N.S.; Steiner, R.A.; Nicholls, R.A.; Winn, M.D.; Long, F.; Vagin, A.A. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D Biol. Crystallogr. 2011, 67, 355–367. [Google Scholar] [CrossRef] [Green Version]
- Krieger, E.; Koraimann, G.; Vriend, G. Increasing the precision of comparative models with YASARA NOVA—a self-parameterizing force field. Proteins Struct. Funct. Bioinforma 2002, 47, 393–402. [Google Scholar] [CrossRef]
- Krivov, G.G.; Shapovalov, M.V.; Dunbrack, R.L. Improved prediction of protein side-chain conformations with SCWRL4. Proteins Struct. Funct. Bioinforma 2009, 77, 778–795. [Google Scholar] [CrossRef] [Green Version]
- Olsson, M.H.M.; Søndergaard, C.R.; Rostkowski, M.; Jensen, J.H. PROPKA3: Consistent Treatment of Internal and Surface Residues in Empirical pKa Predictions. J. Chem. Theory Comput. 2011, 7, 525–537. [Google Scholar] [CrossRef] [PubMed]
- Case, D.A.; Babin, V.; Berryman, J.; Betz, R.M.; Cai, Q.; Cerutti, D.S.; Cheatham, T.E., III; Darden, T.A.; Duke, R.E.; Gohlke, H.; et al. Amber 14; University of California: San Francisco, CA, USA, 2014. [Google Scholar]
- Jorgensen, W.L.; Chandrasekhar, J.; Madura, J.D.; Impey, R.W.; Klein, M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983, 79, 926–935. [Google Scholar] [CrossRef]
- Hornak, V.; Abel, R.; Okur, A.; Strockbine, B.; Roitberg, A.; Simmerling, C. Comparison of multiple Amber force fields and development of improved protein backbone parameters. Proteins Struct. Funct. Bioinforma 2006, 65, 712–725. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cornell, W.D.; Cieplak, P.; Bayly, C.I.; Gould, I.R.; Merz, K.M.; Ferguson, D.M.; Spellmeyer, D.C.; Fox, T.; Caldwell, J.W.; Kollman, P.A. A Second Generation Force Field for the Simulation of Proteins, Nucleic Acids, and Organic Molecules. J. Am. Chem. Soc. 1995, 117, 5179–5197. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Wolf, R.M.; Caldwell, J.W.; Kollman, P.A.; Case, D.A. Development and testing of a general amber force field. J. Comput. Chem. 2004, 25, 1157–1174. [Google Scholar] [CrossRef] [PubMed]
- Davari, M.D.; Kopka, B.; Wingen, M.; Bocola, M.; Drepper, T.; Jaeger, K.-E.; Schwaneberg, U.; Krauss, U. Photophysics of the LOV-Based Fluorescent Protein Variant iLOV-Q489K Determined by Simulation and Experiment. J. Phys. Chem. B 2016, 120, 3344–3352. [Google Scholar] [CrossRef] [PubMed]
- DeLano, W.L. The PyMOL Molecular Graphics System. 2002. Available online: http://www.pymol.org (accessed on 28 March 2020).
- Humphrey, W.; Dalke, A.; Schulten, K. VMD Visual molecular dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef]
- Wingen, M.; Potzkei, J.; Endres, S.; Casini, G.; Rupprecht, C.; Fahlke, C.; Krauss, U.; Jaeger, K.-E.; Drepper, T.; Gensch, T. The photophysics of LOV-based fluorescent proteins – new tools for cell biology. Photochem. Photobiol. Sci. 2014, 13, 875–883. [Google Scholar] [CrossRef] [Green Version]
- Mukherjee, A.; Weyant, K.B.; Agrawal, U.; Walker, J.; Cann, I.K.O.; Schroeder, C.M. Engineering and characterization of new LOV-based fluorescent proteins from Chlamydomonas reinhardtii and Vaucheria frigida. ACS Synth. Biol. 2015, 4, 371–377. [Google Scholar] [CrossRef]
- Tang, K.-H.; Barry, K.; Chertkov, O.; Dalin, E.; Han, C.S.; Hauser, L.J.; Honchak, B.M.; Karbach, L.E.; Land, M.L.; Lapidus, A.; et al. Complete genome sequence of the filamentous anoxygenic phototrophic bacterium Chloroflexus aurantiacus. BMC Genom. 2011, 12, 334. [Google Scholar] [CrossRef] [Green Version]
- Gaisin, V.A.; Kalashnikov, A.M.; Grouzdev, D.S.; Sukhacheva, M.V.; Kuznetsov, B.B.; Gorlenko, V.M. Chloroflexus islandicus sp. nov., a thermophilic filamentous anoxygenic phototrophic bacterium from a geyser. Int. J. Syst. Evol. Microbiol. 2017, 67, 1381–1386. [Google Scholar] [CrossRef] [PubMed]
- Liebschner, D.; Afonine, P.V.; Moriarty, N.W.; Poon, B.K.; Sobolev, O.V.; Terwilliger, T.C.; Adams, P.D. Polder maps: Improving OMIT maps by excluding bulk solvent. Acta Crystallogr. Sect. Struct. Biol. 2017, 73, 148–157. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kottke, T.; Dick, B.; Fedorov, R.; Schlichting, I.; Deutzmann, R.; Hegemann, P. Irreversible photoreduction of flavin in a mutated Phot-LOV1 domain. Biochemistry 2003, 42, 9854–9862. [Google Scholar] [CrossRef] [PubMed]
- Salomon, M.; Christie, J.M.; Knieb, E.; Lempert, U.; Briggs, W.R. Photochemical and mutational analysis of the FMN-binding domains of the plant blue light receptor, phototropin. Biochemistry 2000, 39, 9401–9410. [Google Scholar] [CrossRef] [PubMed]
- Zayner, J.P.; Sosnick, T.R. Factors that control the chemistry of the LOV domain photocycle. PLoS ONE 2014, 9, e87074. [Google Scholar] [CrossRef] [Green Version]
- Raffelberg, S.; Gutt, A.; Gärtner, W.; Mandalari, C.; Abbruzzetti, S.; Viappiani, C.; Losi, A. The amino acids surrounding the flavin 7a-methyl group determine the UVA spectral features of a LOV protein. Biol. Chem. 2013, 394, 1517–1528. [Google Scholar] [CrossRef]
- Fettweiss, T.; Röllen, K.; Granzin, J.; Reiners, O.; Endres, S.; Drepper, T.; Willbold, D.; Jaeger, K.-E.; Batra-Safferling, R.; Krauss, U. Mechanistic Basis of the Fast Dark Recovery of the Short LOV Protein DsLOV from Dinoroseobacter shibae. Biochemistry 2018, 57, 4833–4847. [Google Scholar] [CrossRef]
- Mukherjee, A.; Weyant, K.B.; Walker, J.; Schroeder, C.M. Directed evolution of bright mutants of an oxygen-independent flavin-binding fluorescent protein from Pseudomonas putida. J. Biol. Eng. 2012, 6, 20. [Google Scholar] [CrossRef] [Green Version]
- Consiglieri, E.; Xu, Q.; Bregnhøj, M.; Westberg, M.; Ogilby, P.R.; Losi, A. Single mutation in a novel bacterial LOV protein yields a singlet oxygen generator. Photochem. Photobiol. Sci. 2019, 18, 2657–2660. [Google Scholar] [CrossRef]
- Westberg, M.; Bregnhøj, M.; Etzerodt, M.; Ogilby, P.R. No Photon Wasted: An Efficient and Selective Singlet Oxygen Photosensitizing Protein. J. Phys. Chem. B 2017, 121, 9366–9371. [Google Scholar] [CrossRef]


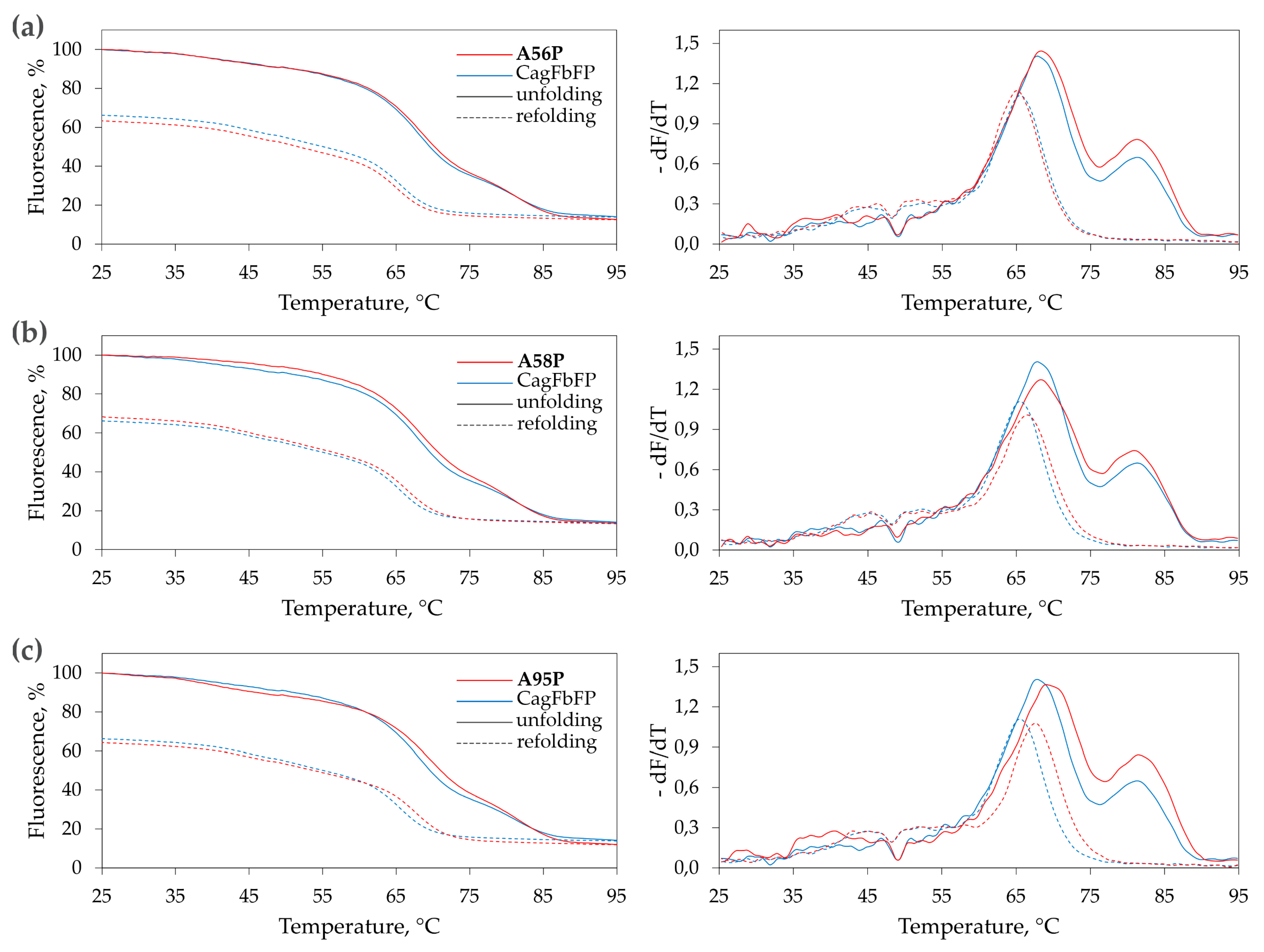

| Variant. | Tm1, °C | Tm2, °C | Tr, °C |
|---|---|---|---|
| Wild type | 67.9 ± 0.3 | 81.3 ± 0.3 | 65.4 ± 0.3 |
| A56P | 68.3 ± 0.3 | 81.3 ± 0.4 | 65.2 ± 0.3 |
| A58P | 68.3 ± 0.3 | 80.9 ± 0.3 | 66.3 ± 0.3 |
| A95P | 69.2 ± 0.5 | 81.5 ± 0.3 | 67.7 ± 0.3 |
| Data Collection | ||
|---|---|---|
| Variant | A56P | A95P |
| Protein Data Bank ID | 6Y7R | 6Y7U |
| Space group | P21212 | P21212 |
| Cell dimensions | - | - |
| a, b, c (Å) | 54.6, 111.3, 39.2 | 54.3, 111.4, 38.9 |
| Wavelength (Å) | 0.9762 | 0.9762 |
| Resolution (Å) | 111.29–1.60 (1.63–1.60) * | 111.36–1.60 (1.63–1.60) * |
| Rpim (%) | 3.3 (46.5) * | 3.2 (21.5) * |
| <I/σI> | 14.5 (1.9) * | 14.0 (3.4) * |
| CC1/2 (%) | 99.9 (76.1) * | 99.9 (92.8) * |
| Completeness (%) | 100.0 (100.0) * | 99.9 (100.0) * |
| Multiplicity | 13.1 (13.3) * | 10.5 (11.1) * |
| Unique reflections | 32,415 (1580) * | 31,994 (1582) * |
| Refinement | ||
| Resolution (Å) | 39.25−1.60 | 38.97−1.60 |
| No. reflections | 30,768 | 30,392 |
| Rwork/Rfree (%) | 18.4/21.4 | 17.8/22.5 |
| No. atoms | - | - |
| Protein | 1717 | 1680 |
| FMN | 62 | 62 |
| Water | 283 | 221 |
| Average B factors (Å2) | - | - |
| Protein | 20.3 | 19.9 |
| FMN | 16.2 | 16.5 |
| Water | 33.5 | 30.6 |
| R.m.s. deviations | - | - |
| Protein bond lengths (Å) | 0.004 | 0.005 |
| Protein bond angles (°) | 1.3 | 1.3 |
| Ramachandran analysis | - | - |
| Favored (%) | 98.7 | 98.7 |
| Outliers (%) | 0 | 0 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Remeeva, A.; Nazarenko, V.V.; Goncharov, I.M.; Yudenko, A.; Smolentseva, A.; Semenov, O.; Kovalev, K.; Gülbahar, C.; Schwaneberg, U.; Davari, M.D.; et al. Effects of Proline Substitutions on the Thermostable LOV Domain from Chloroflexus aggregans. Crystals 2020, 10, 256. https://doi.org/10.3390/cryst10040256
Remeeva A, Nazarenko VV, Goncharov IM, Yudenko A, Smolentseva A, Semenov O, Kovalev K, Gülbahar C, Schwaneberg U, Davari MD, et al. Effects of Proline Substitutions on the Thermostable LOV Domain from Chloroflexus aggregans. Crystals. 2020; 10(4):256. https://doi.org/10.3390/cryst10040256
Chicago/Turabian StyleRemeeva, Alina, Vera V. Nazarenko, Ivan M. Goncharov, Anna Yudenko, Anastasia Smolentseva, Oleg Semenov, Kirill Kovalev, Cansu Gülbahar, Ulrich Schwaneberg, Mehdi D. Davari, and et al. 2020. "Effects of Proline Substitutions on the Thermostable LOV Domain from Chloroflexus aggregans" Crystals 10, no. 4: 256. https://doi.org/10.3390/cryst10040256
APA StyleRemeeva, A., Nazarenko, V. V., Goncharov, I. M., Yudenko, A., Smolentseva, A., Semenov, O., Kovalev, K., Gülbahar, C., Schwaneberg, U., Davari, M. D., Gordeliy, V., & Gushchin, I. (2020). Effects of Proline Substitutions on the Thermostable LOV Domain from Chloroflexus aggregans. Crystals, 10(4), 256. https://doi.org/10.3390/cryst10040256





