tRNA Fusion to Streamline RNA Structure Determination: Case Studies in Probing Aminoacyl-tRNA Sensing Mechanisms by the T-Box Riboswitch
Abstract
:1. Introduction
2. Materials and Methods
3. Results
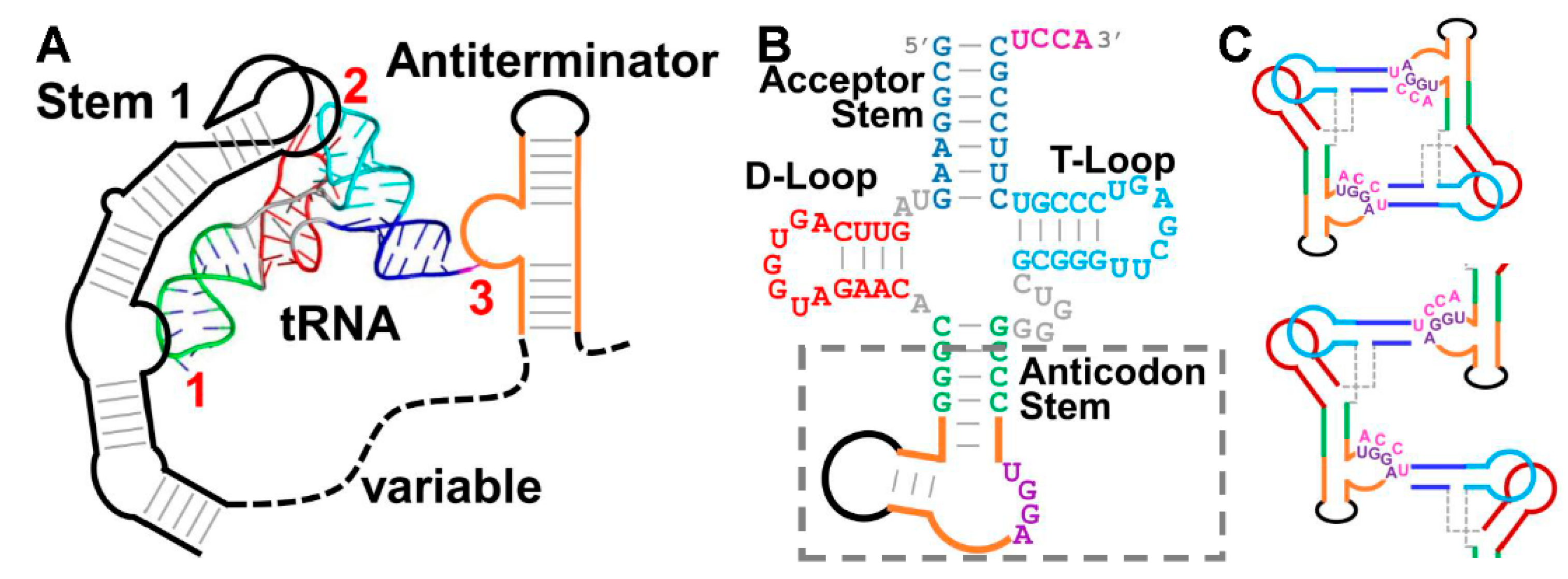
3.1. Challenges in Crystallizing the tRNA/T-Box Antiterminator Complex Inspired the Design of tRNA–Antiterminator Fusions
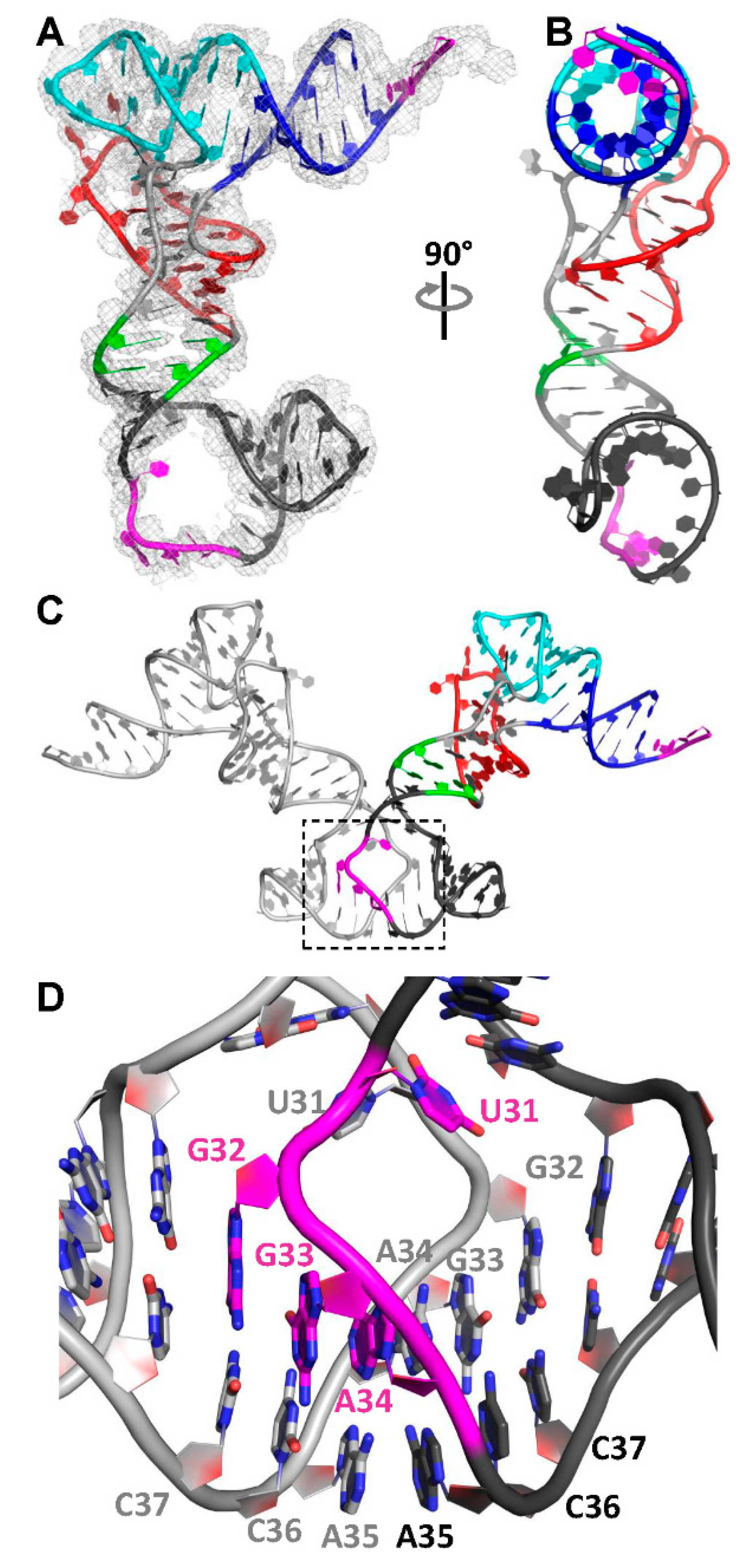
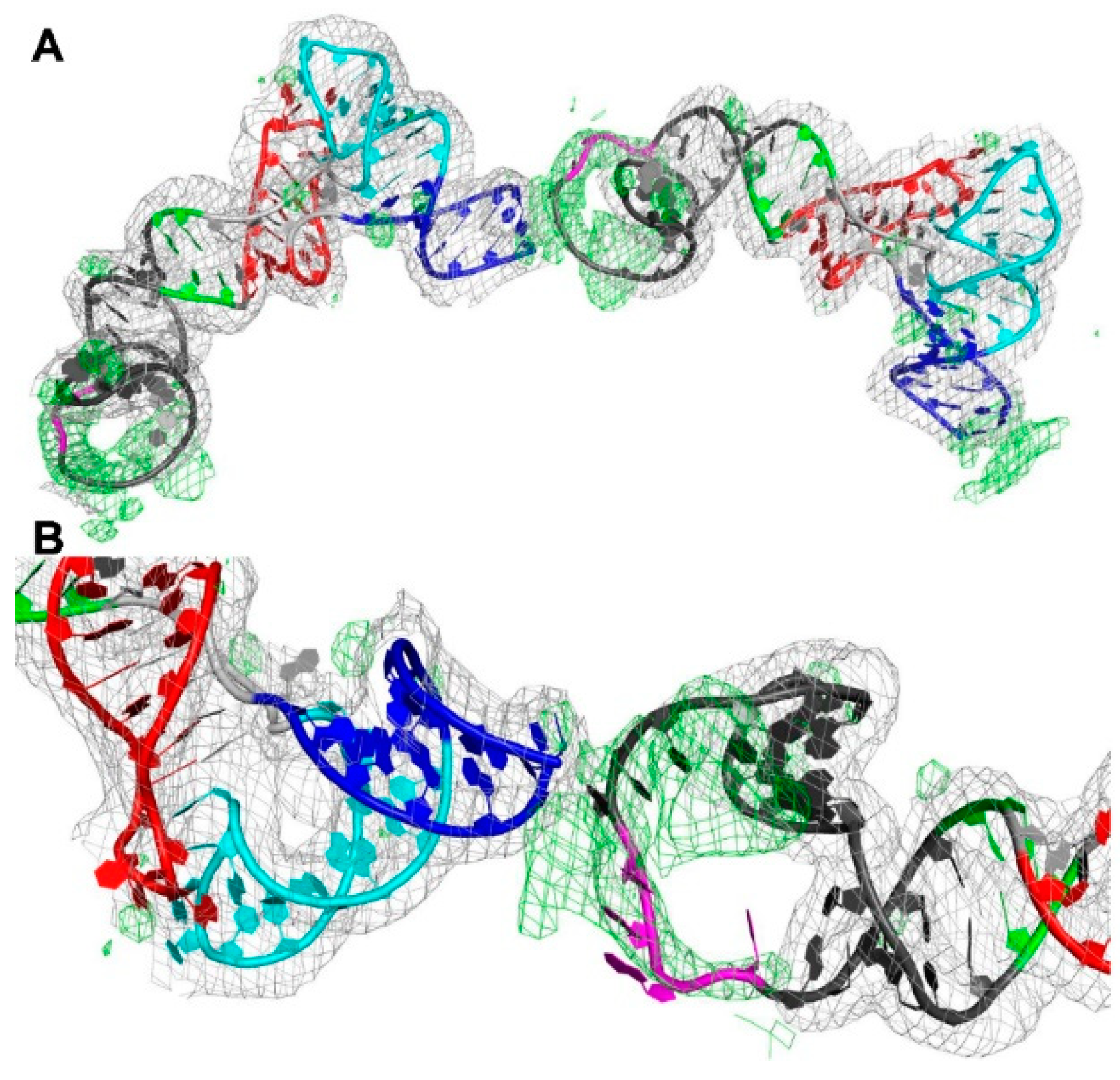
3.2. The Structure of a Minimal tRNA–Antiterminator Fusion (Construct #4)
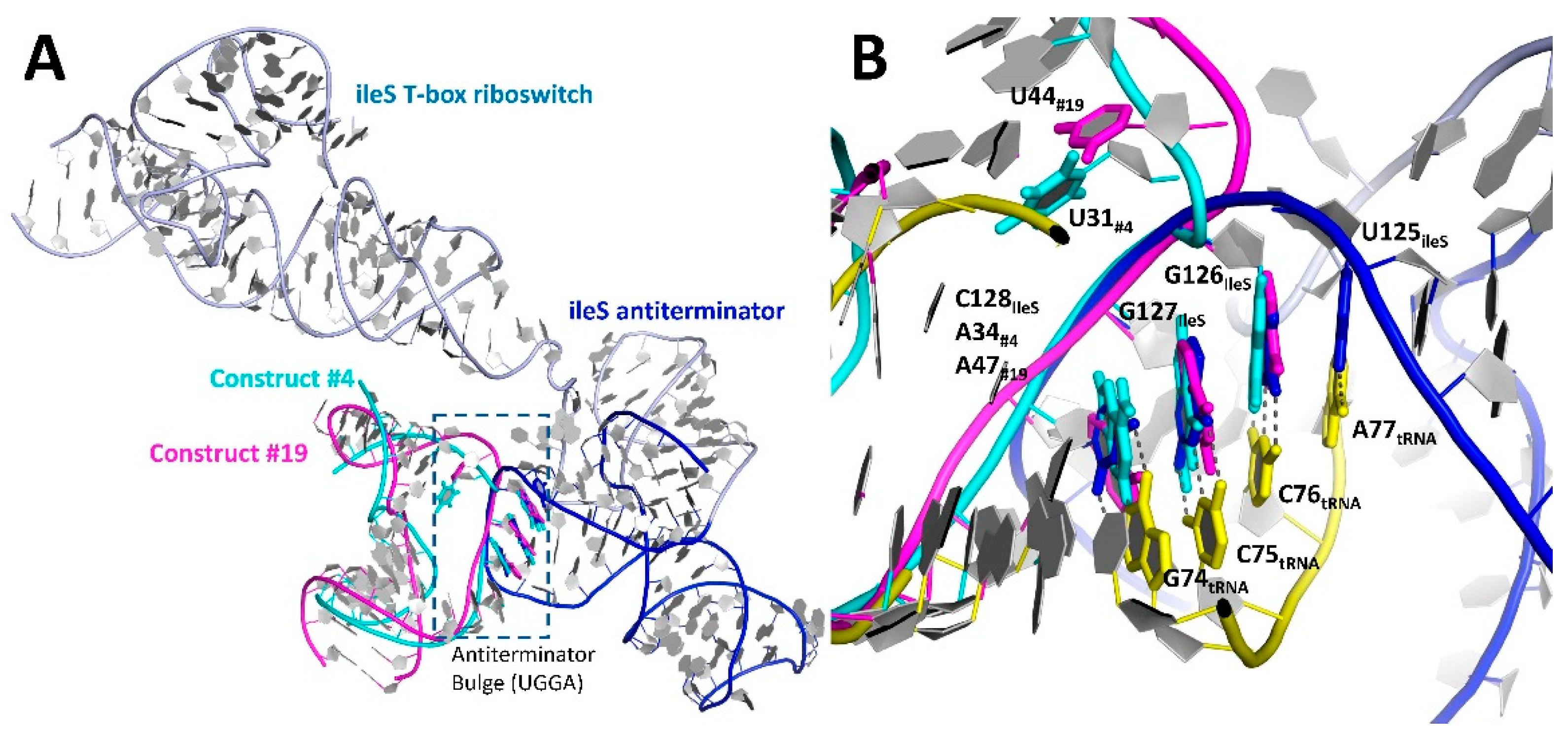
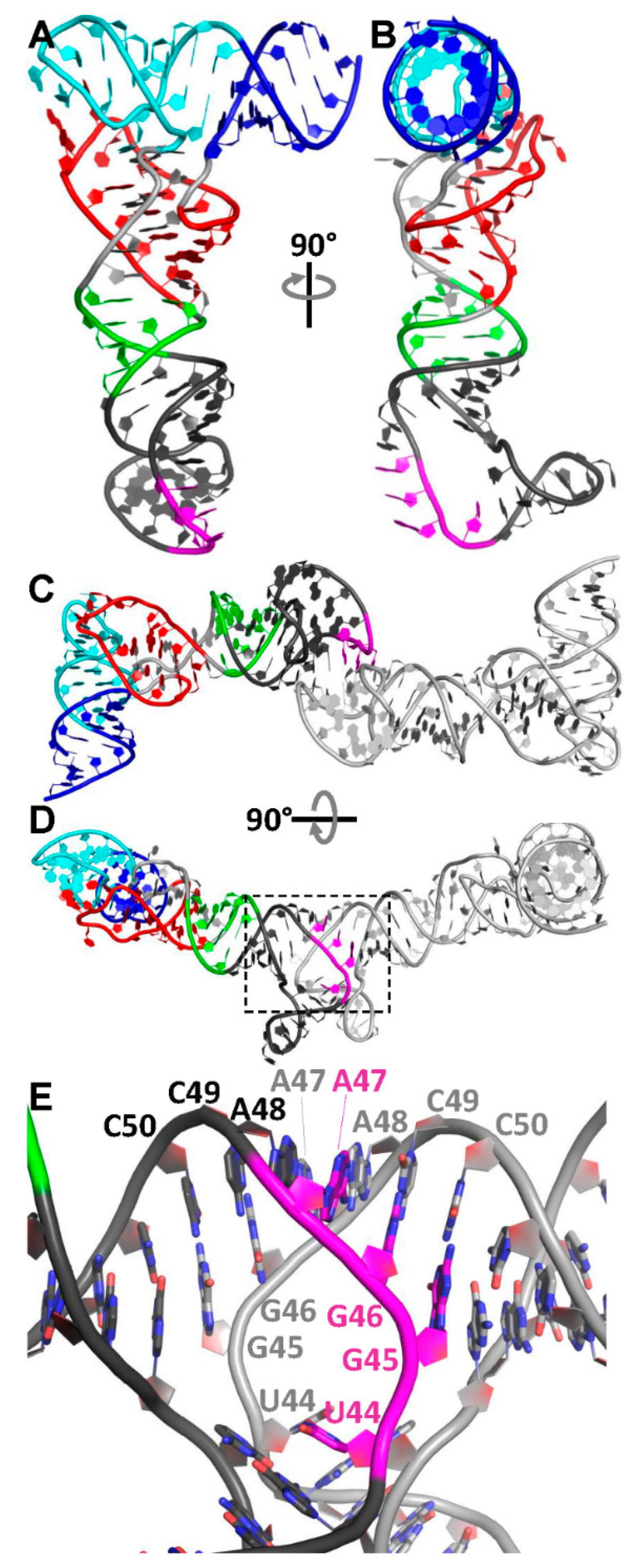
3.3. Relocation of the Antiterminator Bulge to the 3′ Side of the Stem-Loop
3.4. Phasing of a Low-Resolution Interaction between the tRNA Acceptor and Antiterminator
3.5. The tRNA Scaffold Is a Powerful Tool for RNA Structure Determination
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- McCown, P.J.; Corbino, K.A.; Stav, S.; Sherlock, M.E.; Breaker, R.R. Riboswitch diversity and distribution. RNA 2017, 23, 995–1011. [Google Scholar] [CrossRef] [PubMed]
- Garst, A.D.; Edwards, A.L.; Batey, R.T. Riboswitches: Structures and Mechanisms. Cold Spring Harb. Perspect. Biol. 2011, 3, a003533. [Google Scholar] [CrossRef] [PubMed]
- Holcomb, J.; Spellmon, N.; Zhang, Y.; Doughan, M.; Li, C.; Yang, Z. Protein crystallization: Eluding the bottleneck of X-ray crystallography. AIMS Biophys. 2017, 4, 557–575. [Google Scholar] [CrossRef] [PubMed]
- Ke, A.; Doudna, J.A. Crystallization of RNA and RNA-protein complexes. Methods 2004, 34, 408–414. [Google Scholar] [CrossRef]
- Grigg, J.C.; Ke, A. Structures of Large RNAs and RNA-Protein Complexes: Toward Structure Determination of Riboswitches. Methods Enzymol. 2015, 558, 213–232. [Google Scholar] [CrossRef]
- Zhang, J.; Ferré-D’Amaré, A.R. New molecular engineering approaches for crystallographic studies of large RNAs. Curr. Opin. Struct. Biol. 2014, 26, 9–15. [Google Scholar] [CrossRef] [Green Version]
- Peselis, A.; Serganov, A. ykkC riboswitches employ an add-on helix to adjust specificity for polyanionic ligands. Nat. Chem. Biol. 2018, 14, 887–894. [Google Scholar] [CrossRef]
- Ferré-D’Amaré, A.R. Use of the U1A protein to facilitate crystallization and structure determination of large RNAs. Methods Mol. Biol. 2016, 1320, 67–76. [Google Scholar] [CrossRef] [Green Version]
- Ferré-D’Amaré, A.R.; Zhou, K.; Doudna, J.A. Crystal structure of a hepatitis delta virus ribozyme. Nature 1998, 395, 567. [Google Scholar] [CrossRef]
- Baird, N.J.; Zhang, J.; Hamma, T.; Ferré-D’Amaré, A.R. YbxF and YlxQ are bacterial homologs of L7Ae and bind K-turns but not K-loops. RNA 2012, 18, 759–770. [Google Scholar] [CrossRef] [Green Version]
- Huang, L.; Lilley, D.M.J. The molecular recognition of kink-turn structure by the L7Ae class of proteins. RNA 2013, 19, 1703–1710. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, J.; Ferré-D’Amaré, A.R. Co-crystal structure of a T-box riboswitch stem I domain in complex with its cognate tRNA. Nature 2013, 500, 363–366. [Google Scholar] [CrossRef] [PubMed]
- Ravindran, P.P.; Heroux, A.; Ye, J.D. Improvement of the crystallizability and expression of an RNA crystallization chaperone. J. Biochem. 2011, 150, 535–543. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bailey, L.J.; Sheehy, K.M.; Dominik, P.K.; Liang, W.G.; Rui, H.; Clark, M.; Jaskolowski, M.; Kim, Y.; Deneka, D.; Tang, W.J.; et al. Locking the Elbow: Improved Antibody Fab Fragments as Chaperones for Structure Determination. J. Mol. Biol. 2018, 430, 337–347. [Google Scholar] [CrossRef]
- Koirala, D.; Shelke, S.A.; Dupont, M.; Ruiz, S.; DasGupta, S.; Bailey, L.J.; Benner, S.A.; Piccirilli, J.A. Affinity maturation of a portable Fab-RNA module for chaperone-assisted RNA crystallography. Nucleic Acids Res. 2018, 46, 2624–2635. [Google Scholar] [CrossRef] [Green Version]
- Sherman, E.; Archer, J.; Ye, J.D. Fab Chaperone-Assisted RNA Crystallography (Fab CARC). Methods Mol. Biol. 2016, 1320, 77–109. [Google Scholar] [CrossRef]
- Ye, J.D.; Tereshko, V.; Frederiksen, J.K.; Koide, A.; Fellouse, F.A.; Sidhu, S.S.; Koide, S.; Kossiakoff, A.A.; Piccirilli, J.A. Synthetic antibodies for specific recognition and crystallization of structured RNA. Proc. Natl. Acad. Sci. USA 2008, 105, 82–87. [Google Scholar] [CrossRef] [Green Version]
- Koldobskaya, Y.; Duguid, E.M.; Shechner, D.M.; Suslov, N.B.; Ye, J.; Sidhu, S.S.; Bartel, D.P.; Koide, S.; Kossiakoff, A.A.; Piccirilli, J.A. A portable RNA sequence whose recognition by a synthetic antibody facilitates structural determination. Nat. Struct. Mol. Biol. 2011, 18, 100–106. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.; Ferré-D’Amaré, A.R. Dramatic improvement of crystals of large RNAs by cation replacement and dehydration. Structure 2014, 22, 1363–1371. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.; Ferré-D’Amaré, A.R. Post-crystallization Improvement of RNA Crystal Diffraction Quality. Methods Mol. Biol. 2015, 1316, 13–24. [Google Scholar] [CrossRef] [Green Version]
- Stura, E.A.; Taussig, M.J.; Sutton, B.J.; Duquerroy, S.; Bressanelli, S.; Minson, A.C.; Rey, F.A. Scaffolds for protein crystallisation. Acta Crystallogr. Sect. D 2002, 58, 1715–1721. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Center, R.J.; Kobe, B.; Wilson, K.A.; Teh, T.; Howlett, G.J.; Kemp, B.E.; Poumbourios, P. Crystallization of a trimeric human T cell leukemia virus type 1 gp21 ectodomain fragment as a chimera with maltose-binding protein. Protein Sci. A Publ. Protein Soc. 1998, 7, 1612–1619. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ke, A.; Wolberger, C. Insights into binding cooperativity of MATa1/MATα2 from the crystal structure of a MATa1 homeodomain-maltose binding protein chimera. Protein Sci. A Publ. Protein Soc. 2003, 12, 306–312. [Google Scholar] [CrossRef]
- Thorsen, T.S.; Matt, R.; Weis, W.I.; Kobilka, B. Modified T4 lysozyme fusion proteins facilitate G Protein-coupled receptor crystallogenesis. Structure 2014, 22, 1657–1664. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, S.H.; Quigley, G.J.; Suddath, F.L.; McPherson, A.; Sneden, D.; Kim, J.J.; Weinzierl, J.; Rich, A. Three-Dimensional Structure of Yeast Phenylalanine Transfer RNA: Folding of the Polynucleotide Chain. Science 1973, 179, 285. [Google Scholar] [CrossRef]
- Francklyn Christopher, S.; Minajigi, A. tRNA as an active chemical scaffold for diverse chemical transformations. FEBS Lett. 2009, 584, 366–375. [Google Scholar] [CrossRef] [Green Version]
- Ho, P.Y.; Yu, A.-M. Bioengineering of noncoding RNAs for research agents and therapeutics. Wiley Interdiscip. Rev. RNA 2016, 7, 186–197. [Google Scholar] [CrossRef] [Green Version]
- Ponchon, L.; Dardel, F. Recombinant RNA technology: The tRNA scaffold. Nat. Methods 2007, 4, 571. [Google Scholar] [CrossRef] [Green Version]
- Porter, E.B.; Polaski, J.T.; Morck, M.M.; Batey, R.T. Recurrent RNA motifs as scaffolds for genetically encodable small molecule biosensors. Nat. Chem. Biol. 2017, 13, 295–301. [Google Scholar] [CrossRef]
- Cai, R.; Price, I.R.; Ding, F.; Wu, F.; Chen, T.; Zhang, Y.; Liu, G.; Jardine, P.J.; Lu, C.; Ke, A. ATP/ADP modulates gp16–pRNA conformational change in the Phi29 DNA packaging motor. Nucleic Acids Res. 2019, 47, 9818–9828. [Google Scholar] [CrossRef]
- Lu, C.; Cai, R.; Grigg, J.C.; Ke, A. Using tRNA scaffold to assist RNA crystallization. In RNA Scaffolds: Methods and Protocols, Methods in Molecular Biology; Ponchon, L., Ed.; Springer: New York, NY, USA, 2021; Volume 2323, pp. 39–47. [Google Scholar]
- Grigg, J.C.; Ke, A. Structural determinants for geometry and information decoding of tRNA by T box leader RNA. Structure 2013, 21, 2025–2032. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zuker, M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003, 31, 3406–3415. [Google Scholar] [CrossRef] [PubMed]
- Kao, C.; Zheng, M.; Rüdisser, S. A simple and efficient method to reduce nontemplated nucleotide addition at the 3 terminus of RNAs transcribed by T7 RNA polymerase. RNA 1999, 5, 1268–1272. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kabsch, W. XDS. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010, 66, 125–132. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Evans, P. Scaling and assessment of data quality. Acta Crystallogr. Sect. D 2006, 62, 72–82. [Google Scholar] [CrossRef]
- McCoy, A.J.; Grosse-Kunstleve, R.W.; Adams, P.D.; Winn, M.D.; Storoni, L.C.; Read, R.J. Phaser crystallographic software. J. Appl. Crystallogr. 2007, 40, 658–674. [Google Scholar] [CrossRef] [Green Version]
- Adams, P.D.; Afonine, P.V.; Bunkoczi, G.; Chen, V.B.; Davis, I.W.; Echols, N.; Headd, J.J.; Hung, L.-W.; Kapral, G.J.; Grosse-Kunstleve, R.W.; et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. Sect. D 2010, 66, 213–221. [Google Scholar] [CrossRef] [Green Version]
- Emsley, P.; Lohkamp, B.; Scott, W.G.; Cowtan, K. Features and development of Coot. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010, 66, 486–501. [Google Scholar] [CrossRef] [Green Version]
- Keating, K.S.; Pyle, A.M. RCrane: Semi-automated RNA model building. Acta Crystallogr. Sect. D Biol. Crystallogr. 2012, 68, 985–995. [Google Scholar] [CrossRef] [Green Version]
- Afonine, P.V.; Grosse-Kunstleve, R.W.; Echols, N.; Headd, J.J.; Moriarty, N.W.; Mustyakimov, M.; Terwilliger, T.C.; Urzhumtsev, A.; Zwart, P.H.; Adams, P.D. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. Sect. D 2012, 68, 352–367. [Google Scholar] [CrossRef] [Green Version]
- Chou, F.-C.; Sripakdeevong, P.; Dibrov, S.M.; Hermann, T.; Das, R. Correcting pervasive errors in RNA crystallography through enumerative structure prediction. Nat. Methods 2012, 10, 74. [Google Scholar] [CrossRef] [PubMed]
- Green, N.J.; Grundy, F.J.; Henkin, T.M. The T box mechanism: tRNA as a regulatory molecule. FEBS Lett. 2010, 584, 318–324. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grundy, F.J.; Henkin, T.M. tRNA as a positive regulator of transcription antitermination in B. subtilis. Cell 1993, 74, 475–482. [Google Scholar] [CrossRef]
- Suddala, K.C.; Zhang, J. High-affinity recognition of specific tRNAs by an mRNA anticodon-binding groove. Nat. Struct. Mol. Biol. 2019, 26, 1114–1122. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, S.; Su, Z.; Lehmann, J.; Stamatopoulou, V.; Giarimoglou, N.; Henderson, F.E.; Fan, L.; Pintilie, G.D.; Zhang, K.; Chen, M.; et al. Structural basis of amino acid surveillance by higher-order tRNA-mRNA interactions. Nat. Struct. Mol. Biol. 2019, 26, 1094–1105. [Google Scholar] [CrossRef] [PubMed]
- Battaglia, R.A.; Grigg, J.C.; Ke, A. Structural basis for tRNA decoding and aminoacylation sensing by T-box riboregulators. Nat. Struct. Mol. Biol. 2019, 26, 1106–1113. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J. Unboxing the T-box riboswitches-A glimpse into multivalent and multimodal RNA-RNA interactions. Wiley Interdiscip. Rev. RNA 2020, 11, e1600. [Google Scholar] [CrossRef]
- Grigg, J.C.; Chen, Y.; Grundy, F.J.; Henkin, T.M.; Pollack, L.; Ke, A. T box RNA decodes both the information content and geometry of tRNA to affect gene expression. Proc. Natl. Acad. Sci. USA 2013, 110, 7240–7245. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Henkin, T.M.; Nikonowicz, E.P. NMR structure and dynamics of the Specifier Loop domain from the Bacillus subtilis tyrS T box leader RNA. Nucleic Acids Res. 2010, 38, 3388–3398. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Nikonowicz, E.P. Solution structure of the K-turn and Specifier Loop domains from the Bacillus subtilis tyrS T-box leader RNA. J. Mol. Biol. 2011, 408, 99–117. [Google Scholar] [CrossRef] [Green Version]
- Grundy, F.J.; Winkler, W.C.; Henkin, T.M. tRNA-mediated transcription antitermination in vitro: Codon–anticodon pairing independent of the ribosome. Proc. Natl. Acad. Sci. USA 2002, 99, 11121–11126. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yousef, M.R.; Grundy, F.J.; Henkin, T.M. Structural Transitions Induced by the Interaction between tRNAGly and the Bacillus subtilis glyQS T Box Leader RNA. J. Mol. Biol. 2005, 349, 273–287. [Google Scholar] [CrossRef] [PubMed]
- Vitreschak, A.G.; Mironov, A.A.; Lyubetsky, V.A.; Gelfand, M.S. Comparative genomic analysis of T-box regulatory systems in bacteria. RNA 2008, 14, 717–735. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gerdeman, M.S.; Henkin, T.M.; Hines, J.V. Solution structure of the Bacillus subtilis T-box antiterminator RNA: Seven nucleotide bulge characterized by stacking and flexibility. J. Mol. Biol. 2003, 326, 189–201. [Google Scholar] [CrossRef]
- Suddala, K.C.; Cabello-Villegas, J.; Michnicka, M.; Marshall, C.; Nikonowicz, E.P.; Walter, N.G. Hierarchical mechanism of amino acid sensing by the T-box riboswitch. Nat. Commun. 2018, 9, 1896. [Google Scholar] [CrossRef]
- Fang, X.; Michnicka, M.; Zhang, Y.; Wang, Y.X.; Nikonowicz, E.P. Capture and Release of tRNA by the T-Loop Receptor in the Function of the T-Box Riboswitch. Biochemistry 2017, 56, 3549–3558. [Google Scholar] [CrossRef] [Green Version]
- Chetnani, B.; Mondragon, A. Molecular envelope and atomic model of an anti-terminated glyQS T-box regulator in complex with tRNAGly. Nucleic Acids Res. 2017, 45, 8079–8090. [Google Scholar] [CrossRef] [Green Version]
- Grigg, J.C.; Ke, A. Sequence, structure, and stacking: Specifics of tRNA anchoring to the T box riboswitch. RNA Biol. 2013, 10, 1761–1764. [Google Scholar] [CrossRef] [Green Version]
- Lehmann, J.; Jossinet, F.; Gautheret, D. A universal RNA structural motif docking the elbow of tRNA in the ribosome, RNAse P and T-box leaders. Nucleic Acids Res. 2013, 41, 5494–5502. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.; Ferré-D’Amaré, A.R. Direct evaluation of tRNA aminoacylation status by the T-box riboswitch using tRNA-mRNA stacking and steric readout. Mol. Cell 2014, 55, 148–155. [Google Scholar] [CrossRef] [Green Version]
- Fauzi, H.; Agyeman, A.; Hines, J.V. T box transcription antitermination riboswitch: Influence of nucleotide sequence and orientation on tRNA binding by the antiterminator element. Biochim. Biophys. Acta 2009, 1789, 185–191. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gerdeman, M.S.; Henkin, T.M.; Hines, J.V. In vitro structure-function studies of the Bacillus subtilis tyrS mRNA antiterminator: Evidence for factor-independent tRNA acceptor stem binding specificity. Nucleic Acids Res. 2002, 30, 1065–1072. [Google Scholar] [CrossRef] [PubMed]
- Grundy, F.J.; Rollins, S.M.; Henkin, T.M. Interaction between the acceptor end of tRNA and the T box stimulates antitermination in the Bacillus subtilis tyrS gene: A new role for the discriminator base. J. Bacteriol. 1994, 176, 4518–4526. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yousef, M.R.; Grundy, F.J.; Henkin, T.M. tRNA requirements for glyQS antitermination: A new twist on tRNA. RNA 2003, 9, 1148–1156. [Google Scholar] [CrossRef] [Green Version]
- Giegé, R. Toward a more complete view of tRNA biology. Nat. Struct. Mol. Biol. 2008, 15, 1007. [Google Scholar] [CrossRef]
- Maizels, N.; Weiner, A.M. Phylogeny from function: Evidence from the molecular fossil record that tRNA originated in replication, not translation. Proc. Natl. Acad. Sci. USA 1994, 91, 6729–6734. [Google Scholar] [CrossRef] [Green Version]
- Pütz, J.; Giegé, R.; Florentz, C. Diversity and similarity in the tRNA world: Overall view and case study on malaria-related tRNAs. FEBS Lett. 2010, 584, 350–358. [Google Scholar] [CrossRef] [Green Version]
- Gutmann, S.; Haebel, P.W.; Metzinger, L.; Sutter, M.; Felden, B.; Ban, N. Crystal structure of the transfer-RNA domain of transfer-messenger RNA in complex with SmpB. Nature 2003, 424, 699. [Google Scholar] [CrossRef]
- Colussi, T.M.; Costantino, D.A.; Hammond, J.A.; Ruehle, G.M.; Nix, J.C.; Kieft, J.S. The structural basis of tRNA mimicry and conformational plasticity by a viral RNA. Nature 2014, 511, 366–369. [Google Scholar] [CrossRef] [Green Version]
- Wang, W.; Chen, X.; Wolin, S.L.; Xiong, Y. Structural basis for transfer RNA mimicry by a bacterial Y RNA. Structure 2018, 26, 1635–1644. [Google Scholar] [CrossRef] [Green Version]
- Reiter, N.J.; Osterman, A.; Torres-Larios, A.; Swinger, K.K.; Pan, T.; Mondragón, A. Structure of a bacterial ribonuclease P holoenzyme in complex with tRNA. Nature 2010, 468, 784–789. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Ferré-D’Amaré, A.R. A flexible, scalable method for preparation of homogeneous aminoacylated tRNAs. Methods Enzymol. 2014, 549, 105–113. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bashan, A.; Agmon, I.; Zarivach, R.; Schluenzen, F.; Harms, J.; Berisio, R.; Bartels, H.; Franceschi, F.; Auerbach, T.; Hansen, H.A.S.; et al. Structural Basis of the Ribosomal Machinery for Peptide Bond Formation, Translocation, and Nascent Chain Progression. Mol. Cell 2003, 11, 91–102. [Google Scholar] [CrossRef]
- Schmeing, T.M.; Moore, P.B.; Steitz, T.A. Structures of deacylated tRNA mimics bound to the E site of the large ribosomal subunit. RNA 2003, 9, 1345–1352. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hansen, J.L.; Schmeing, T.M.; Moore, P.B.; Steitz, T.A. Structural insights into peptide bond formation. Proc. Natl. Acad. Sci. USA 2002, 99, 11670. [Google Scholar] [CrossRef] [Green Version]
- Morais, M.C.; Koti, J.S.; Bowman, V.D.; Reyes-Aldrete, E.; Anderson, D.L.; Rossmann, M.G. Defining Molecular and Domain Boundaries in the Bacteriophage ϕ29 DNA Packaging Motor. Structure 2008, 16, 1267–1274. [Google Scholar] [CrossRef] [Green Version]
- Ding, F.; Lu, C.; Zhao, W.; Rajashankar, K.R.; Anderson, D.L.; Jardine, P.J.; Grimes, S.; Ke, A. Structure and assembly of the essential RNA ring component of a viral DNA packaging motor. Proc. Natl. Acad. Sci. USA 2011, 108, 7357. [Google Scholar] [CrossRef] [Green Version]
- Byrne, R.T.; Konevega, A.L.; Rodnina, M.V.; Antson, A.A. The crystal structure of unmodified tRNA(Phe) from Escherichia coli. Nucleic Acids Res. 2010, 38, 4154–4162. [Google Scholar] [CrossRef] [Green Version]
- Moras, D.; Bergdoll, M. Packing and molecular interactions in tRNA crystals. J. Cryst. Growth 1988, 90, 283–294. [Google Scholar] [CrossRef]

| / | AntiTtRNA Construct 4 UCCA -2bp | AntiT-tRNA Construct 19 Flip UCCA | AntiT-tRNA Construct 5 MOD FL |
|---|---|---|---|
| Data collection a | / | / | / |
| Beamline | APS 24ID-E | APS 24ID-E | APS 24ID-E |
| Wavelength (Å) | 0.979 | 0.979 | 0.978 |
| Resolution range (Å) | 32–2.95 (3.11–2.95) | 33.6–3.03 (3.14–3.03) | 45.8–6.1 (6.45–6.12) |
| Space group | C2221 | C2221 | P6122 |
| Unit cell dimensions (Å) | a = 88.6, b = 165.2, c = 57.1 | a = 46.2, b = 56.7, c = 268.9 | a = 168.5, b = 168.5, c = 136.3 |
| Unique reflections | 9103 (1263) | 7208 (688) | 2948 |
| Completeness (%) | 99.5 (97.5) | 99.2 (98.6) | 99.3 (99.7) |
| Multiplicity | 6.2 (4.9) | 7.1 (7.3) | 9.8 (10.1) |
| Average I/σI | 18.9 (3.2) | 12.9 (1.6) | 13.6 (1.3) |
| Rmerge | 0.115 (0.552) | 0.131 (1.135) | 0.136 (1.991) |
| Rp.i.m. | 0.055 (0.277) | 0.053 (0.446) | 0.047 (0.673) |
| CC1/2 | 0.942 (0.808) | 0.997 (0.991) | 0.998 (0.351) |
| Refinement | / | / | / |
| Rwork (Rfree) | 24.7 (26.5) | 24.6 (27.6) | / |
| B-factors (Å2) | / | / | / |
| All atoms | 63.3 | 100.7 | / |
| RNA | 63.0 | 100.3 | / |
| Ions | 100.2 | 144.57 | / |
| r.m.s.d. bond length (Å) | 0.002 | 0.005 | / |
| r.m.s.d. bond angle (°) | 0.536 | 0.95 | / |
| PDB Accession Codes | 7UQ6 | 7UZ0 | / |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Grigg, J.C.; Price, I.R.; Ke, A. tRNA Fusion to Streamline RNA Structure Determination: Case Studies in Probing Aminoacyl-tRNA Sensing Mechanisms by the T-Box Riboswitch. Crystals 2022, 12, 694. https://doi.org/10.3390/cryst12050694
Grigg JC, Price IR, Ke A. tRNA Fusion to Streamline RNA Structure Determination: Case Studies in Probing Aminoacyl-tRNA Sensing Mechanisms by the T-Box Riboswitch. Crystals. 2022; 12(5):694. https://doi.org/10.3390/cryst12050694
Chicago/Turabian StyleGrigg, Jason C., Ian R. Price, and Ailong Ke. 2022. "tRNA Fusion to Streamline RNA Structure Determination: Case Studies in Probing Aminoacyl-tRNA Sensing Mechanisms by the T-Box Riboswitch" Crystals 12, no. 5: 694. https://doi.org/10.3390/cryst12050694
APA StyleGrigg, J. C., Price, I. R., & Ke, A. (2022). tRNA Fusion to Streamline RNA Structure Determination: Case Studies in Probing Aminoacyl-tRNA Sensing Mechanisms by the T-Box Riboswitch. Crystals, 12(5), 694. https://doi.org/10.3390/cryst12050694






