Abstract
Magnetotactic bacteria (MTB) biomineralize intracellular magnetite (Fe3O4) crystals surrounded by a magnetosome membrane (MM). The MM contains membrane-specific proteins that control Fe3O4 mineralization in MTB. Previous studies have demonstrated that Mms13 is a critical protein within the MM. Mms13 can be isolated from the MM fraction of Magnetospirillum magneticum AMB-1 and a Mms13 homolog, MamC, has been shown to control the size and shape of magnetite nanocrystals synthesized in-vitro. The objective of this study was to use several independent methods to definitively determine the localization of native Mms13 in M. magneticum AMB-1. Using Mms13-immunogold labeling and transmission electron microscopy (TEM), we found that Mms13 is localized to the magnetosome chain of M. magneticum AMB-1 cells. Mms13 was detected in direct contact with magnetite crystals or within the MM. Immunofluorescence detection of Mms13 in M. magneticum AMB-1 cells by confocal laser scanning microscopy (CLSM) showed Mms13 localization along the length of the magnetosome chain. Proteins contained within the MM were resolved by SDS-PAGE for Western blot analysis and LC-MS/MS (liquid chromatography with tandem mass spectrometry) protein sequencing. Using Anti-Mms13 antibody, a protein band with a molecular mass of ~14 kDa was detected in the MM fraction only. This polypeptide was digested with trypsin, sequenced by LC-MS/MS and identified as magnetosome protein Mms13. Peptides corresponding to the protein’s putative MM domain and catalytic domain were both identified by LC-MS/MS. Our results (Immunogold TEM, Immunofluorescence CLSM, Western blot, LC-MS/MS), combined with results from previous studies, demonstrate that Mms13 and homolog proteins MamC and Mam12, are localized to the magnetosome chain in MTB belonging to the class Alphaproteobacteria. Because of their shared localization in the MM and highly conserved amino acid sequences, it is likely that MamC, Mam12, and Mms13 share similar roles in the biomineralization of Fe3O4 nanocrystals.
Keywords:
bacteria; biomineralization; magnetite; magnetotactic; magnetosome; nanocrystal; protein; TEM 1. Introduction
Magnetotactic bacteria (MTB) are a group of prokaryotes, which biomineralize magnetic crystals of magnetite (Fe3O4) and/or greigite (Fe3S4) surrounded by a lipid-bilayer, called the magnetosome membrane (MM) [1,2,3,4]. The magnetosome membrane (MM) originates as an invagination of the cytoplasmic membrane creating a vesicle in which the magnetite crystal nucleates and subsequently grows [5,6,7,8,9]. Proteins associated with the MM are believed to control the biomineralization of magnetosome crystals [10,11,12,13,14,15,16,17,18,19,20,21]. Individual magnetosomes are arranged in a chain(s) within a bacterium to maximize the magnetic dipole moment of the cell therefore allowing MTB to passively align and swim along Earth’s magnetic field lines [1,2,3,4]. Presumably MTB use magnetotaxis in conjunction with chemotaxis to locate and maintain an optimum position in a water column for growth and survival [1,2,3,4].
Previous studies have shown that the MM contains proteins required for the biogenesis of intracellular magnetosomes and that the MM appears to be derived from the cytoplasmic membrane [9,12,13,15,16,17,18,19]. Some of these MM proteins are also found in the cytoplasmic membrane and fatty acid analysis suggests that the MM is derived from the cytoplasmic membrane [13,16,19]. Several MM proteins appear to control the formation of the magnetosome membrane and the biomineralization of the magnetosome crystals [2,6,7,10,11,12,13,14,15,16,17]. For example, in Magnetospirillum magneticum AMB-1, genomic and proteomic analysis have identified almost 100 magnetosome specific proteins, and at least a dozen of these proteins are believed to be involved in magnetosome biomineralization [3,4,16,22].
It has been demonstrated in Magnetospirillum species that several proteins, including Mms5, Mms6, Mms7 (MamD), Mms12 (MamF) and Mms13 (MamC) are tightly associated with the MM [3,4,6,9,12,13,14,15,16,18]. To date, inorganic synthesis of magnetite in the presence of some of these purified proteins has failed to yield crystals that are identical to those found within the magnetosomes of MTB [17,21]. The morphology of the magnetite crystals in MTB appears to be strain specific [2,3,4,23]. This suggests that the mineralization process and the function of individual proteins may vary by microbial strain. Therefore, it is vital to study individual proteins in all MTB species to determine similarities and differences between different bacterial species. There is great interest in understanding the protein-catalyzed mineralization process involved in magnetosome synthesis and the function of specific MM proteins in controlling the number, size, and morphology of nascent Fe3O4 nanocrystals.
Among MM proteins, Mms13 and homologous proteins MamC and Mam12, are small (e.g., Mms13 is 145 amino acids) highly conserved protein within the magnetotactic Alphaproteobacteria class and no obvious homologs have been identified in non-magnetotactic bacteria [3,4,9,12,13,16,18,22]. The secondary structure of MamC (a Mms13 homolog) reveals two helicoidal transmembrane domains connected by a α-helix structured loop in the magnetosome lumen that contains several charged residues predicted to interact with the magnetite crystal [2,3,4,11,21,24]. Furthermore, it has been demonstrated that MamC plays a role in regulating the size and shape of magnetite crystals in MTB [2,4,14,21,25,26,27] and in in-vitro mineralization experiments [6,24,25,26,27].
Previous studies [12,13,15,16,18,28,29,30,31,32] have employed different methods and techniques to help define the localization of Mms13 and homologous proteins, MamC and Mam12, in MTB species belonging to the class Alphaproteobacteria. These studies used several techniques including plasmid-overexpressed fusion proteins, immunogold electron microscopy, immunofluorescence microscopy, and biochemical and proteomic analysis of purified cell fractions. Indeed, the use of different methods and techniques to determine the localization of a protein within a cell is critical to validate results and avoid bias that can lead to false conclusions.
The objective of this study was to use multiple independent methods on the same organism (M. magneticum AMB-1) to determine the subcellular location of native Mms13 with micron to nanometer scale resolution. We used (1) protein fractionation followed by Western blot analysis, (2) LC-MS/MS protein sequencing, (3) immunogold-Mms13 labeling followed by transmission electron microscopy (TEM) of cells, and (4) immunofluorescence detection of Mms13 by confocal laser scanning microscopy (CLSM) on whole cells. To our knowledge, this is the first time that immunogold TEM and immunofluorescence microscopy have be used to examine the localization of native Mms13 in M. magneticum AMB-1 cells and purified magnetosomes. This work provides for a more complete understanding of the subcellular localization of homologous proteins MamC, Mam12, and Mms13 in members of the magnetotactic Alphaproteobacteria group. This information is important to understanding protein function and identifying shared biomineralization pathways among MTB.
2. Materials and Methods
To purify magnetosomes and prepare cell soluble and membrane fractions of M. magneticum AMB-1 (ATCC 700264), the protocol described in [33] was followed. To clone mms13 (amb0951 in M. magneticum AMB-1; [22]), genomic DNA from M. magneticum AMB-1 was obtained from the American Type Culture Collection (ATCC #700264). The gene amb0951 was amplified by the polymerase chain reaction (PCR) with the primers: fw1 (5′-CACCATGCGCTCCTGGCTG-3′) and rev2 (5′-TCAGGCCAGTTCGTCCCGC-3′). The PCR product was cloned into pET160/GW/D-TOPO according to the manufacturer’s instructions (Invitrogen, Carlsbad, CA, USA). PCR products and the vector containing mms13 (pET160/GW/D-TOPO/mms13) were sequenced at the Plant-Microbe Genomics Facility at The Ohio State University.
Escherichia coli TOP10 (Invitrogen, Carlsbad, CA, USA) and E. coli BL21 (Invitrogen, Carlsbad, CA, USA) cells were used for protein expression. Cells were cultured in 250 mL of Luria broth (LB) at 37 °C and induced with 1 mM isopropyl-1-thio-β-D galactopyranoside for 5.5 h. The recombinant Mms13 protein (rMms13) was insoluble and found in inclusion bodies. Therefore, 8 M urea was used to solubilize the inclusion bodies and rMms13 was purified under denaturing conditions using a nickel-nitrilotriacetic acid column (Qiagen, Hilden, Germany). rMms13 was renatured at 4 °C by dialysis against 1 L of buffer containing 0.02 M Tris-HCl, 5 M NaCl, and 1 mM phenylmethanesulfonylfluoride (PMSF), pH 7.5. This buffer also contained 4 M, 2 M, 1 M, and 0.5 M urea for 2 h each. The final dialysis was performed overnight at 4 °C in 1 L of buffer without urea. The protein was dialyzed several more times for 3 h each, at 4 °C against 1 L of fresh buffer (0.02 M Tris-HCl, 0.5 M NaCl, pH 7.5). This was the rMms13-protein fraction that was used for antibody production.
To determine that the purified recombinant protein was indeed rMms13, the final protein fraction was resolved by SDS-PAGE and stained with Coomassie R-250. The protein band corresponding to the molecular mass of rMms13 was excised and subjected to peptide mass fingerprinting at the Mass Spectrometry & Proteomics Facility at The Ohio State University.
Polyclonal antibodies against rMms13 were produced in a rabbit by ProSci Incorporated (Poway, CA, USA). The final concentration of the antibody (anti-Mms13) determined by direct ELISA was 1.4 mg/mL. These antibodies were used for Western blot and immunogold labeling. Pre-immune serum was removed from the rabbit before injecting with antigen (rMms13) into the rabbit, which was later used as a control for immunogold labeling. For the Western blots, the protocol described in [33] was followed, using anti-Mms13 antibody (1:50,000) as primary antibody and goat anti-rabbit HRP (horseradish peroxidase) antibody (1:200) as secondary antibody and the Clean-Blot IP Detection Kit HRP (Thermo-Pierce, Rockford, IL, USA). PVDF membrane (Invitrogen) was blocked with 5% BSA, imaged using a Kodak Gel Logic 1500 Imager (Rochester, New York, NY, USA) and image processing (i.e., exposure, contrast) was applied to every pixel of the image using Adobe Photoshop.
For protein identification, protein bands were excised from polyacrylamide gel, digested with trypsin and sequenced by liquid chromatographic tandem mass spectrometry (LC-MS/MS). Protein identification was performed by staff at The Ohio State University, Mass Spectrometry & Proteomics Facility. Proteins with a Mascot score of 50 or higher and a minimum of two unique peptides from one protein with a -b or -y ion sequence tag of five residues or better were accepted.
TEM grid preparation and immunogold labeling experiments of M. magneticum AMB-1 cells and purified magnetosomes were done following the procedure described in [33,34]. Cells were placed on a TEM grid and incubated first with anti-Mms13 (1:2000), and second with goat anti-rabbit IgG antibody conjugated with 10 nm gold (Sigma-Aldrich, St. Louis, MO, USA) diluted 1:100 in 0.5% BSA in Tris-HCl. Purified magnetosomes (1:1250 diluted) were incubated with anti-Mms13 diluted 1:4000 and goat anti-rabbit IgG antibody (1:100) conjugated with 10 nm colloidal gold (Sigma-Aldrich, St. Louis, MO, USA).
Fluorescence CLSM of whole, intact M. magneticum AMB-1 cells was performed as described by Oestreicher et al., 2016 [33,34]. Cells were harvested by centrifuging at 10,000× g for 10 min at 4 °C and fixed in 4% paraformaldehyde for 5 min, placed on a glass slide and washed with PBS (phosphate buffered saline, pH 7.4). Cells were incubated and labeled with primary antibody (Anti-Mms13) at 1:400 dilution for 1 h at room temperature. Cells were washed 3X with PBS and labeled with secondary antibody, goat anti-rabbit IgG antibody conjugated to DyLight 488 (1:100; Thermo Fisher Scientific, Waltham, MA, USA). Cells were washed with PBS and analyzed using an Olympus FV 1000 CLSM (Olympus Corporation).
3. Results
Cloning the PCR product into the pET160/GW/D-TOPO vector yielded a heterologous expressed protein (rMms13), which preferentially accumulated in E. coli cells as inclusion bodies and had to be solubilized and purified using urea. The urea was removed and rMms13 renatured by extensive dialysis. The maximal yield in total protein, monitored by SDS-PAGE, was achieved using BL21 CodonPlus E. coli cells, under the following induction conditions: 1 mM IPTG, at 37 °C for 5.5 h.
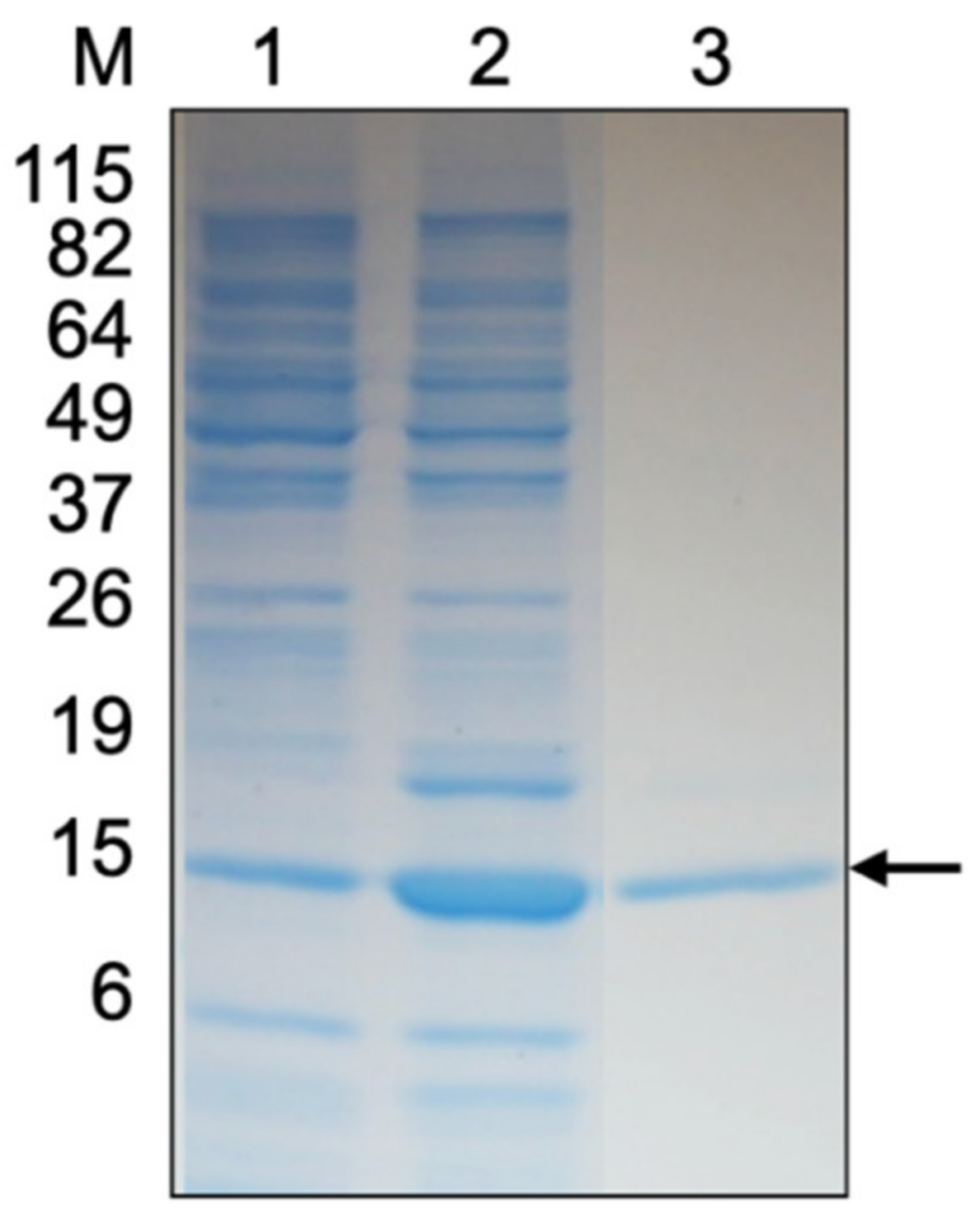
The rMms13 protein was purified and renatured by extensive dialysis in buffer containing progressively less urea until the dialysis buffer contained no urea. The molecular mass of rMms13 was found to be approximately 18 kDa by SDS-PAGE (Figure 1) corresponding to the expected size of the Mms13 protein (~14.5 kDa) plus an additional ~4 kDa resulting from the pET160 vector. The ~18 kDa Coomassie-stained protein band (Figure 1, lane 3) was identified using LC-MS/MS. The polypeptide was found to correspond (p < 0.05) to the Mms13 protein from M. magneticum AMB-1.
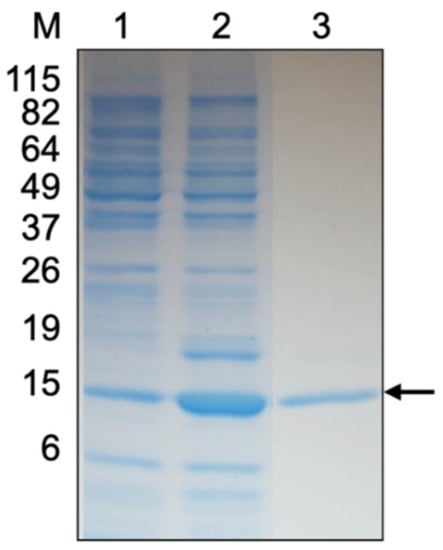
Figure 1.
SDS-PAGE gel of purified recombinant Mms13 (rMms13) stained with Coomassie R-250. The lanes correspond to 10 μg of protein from non-induced Escherichia coli cells (lane 1), 10 μg of protein from induced E. coli cells (lane 2), and 5 μg of rMms13 protein purified using IMAC (lane 3). The black arrow shows the protein band that was excised from the gel, digested with trypsin, sequenced using LC-MS/MS to confirm its identity as rMms13. The positions and molecular masses (in kilodaltons) of protein standards are indicated at the left (lane M).
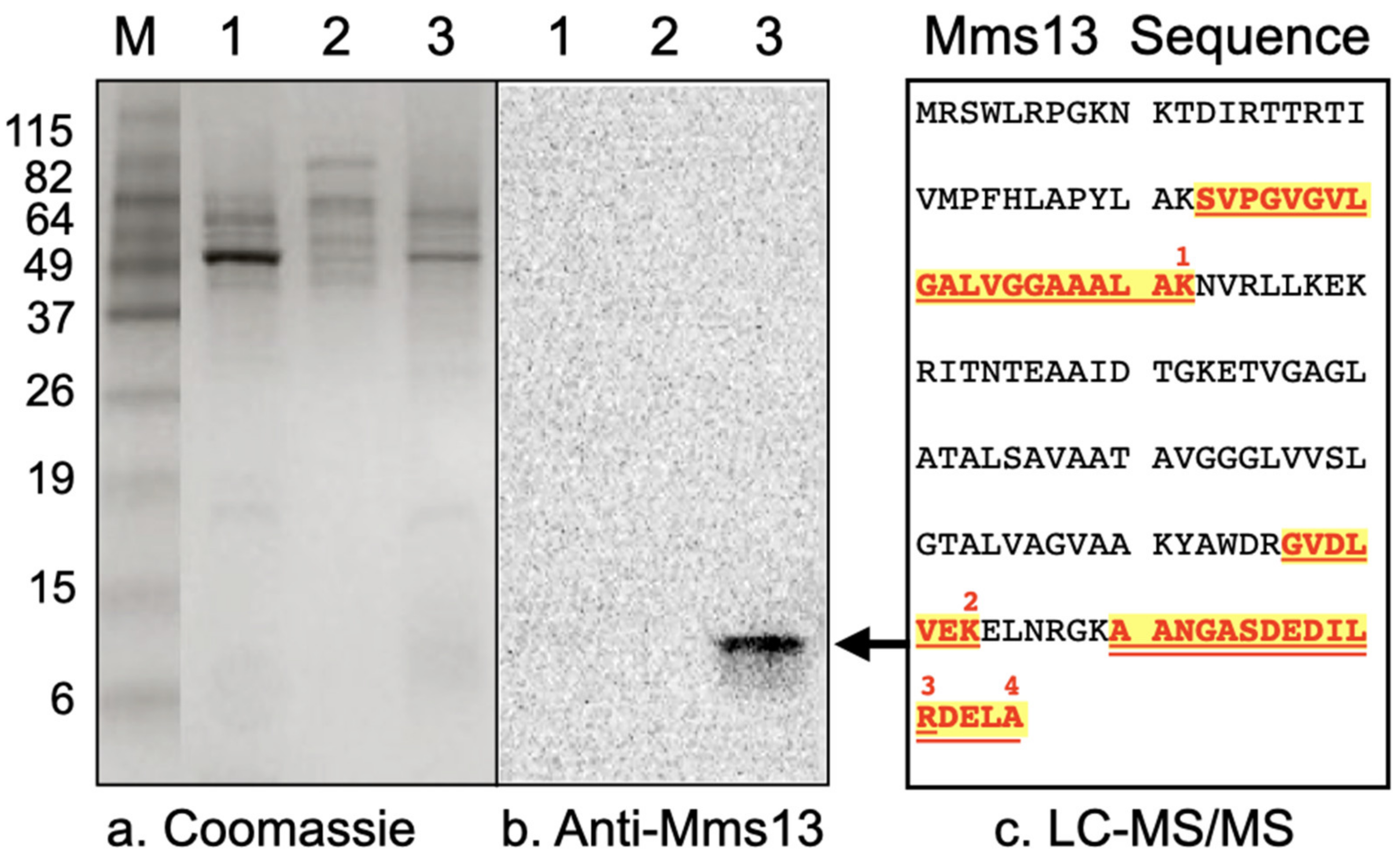
Western blot analysis was performed to determine whether Mms13 was present in the soluble fraction, cytoplasmic membrane fraction, or magnetosome membrane fraction of M. magneticum AMB-1. A band corresponding to the expected molecular mass of Mms13 (~14.5 kDa) was only present in the magnetosome membrane fraction (Figure 2b, lane 3; Figure S1). No band was observed in the soluble fraction (Figure 2b, lane 1). No band was observed in the cytoplasmic membrane fraction (Figure 2b, lane 2).
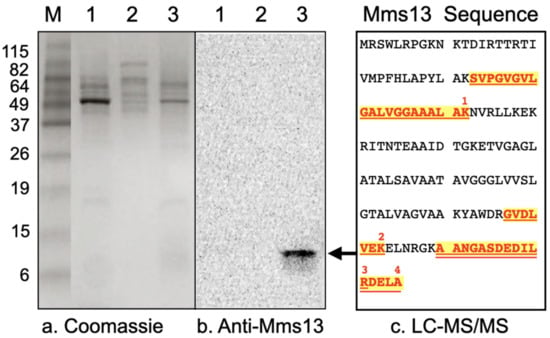
Figure 2.
(a) Coomassie R-250 stained protein fraction of M. magneticum AMB-1 as resolved by SDS-PAGE. (b) Western blot of proteins solubilized from different fractions of M. magneticum AMB-1, resolved by SDS-PAGE and incubated with anti-Mms13 antibody. Lanes correspond to 10 μg soluble proteins (lane 1), 10 μg of cytoplasmic membrane proteins (lane 2), and 10 μg of magnetosome membrane proteins (lane 3). For comparison, molecular weight markers (lane M) were also resolved by SDS-PAGE. The positions and molecular masses (in kilodaltons) of protein standards are indicated using numbers down the left-side of the gel. The black arrow shows protein band that was excised, digested with trypsin and identified by LC-MS/MS peptide sequencing. (c) Mms13 amino acid sequence and locations of the four peptides that were identified by LC-MS/MS (numbered 1–4, red-colored, bold font that is highlighted and underlined). Mms13 was not detected by LC-MS/MS peptide sequencing in lanes 1 or 2.
This ~14 kDa band (Figure 2b, lane 3, black arrow) was excised from a polyacrylamide gel, digested with trypsin, and its amino acid sequence determined by LC-MS/MS. Four peptides were isolated and their amino acid sequences are shown in Figure 2c. LC-MS/MS analysis identified the ~14 kDa protein band contained within the magnetosome membrane fraction as the 145 amino acid magnetic particle protein Mms13 from Magnetospirillum magneticum AMB-1 (Figure 2c). Gel slices from lanes 1 and 2 (Figure 2), corresponding to ~14 kDa were excised and subjected to in-gel trypsin digestion followed by LC-MS/MS. Peptides corresponding to Mms13 were not detected by LC-MS/MS for the soluble fraction (Figure 2, lane 1) or for the cytoplasmic membrane fraction (Figure 2, lane 2).
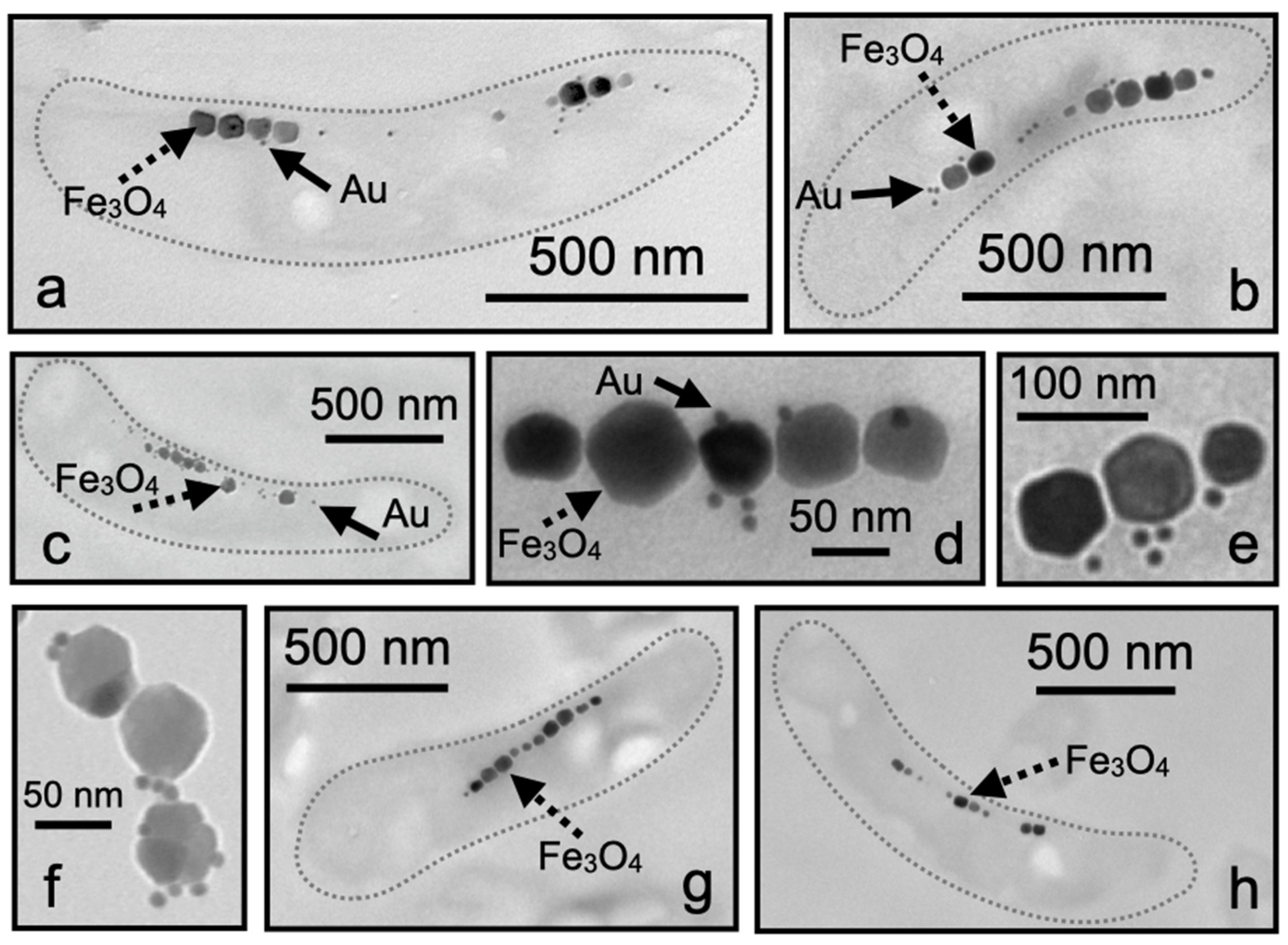
TEM analysis showed that the M. magneticum AMB-1 cells displayed Mms13 labeling along the magnetosome chain (Figure 3a–c). The nanogold particles were either detected within the matrix surrounding the magnetosomes or directly touching the Fe3O4 crystal (Figure 3d–f). It was also observed that some labeling appeared to occur distal to the Fe3O4 particles but typically nanogold particles were found to be in-line with the magnetosome chain (Figure 3a–c). In many instances, we observed that individual magnetosome particles were coated with numerous nanogold particles (Figure 3d–f). No nanogold labeling of the magnetosome chain was observed in either negative control when substituting 0.5% BSA for primary antibody (Figure 3g) or when 5% pre-immune serum was used in place of anti-Mms13 (Figure 3h).
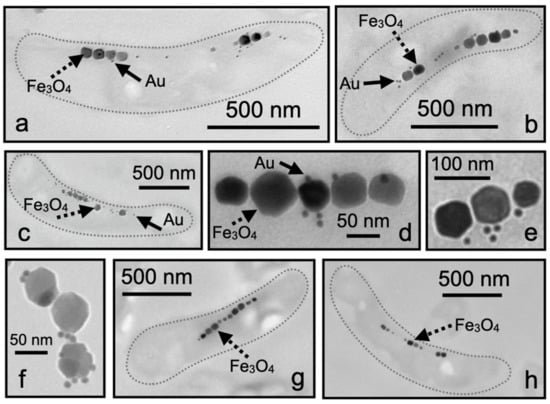
Figure 3.
Ultrathin sections of M. magneticum AMB-1. (a–c) A single cell showing magnetosomes labeled with Mms13 antibody and then labeled with goat anti-rabbit antibody conjugated with 10 nm colloidal gold. (b) Three individual magnetosome particles showing multiple bound nanogold particles. (d–f) Magnetosome chain from M. magneticum AMB-1 cells showing nanogold particles bound directly to magnetosome crystal or observed within magnetosome membrane. (g) Negative control for immunolabeling showing an ultrathin section of M. magneticum AMB-1 treated exactly as in (a), but with 0.5% BSA substituted in place of the primary antibody. (h) Negative control treated exactly as in (a) but with 5% pre-immune serum substituted in place of the primary antibody. Dotted gray line (shown in a–c,g,h) is outline of bacterium. Scale bars provided for each image. Magnetosomes are large (i.e., ~70 nm) black particles arranged in a linear chain. Several individual magnetosomes are indicated with dotted black arrow and labeled Fe3O4. Colloidal gold particles are very small (i.e., ~10 nm) black spheres. Several individual nanogold particles are indicated with solid black arrow and labeled Au.
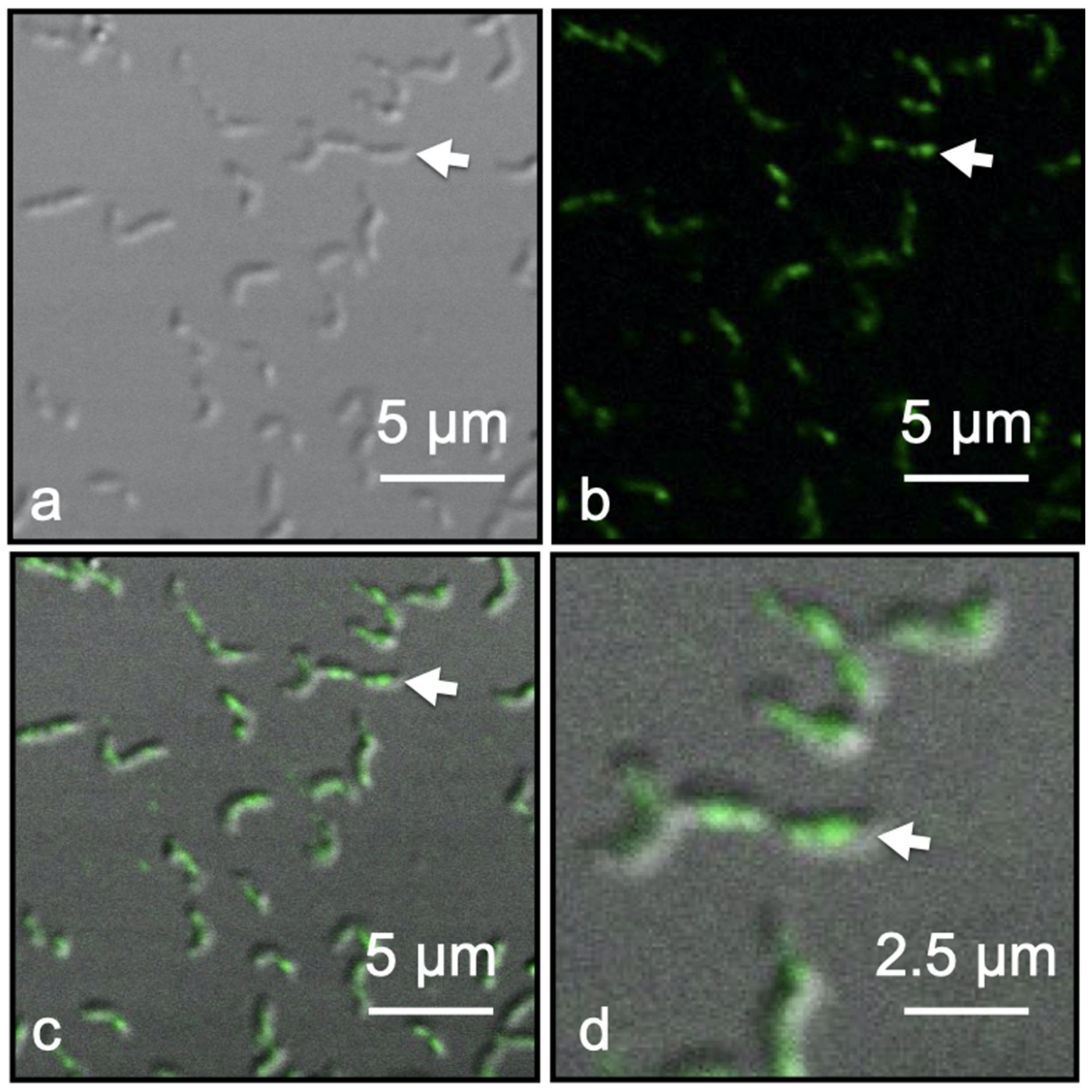
Immunofluorescence of Dylight 488 tagged Anti-Mms13 allowed for the detection and localization of Mms13 within intact M. magneticum AMB-1 cells by CLSM coupled with Nomarski interference contrast technique. Mms13-labeling was observed only along the magnetosome chain of M. magneticum AMB-1, occurring in the center and along the long axis of each cell (Figure 4d). No fluorescence labeling occurred distal to the long axis of each cell (Figure 4). Negative controls were conducted on M. magneticum AMB-1 cells using buffer that lacked primary antibody (i.e., Anti-Mms13) or lacked secondary antibody (i.e., goat anti-rabbit IgG antibody conjugated to DyLight 488). No fluorescence was observed in these two control experiments.
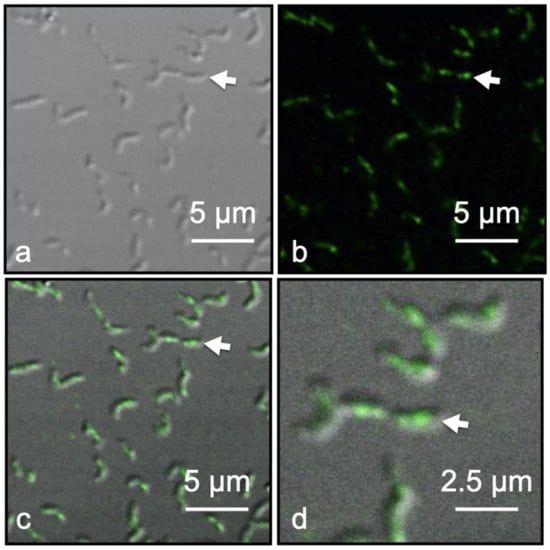
Figure 4.
Fluorescent labeled Magnetospirillum magneticum AMB-1 cells using Anti-Mms13 (1:400) as the primary antibody and goat anti-rabbit Dylight 488 (1:100) as the secondary antibody. (a) Nomarski image of M. magneticum AMB-1 cells. (b) The same view field as (a) showing only fluorescent image. (c) Merged Nomarski and fluorescent images showing the outline of M. magneticum AMB-1 cells with green fluorescent tag along the major axis of the cells co-localized with the magnetosome chain. (d) Enlargement of view field shown in previous three panels. White arrow in each panel identifies the same cell. Scale bars provided in each panel.
4. Discussion
As shown in Table 1, previous reports have employed different methods to help define the localization of Mms13 and its homologs, MamC and Mam12, in four MTB species belonging to the class Alphaproteobacteria. In M. gryphiswaldense and M. magneticum AMB-1, the localization of MamC and Mms13, was examined using an inducible expression plasmid to overexpress recombinant GFP-MamC fusion protein in MTB and examine cells and purified magnetosomes using fluorescence microscopy and spectroscopy [28,29]. Similar studies have not yet been employed on Mam12 in M. magnetotacticum MS-1 or on MamC in M. marinus MC-1. In other reports, immunogold labeling TEM of whole cells and purified magnetosomes was done with M. magnetotacticum MS-1 and M. marinus MC-1 [18,30]. Immunogold labeling TEM has not yet been conducted for MamC in M. gryphiswaldense or Mms13 in M. magneticum AMB-1.

Table 1.
Studies conducted to examine subcellular localization of homologous proteins MamC, Mam12 and Mms13 in MTB belonging to Alphaproteobacteria [12,13,15,16,18,28,29,30,31,32,35,36,37,38,39,40].
In addition to microscopy, biochemical and proteomic analysis of the magnetosome membrane has been conducted on MamC, Mam12 and Mms13 in all four magnetotactic Alphaproteobacteria (Table 1) [12,13,15,16,18,28,29,30,31,32,35,36,37,38,39,40]. MamC is a highly conserved protein and one of the most abundant magnetosome-associated proteins [14,30]. When extracted from the cell, MamC, Mam12 and Mms13 have been found to associate with the MM fraction [12,13,15,16,18]. In-vivo genetic studies demonstrated that MamC regulates the size of magnetosome crystals in M. gryphiswaldense [14]. In-vitro biomineralization experiments using recombinant MamC demonstrated that MamC plays a role in controlling the size and shape of magnetite crystals in Magnetococcus marinus strain MC-1 [21,24,26].
By establishing the location of MamC, Mam12, and Mms13 within a bacterium and relative to the magnetosome chain, we gain significant insight into if and how these proteins function to control the mineralization of Fe3O4 nanocrystals within the magnetosome. Consequently, this information is important for the in-vitro synthesis of Fe3O4 nanocrystals in the presence of purified proteins [15]. Moreover, establishing the subcellular localization of these three homologous proteins in members of the magnetotactic Alphaproteobacteria group allows for a better understanding of protein function and helps to identify shared biomineralization pathways among MTB (Table 1). Using immunogold stained ultrathin sections of bacterial cells and purified magnetosomes, native Mam12 was found to be localized to the magnetosome membrane of M. magnetotacticum [18]. Recombinant MamC-GFP was detected within the magnetosome chain of M. gryphiswaldense using immunoblotting and fluorescence microscopy [28]. In addition, finally, immunogold labeling combined with TEM, showed that MamC was localized exclusively to the magnetosome chain of M. marinus strain MC-1 [30].
Previous studies, as summarized in Table 1, provide compelling, but not conclusive evidence, for the localization of Mms13 in M. magneticum AMB-1. Arakaki et al., 2003 conducted experiments with M. magneticum AMB-1, where they extracted proteins tightly bound to magnetic particles using 1% SDS (sodium dodecyl sulfate), separated them by two-dimensional gel electrophoresis and sequenced the stained proteins by Edman degradation [15]. They identified four proteins that were tightly bound to the purified magnetic particles including Mms13 [15]. Tanaka et al., 2006 conducted similar proteomic analysis of MM fractions isolated from M. magneticum AMB-1 [16]. They identified a total of 78 proteins associated with the MM fraction, including Mms13 [16].
Yoshino and Matsunaga, 2006 [31], Yoshino et al., 2010 [29], and Yoshino et al., 2021 [36] used a tetracycline-controlled inducible expression plasmid for M. magneticum AMB-1, using Mms13 as an anchor protein, to overexpress heterologous proteins, including green fluorescent protein (GFP, molecular mass ~27 kDa), on the surface of magnetic particles. The plasmid containing mms13-GFP was overexpressed in M. magneticum AMB-1 and magnetic particles were extracted and collected from lysed M. magneticum AMB-1 cells using a neodymium-boron (Nd-B) magnet [29,31]. Yoshino et al., 2010 [29] used fluorescence spectroscopy to demonstrate that magnetosomes extracted from M. magneticum AMB-1 were coated with the recombinant Mms13-GFP fusion protein [29]. M. magneticum AMB-1 cells transformed with a tetracycline-inducible Mms13-GFP plasmid were also observed by fluorescence microscopy in the presence of anhydrotetracycline hydrochloride [29]. Yoshino and Matsunaga, 2006 [31] and Yoshino et al., 2010 [29] expressed two other recombinant Mms-13 fusion proteins (i.e., Mms13-Luciferase [31] and Mms13-FLAG-CD81 [29]) in M. magneticum AMB-1. They extracted the proteins from purified magnetic particles using 1% SDS and determined that both Mms13-Luciferase and Mms13-FLAG-CD81 fusion proteins were found attached to the magnetic particles using Western blotting and anti-luciferase antibody or anti-FLAG antibody [29,31]. Quinlan et al., 2011 [32] also developed an inducible plasmid expression system that used IPTG (isopropylthio-β-galactoside) to overexpress recombinant Mms13-GFP in M. magneticum AMB-1. When the mms13-GFP was overexpressed in M. magneticum AMB-1 and visualized by fluorescence microscopy, green fluorescence was observed along the length of the cell [32].
The previous studies [15,16,29,31,32,36] on Mms13 expression and localization in M. magneticum AMB-1 (Table 1) focused on two types of experiments: (1) the use of fluorescence microscopy and spectroscopy or enzyme activity to examine plasmid-overexpressed recombinant fusion proteins (i.e., Mms13-GFP, Mms13-FLAG-CD81, Mms13-Luciferase, Mms13-TSHR) and (2) cellular fractionation, purification of MM proteins, followed by amino acid sequencing or enzyme assays. While important, these experiments have limitations. For example, previous studies have demonstrated that fusion proteins can perturb a native protein’s activity, expression, function, and localization [41,42,43,44]. In Yoshino et al., 2010 [29], Yoshino and Matsunaga, 2006 [31], Quinlan et al., 2011 [32] and Yoshino et al., 2021 [36] an inducible plasmid expression system was used in M. magneticum AMB-1 to overexpress comparatively large non-native proteins (i.e., GFP is ~27 kDa; CD81 is ~25 kDa; Luciferase is ~61kDa; TSHR, thyroid-stimulating hormone receptor is ~87 kDa) fused to the much small native Mms13 protein (i.e., Mms13 is ~12 kDa). The considerably larger size of the non-native protein (e.g., GFP is twice as large as Mms13, Luciferase is five-times larger than Mms13, TSHR is seven-times larger than Mms13) has steric consequences for protein folding, function and targeting that could alter expression and/or localization of the native protein [41,42,43,44]. In addition, the resolution limit of conventional fluorescence microscopy is approximately 250 nm due to its dependance on the wavelength of the excitation light and microscope optics. This presents a challenge when imaging magnetosome particles, which are approximately 2–6 times smaller than the resolution offered by fluorescence microscopy. The spatial resolution of the M. magneticum AMB-1 fluorescence images provided in Yoshino et al., 2010 [29] and Quinlan et al., 2011 [32] was not sufficient to resolve magnetosome particles contained within the bacteria. TEM, on the other hand, which was conducted as part of our study on M. magneticum AMB-1, is ideally suited for obtaining the nm resolution required to image individual magnetosomes and Mms13 molecules within a bacterium.
In the present study, we used Western blot analysis, LC-MS/MS protein sequencing, immunofluorescence microscopy, and immunogold labeling TEM to establish the subcellular location of native Mms13 in M. magneticum AMB-1. Western blot analysis showed that a polypeptide with a molecular mass similar to Mms13 (~14.5 kDa) was present in the MM fraction of M. magneticum AMB-1 (Figure 2b, lane 3). This ~14.5 kDa polypeptide was not detected in either the soluble fraction or cytoplasmic membrane fraction (Figure 2b, lanes 1 and 2). To determine the identity of this protein, LC-MS/MS peptide sequence analysis was performed on the protein band observed in the MM fraction (Figure 2b, lane 3, black arrow). LC-MS/MS analysis matched four peptides to Mms13 (Figure 2c). Peptide 1 was from the N-terminal end of Mms13 and peptides 2–4 were from the C-terminal end of Mms13 (Figure 2c). These results show that the mature Mms13 protein in M. magneticum AMB-1 contains at the very least amino acid residues 33–145 (Figure 2c). Thus, the mature Mms13 protein in M. magneticum AMB-1 contains both its hydrophobic, N-terminal putative magnetosome membrane domain and its hydrophilic, C-terminal putative catalytic domain. These results are consistent with Arakaki et al., 2003 [15] and Tanaka et al., 2006 [16], which showed that Mms13 could be extracted from purified magnetosomes.
Western blot analysis and LC-MS/MS protein identification agreed with TEM analysis of M. magneticum AMB-1 (Figure 3). Immunogold electron microscopy showed Mms13 labeling along a bacterium’s magnetosome chain (Figure 3a–c). Nanogold particles were observed to be directly touching the Fe3O4 particles, within the magnetosome membrane surrounding the Fe3O4 particles or very close (<10 nm) to the magnetosome chain (Figure 3a–c). In many instances, multiple nanogold particles were found to be attached to a single magnetosome crystal (Figure 3d–f). For example, in Figure 3f, we see a binding ratio of 3:1 (gold particle to magnetosome particle), with all nanogold particles touching or <10 nm from a magnetosome. These observations indicated that there was a strong affinity of the Anti-Mms13-nanogold-label for the magnetosome chain.
Control experiments were also performed using 5% pre-immune serum and no primary antibody to confirm the specificity of Anti-Mms13 antibody for Mms13 (Figure 3h). The pre-immune serum does not contain antibodies for Mms13 and thus nanogold particles should not be detected in TEM images using pre-immune serum. Occasionally a small number of nanogold particles were observed outside the bacterial cell; however, no nanogold labeling occurred within or near the magnetosome chain (Figure 3g,h). This confirmed that the results were not due to background labeling by the secondary antibody.
Fluorescence microscopy was used to confirm the TEM analysis, which showed Mms13 localized to the MM and magnetite crystal surface. CLSM revealed fluorescently labeled Mms13 molecules along the major axis of the bacterium (Figure 4). Green fluorescence DyLight 488 molecules were observed co-localized with the magnetosome chain (Figure 4). No fluorescent labeling was observed outside the bacteria or distal to the magnetosome chain (Figure 4). Thus, our fluorescence measurements were consistent with TEM analysis that showed native Mms13 molecules localized to the magnetosome chain of M. magneticum AMB-1. These results were also consistent with our biochemical analyses that used LC-MS/MS and Western blotting to identify native Mms13 molecules within the MM fraction M. magneticum AMB-1.
5. Conclusions
Defining the localization of magnetosome proteins in MTB is a necessary step in determining their function(s) in magnetosome biomineralization. By understanding the mineralization process of Fe3O4 crystals in all MTB we can better understand the evolution of magnetotaxis and learn to reproduce the synthesis of Fe3O4 nanocrystals in-vitro. Such information could be explored in attempts to obtain tailored Fe3O4 magnets with specific morphology and size. By combining the results presented here with previous [12,13,14,15,16,18,21,24,26,27,28,29,30,31,32,35,36,37,38,39,40], we can definitively conclude that homologous proteins MamC, Mam12, and Mms13 are localized to the magnetosome chain of all magnetotactic Alphaproteobacteria where the proteins likely function in the biomineralization of Fe3O4 nanocrystals.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/cryst11080874/s1, Figure S1. This Western blot is the same Western blot shown in Figure 2b, except that this image has not been modified (e.g., contrast, exposure, brightness) using image processing software (i.e., Adobe Photoshop). Lanes correspond to 10 μg soluble proteins (lane 1), 10 μg of cytoplasmic membrane proteins (lane 2), and 10 μg of magnetosome membrane proteins (lane 3). For comparison, molecular weight markers (lane M) are provided with the molecular masses (in kilodaltons) of protein standards indicated using numbers down the left-side of the gel. The black arrow shows protein band corresponding to Mms13.
Author Contributions
Conceptualization, B.H.L., S.K.L., D.A.B. and C.J.-L.; methodology, Z.O., C.V.-T. and L.P.-G.; investigation, Z.O., C.V.-T., E.M., N.N.C.-I. and L.P.-G.; resources, B.H.L., S.K.L., D.A.B. and C.J.-L.; writing—original draft preparation, B.H.L., S.K.L., D.A.B., E.M. and C.J.-L.; writing—review and editing, B.H.L., N.N.C.-I. and S.K.L.; supervision, B.H.L., S.K.L., D.A.B. and C.J.-L.; project administration, B.H.L., S.K.L., D.A.B. and C.J.-L.; funding acquisition, B.H.L., S.K.L., D.A.B. and C.J.-L. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by U.S. National Science Foundation, grant number EAR-2038207 and EAR-1423939; and Ministerio de Economía y Competitividad, SPAIN and Fondo Europeo de Desarrollo Regional, FEDER grant numbers CGL2010-18274 and CGL2013-46612.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data sharing not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Gorby, Y.A.; Beveridge, T.J.; Blakemore, R. Characterization of the bacterial magnetosome membrane. J. Bacteriol. 1988, 170, 834–841. [Google Scholar] [CrossRef] [Green Version]
- Bazylinski, D.A.; Frankel, R.B. Magnetosome formation in prokaryotes. Nat. Rev. Microbiol. 2004, 2, 217–230. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lower, B.H.; Bazylinski, D.A. The bacterial magnetosome: A unique prokaryotic organelle. J. Mol. Microbiol. Biotechnol. 2013, 23, 63–80. [Google Scholar] [CrossRef] [PubMed]
- Uebe, R.; Schüler, D. Magnetosome biogenesis in magnetotactic bacteria. Nat. Rev. Microbiol. 2016, 14, 621–637. [Google Scholar] [CrossRef]
- Komeili, A.; Li, Z.; Newman, D.K.; Jensen, G.I. Magnetosomes are cell membrane invaginations organized by the actin-like protein MamK. Science 2006, 311, 242–245. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schüler, D. Genetics and cell biology of magnetosome formation in magnetotactic bacteria. FEMS Microbiol. Rev. 2008, 32, 654–672. [Google Scholar] [CrossRef] [Green Version]
- Komeili, A. Molecular mechanisms of compartmentalization and biomineralization in magnetotactic bacteria. FEMS Microbiol. Rev. 2012, 36, 232–255. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schüler, D.; Baeuerlein, E. Dynamics of iron uptake and Fe3O4 biomineralization during aerobic and microaerobic growth of Magnetospirillum gryphiswaldense. J. Bacteriol. 1998, 180, 159–162. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schüler, D. Molecular analysis of a subcellular compartment: The magnetosome membrane in Magnetospirillum gryphiswaldense. Arch. Microbiol. 2004, 181, 1–7. [Google Scholar] [CrossRef]
- Matsunaga, T.; Sakaguchi, T. Molecular mechanism of magnet formation in bacteria. J. Biosci. Bioeng. 2000, 90, 1–13. [Google Scholar] [CrossRef]
- Arakaki, A.; Tanaka, M.; Matsunaga, T. Molecular mechanism of magnetic crystal formation in magnetotactic bacteria. In Biologic Magnetic Materials and Applications; Matsunaga, T., Tanaka, T., Kisailus, D., Eds.; Springer: Singapore, 2018; pp. 23–51. [Google Scholar]
- Grünberg, K.; Wawer, C.; Tebo, B.M.; Schüler, D. A large cluster encoding several magnetosome proteins is conserved in different species of magnetotactic bacteria. Appl. Environ. Microbiol. 2001, 67, 4573–4582. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grünberg, K.; Müller, E.-C.; Otto, A.; Reszka, R.; Linder, D.; Kube, M.; Reinhardt, R.; Schüler, D. Biochemical and proteomic analysis of the magnetosome membrane in Magnetospirillum gryphiswaldense. Appl. Environ. Microbiol. 2004, 70, 1040–1050. [Google Scholar] [CrossRef] [Green Version]
- Scheffel, A.; Gärdes, A.; Grünberg, K.; Wanner, G.; Schüler, D. The major magnetosome proteins MamGFDC are not essential for magnetite biomineralization in Magnetospirillum gryphiswaldense, but regulate the size of magnetosome crystals. J. Bacteriol. 2008, 190, 377–386. [Google Scholar] [CrossRef] [Green Version]
- Arakaki, A.; Webb, J.; Matsunaga, T. A novel protein tightly bound to bacterial magnetic particles in Magnetospirillum magneticum strain AMB-1. J. Biol. Chem. 2003, 278, 8745–8750. [Google Scholar] [CrossRef] [Green Version]
- Tanaka, M.; Okamura, Y.; Arakaki, A.; Tanaka, T.; Takeyama, H.; Matsunaga, T. Origin of magnetosome membrane: Proteomic analysis of magnetosome membrane and comparison with cytoplasmic membrane. Proteomics 2006, 6, 5234–5247. [Google Scholar] [CrossRef] [PubMed]
- Kashyap, S.; Woehl, T.J.; Liu, X.; Mallapragada, S.K.; Prozorov, T.; Frankel, R.B. Nucleation of iron oxide nanoparticles mediated by Mms6 protein in situ. ACS Nano 2014, 8, 9097–9106. [Google Scholar] [CrossRef] [PubMed]
- Taoka, A.; Asada, R.; Sasaki, H.; Anzawa, K.; Wu, L.F.; Fukumori, Y. Spatial localizations of Mam22 and Mam12 in the magnetosomes of Magnetospirillum magnetotacticum. J. Bacteriol. 2006, 188, 3805–3812. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Toro-Nahuelpan, M.; Giacomelli, G.; Raschdorf, O.; Borg, S.; Plitzko, J.M.; Bramkamp, M.; Schuler, D.; Müller, F.-D. MamY is a membrane-bound protein that aligns magnetosomes and the motility axis of helical magnetotactic bacteria. Nat. Microb. 2019, 4, 1978–1989. [Google Scholar] [CrossRef] [PubMed]
- Ren, E.; Lei, Z.; Wang, J.; Zhang, Y.; Liu, G. Magnetosome modification: From bio-nano engineering toward nanomedicine. Adv. Therap. 2018, 1, 1800080. [Google Scholar] [CrossRef]
- Valverde-Tercedor, C.; Montalbán-Lopez, M.; Perez-Gonzalez, T.; Sanchez-Quesada, M.S.; Prozorov, T.; Pineda-Molina, E.; Fernandez-Vivas, M.A.; Rodriguez-Navarro, A.B.; Trubitsyn, D.; Bazylinski, D.A.; et al. Size control of in vitro synthesized magnetite crystals by the MamC protein of Magnetococcus marinus strain MC-1. Appl. Microbiol. Biotechnol. 2015, 99, 5109–5121. [Google Scholar] [CrossRef]
- Matsunaga, T.; Okamura, Y.; Fukuda, Y.; Wahyudi, A.T.; Murase, Y.; Takeyama, H. Complete genome sequence of the facultative anaerobic magnetotactic bacterium Magnetospirillum sp. strain AMB-1. DNA Res. 2005, 12, 157–166. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pósfai, M.; Lefèvre, C.T.; Trubitsyn, D.; Bazylinski, D.A.; Frankel, R.B. Phylogenetic significance of composition and crystal morphology of magnetosome minerals. Front. Microbiol. 2013, 4, 1–15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nudelman, H.; Valverde-Tercedor, C.; Kolusheva, S.; Perez-Gonzalez, T.; Widdrat, M.; Grimberg, N.; Levi, H.; Nelkenbaum, O.; Davidov, G.; Faivre, D.; et al. Structure-function studies of the magnetite-biomineralizing magnetosome-associated protein MamC. J. Struct. Biol. 2016, 194, 244–252. [Google Scholar] [CrossRef]
- Rawlings, A.E.; Somner, L.A.; Fitzpatrick-Milton, M.; Roebuck, T.P.; Gwyn, C.; Liravi, P.; Seville, V.; Neal, T.J.; Mykhaylyk, O.O.; Baldwin, S.A.; et al. Artificial coiled coil biomineralization protein for the synthesis of magnetic nanoparticles. Nat. Commun. 2019, 10, 2873. [Google Scholar] [CrossRef] [PubMed]
- Jabalera, Y.; Casares Atienza, S.; Fernández-Vivas, A.; Peigneux, A.; Azuaga Fortes, A.I.; Jimenez-Lopez, C. Protein conservation method affects MamC-mediated biomineralization of magnetic nanoparticles. Cryst. Growth Des. 2019, 19, 1064–1071. [Google Scholar] [CrossRef]
- Nudelman, H.; Perez-Gonzalez, T.; Kolushiva, S.; Widdrat, M.; Reichel, V.; Peigneux, A.; Davidov, G.; Bitton, R.; Faivre, D.; Jimenez-Lopez, C.; et al. The importance of the helical structure of a MamC-derived magnetite-interacting peptide for its function in magnetite formation. Acta Crystallogr. D Struct. Biol. 2018, 74, 10–20. [Google Scholar] [CrossRef]
- Lang, C.; Schüler, D. Expression of green fluorescent protein fused to magnetosome proteins in microaerophilic magnetotactic bacteria. Appl. Environ. Microbiol. 2008, 74, 4944–4953. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yoshino, T.; Shimojo, A.; Maeda, T.; Matsunaga, T. Inducible expression of transmembrane proteins on bacterial magnetic particles in Magnetospirillum magneticum AMB-1. Appl. Environ. Microbiol. 2010, 76, 1152–1157. [Google Scholar] [CrossRef] [Green Version]
- Valverde-Tercedor, C.; Abadía-Molina, F.; Martinez-Bueno, M.; Pineda-Molina, E.; Chen, L.; Oestreicher, Z.; Lower, B.H.; Lower, S.K.; Bazylinski, D.A.; Jimenez-Lopez, C. Subcellular localization of the magnetosome protein MamC in the marine magnetotactic bacterium Magnetococcus marinus strain MC-1 using immunoelectron microscopy. Arch. Microbiol. 2014, 196, 481–488. [Google Scholar] [CrossRef]
- Yoshino, T.; Matsunaga, T. Efficient and stable display of functional proteins on bacterial magnetic particles using Mms13 as a novel anchor molecule. Appl. Environ. Microbiol. 2006, 72, 465–471. [Google Scholar] [CrossRef] [Green Version]
- Quinlan, A.; Murat, D.; Vali, H.; Komeili, A. The HtrA/DegP family protease MamE is a bifunctional protein with roles in magnetosome protein localization and magnetite biomineralization. Mol. Microbiol. 2011, 80, 1075–1087. [Google Scholar] [CrossRef] [Green Version]
- Oestreicher, Z.; Mumper, E.; Gassman, C.; Bazylinski, D.A.; Lower, S.K.; Lower, B.H. Spatial localization of Mms6 during biomineralization of Fe3O4 nanocrystals in Magnetospirillum magneticum AMB-1. J. Mater. Res. 2016, 31, 527–535. [Google Scholar] [CrossRef]
- Oestreicher, Z.; Valverde-Tercedor, C.; Chen, L.; Jimenez-Lopez, C.; Bazylinski, D.A.; Casillas-Ituarte, N.N.; Lower, S.K.; Lower, B.H. Magnetosmoes and magnetite crystals produced by magnetotactic bacteria as resolved by atomic force microscopy and transmission electron microscopy. Micron 2012, 43, 1331–1335. [Google Scholar] [CrossRef] [PubMed]
- Raschdorf, O.; Bonn, F.; Zeytuni, N.; Zarivach, R.; Becher, D.; Schüler, D. A quantitative assessment of the membrane-integral sub-proteome of a bacterial magnetic organelle. J. Proteom. 2018, 172, 89–99. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yoshino, T.; Tayama, S.; Maeda, Y.; Fujimoto, K.; Ota, S.; Wake, S.; Kisailus, D.; Tanaka, T. Magnetosome membrane engineering to improve G protein-coupled receptor activities in the magnetosome display system. Metabol. Eng. 2021, 67, 125–132. [Google Scholar] [CrossRef] [PubMed]
- Mickoleit, F.; Lanzloth, C.; Schüler, D. A Versatile Toolkit for Controllable and Highly Selective Multifunctionalization of Bacterial Magnetic Nanoparticles. Small 2020, 16, 1906922. [Google Scholar] [CrossRef]
- Ohuchi, S.; Schüler, D. In Vivo Display of a Multisubunit Enzyme Complex on Biogenic Magnetic Nanoparticles. Appl. Environ. Microbiol. 2009, 75, 7734–7738. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Uebe, R.; Junge, K.; Henn, V.; Poxieitner, G.; Katzmann, E.; Plitzko, J.M.; Zarivach, R.; Kasama, T.; Wanner, G.; Pósfai, M.; et al. The cation diffusion facilitator proteins MamB and MamM of Magnetospirillum gryphiswaldense have distinct and complex functions, and are involved in magnetite biomineralization and magnetosome membrane assembly. Mol. Microbiol. 2011, 82, 818–835. [Google Scholar] [CrossRef]
- Lohße, A.; Uilrich, S.; Katzmann, E.; Borg, S.; Wanner, G.; Richter, M.; Boigt, B.; Schweder, T.; Schüler, D. Functional Analysis of the Magnetosome Island in Magnetospirillum gryphiswaldense: The mamAB Operon Is Sufficient for Magnetite Biomineralization. PLoS ONE 2011, 6, e25561. [Google Scholar] [CrossRef] [Green Version]
- Heo, M.; Nord, A.L.; Chamousset, D.; van Rijn, E.; Beaumont, H.J.E.; Pedaci, F. Impact of fluorescent protein fusion on the bacterial flagellar motor. Sci. Rep. 2017, 7, 12583. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marshall, A.P.; Shirley, J.D.; Carlson, E.E. Enzyme-targeted fluorescent small-molecule probes for bacterial imaging. Curr. Opin. Chem. Biol. 2020, 57, 155–165. [Google Scholar] [CrossRef] [PubMed]
- Snapp, E. Design and use of fluorescent fusion proteins in cell biology. Curr. Protoc. Cell Biol. 2005, 27, 21–24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Snapp, E.L. Fluorescent proteins: A cell biologist’s user guide. Trends Cell Biol. 2009, 19, 649–655. [Google Scholar] [CrossRef] [PubMed] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).