Selenourea for Experimental Phasing of Membrane Protein Crystals Grown in Lipid Cubic Phase
Abstract
:1. Introduction

2. Materials & Methods
2.1. Materials
2.2. Purification of PlsY
2.3. Crystallization of Lysozyme in Lipid Cubic Phase
2.4. Crystallization of PlsY in Lipid Cubic Phase
2.5. Soaking Lipid Cubic Phase Crystals with Se-Urea
2.6. X-ray Diffraction Data Collection
2.7. Data Processing and Structure Determination
3. Results & Discussions
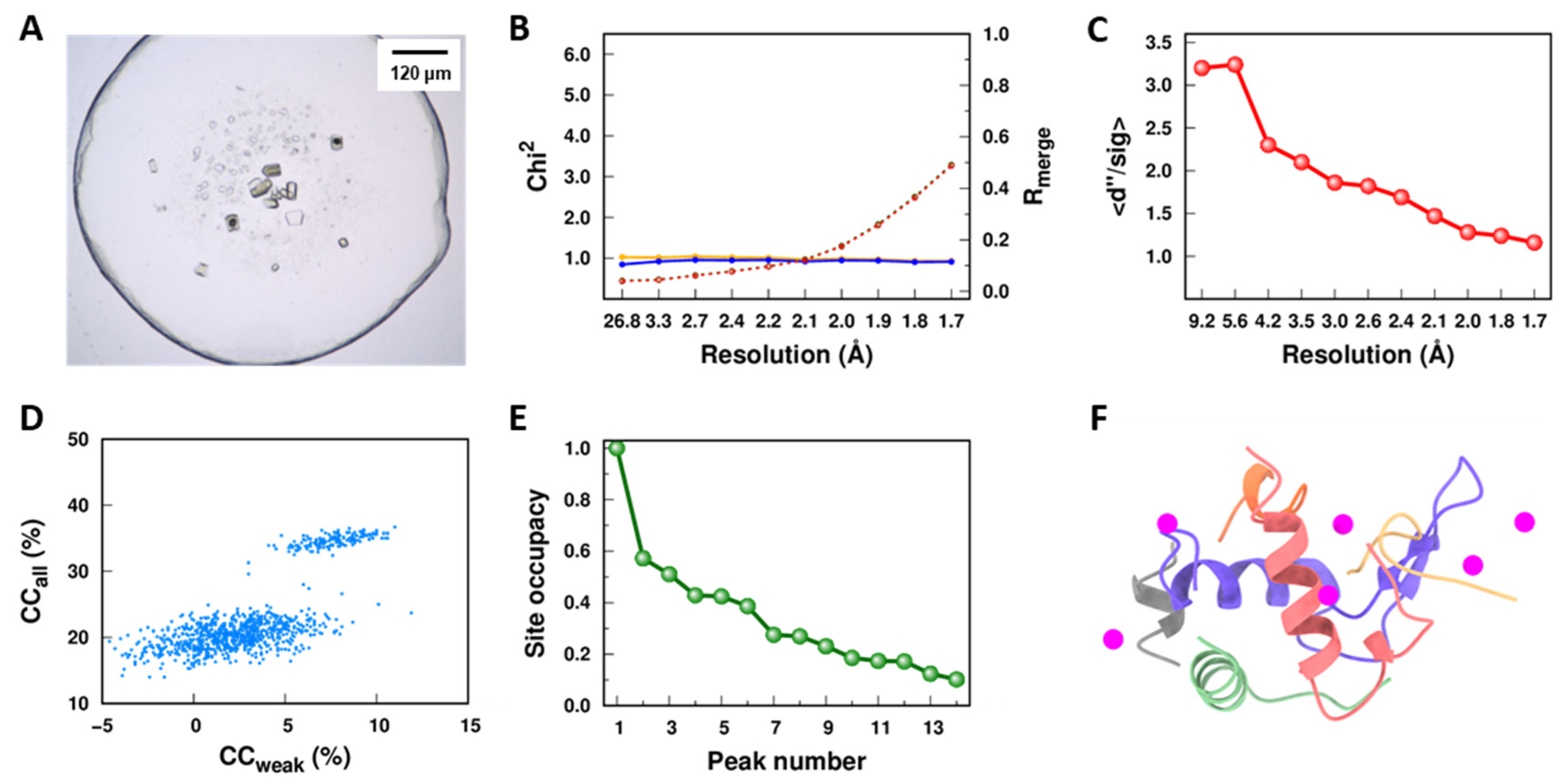
3.1. Both Lipid Cubic Phase and PlsY Tolerated Se-Urea Soaking
3.2. Prove-of-Principle Experiment with Lysozyme
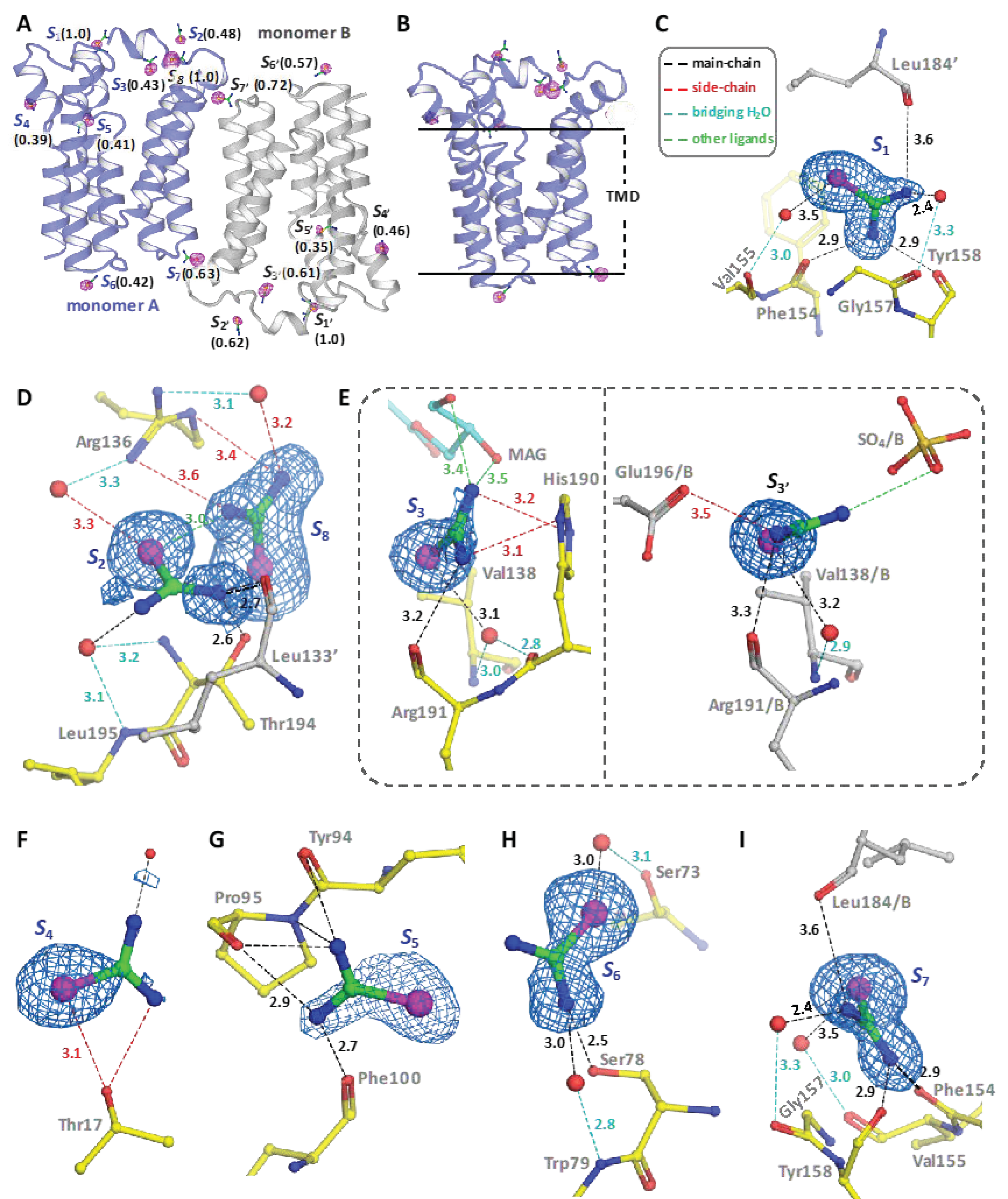
3.3. Experimental Phasing of PlsY
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef] [PubMed]
- Baek, M.; DiMaio, F.; Anishchenko, I.; Dauparas, J.; Ovchinnikov, S.; Lee, G.R.; Wang, J.; Cong, Q.; Kinch, L.N.; Baker, D.; et al. Accurate prediction of protein structures and interactions using a three-track neural network. Science 2021, 373, 871–876. [Google Scholar] [CrossRef] [PubMed]
- McCoy, A.J.; Sammito, M.D.; Read, R.J. Implications of AlphaFold2 for crystallographic phasing by molecular replacement. Acta Crystallogr. Sect. D Struct. Biol. 2022, 78, 1–13. [Google Scholar] [CrossRef]
- Pándy-Szekeres, G.; Munk, C.; Tsonkov, T.M.; Mordalski, S.; Harpsøe, K.; Hauser, A.S.; Bojarski, A.J.; Gloriam, D.E. GPCRdb in 2018: Adding GPCR structure models and ligands. Nucleic Acids Res. 2017, 46, D440–D446. [Google Scholar] [CrossRef] [Green Version]
- Chang, Y.; Bruni, R.; Kloss, B.; Assur, Z.; Kloppmann, E.; Rost, B.; Hendrikson, W.A.; Liu, Q. Structural basis for a pH-sensitive calcium leak across membranes. Science 2014, 344, 1131–1135. [Google Scholar] [CrossRef] [Green Version]
- Taylor, G.L. Introduction to phasing. Acta Crystallogr. D 2010, 66, 325–338. [Google Scholar] [CrossRef] [PubMed]
- Hendrickson, W.A.; Horton, J.R.; LeMaster, D.M. Selenomethionyl proteins produced for analysis by multiwavelength anomalous diffraction (MAD): A vehicle for direct determination of three-dimensional structure. EMBO J. 1990, 9, 1665–1672. [Google Scholar] [CrossRef]
- Larsson, A.M.; Jones, T.A. Selenomethionine labeling of recombinant proteins. Methods Mol. Biol. 2007, 389, 165–174. [Google Scholar]
- Bellizzi, J.J., III; Widom, J.; Kemp, C.W.; Clardy, J. Producing selenomethionine-labeled proteins with a baculovirus expression vector system. Structure 1999, 7, R263–R267. [Google Scholar] [CrossRef] [Green Version]
- Kigawa, T.; Yamaguchi-Nunokawa, E.; Kodama, K.; Matsuda, T.; Yabuki, T.; Matsuda, N.; Ishitani, R.; Nureki, O.; Yokoyama, S. Selenomethionine incorpora-tion into a protein by cell-free synthesis. J. Struct. Funct. Genom. 2002, 2, 29–35. [Google Scholar] [CrossRef]
- Vogeley, L.; El Arnaout, T.; Bailey, J.; Stansfeld, P.J.; Boland, C.; Caffrey, M. Structural basis of lipoprotein signal peptidase II action and inhibition by the antibiotic globomycin. Science 2016, 351, 876–880. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hendrickson, W.A.; Pähler, A.; Smith, J.L.; Satow, Y.; Merritt, E.A.; Phizackerley, R.P. Crystal structure of core streptavidin determined from multiwavelength anomalous diffraction of synchrotron radiation. Proc. Natl. Acad. Sci. USA 1989, 86, 2190–2194. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Du, Q.; Carrasco, N.; Teplova, M.; Wilds, C.J.; Egli, M.; Huang, Z. Internal Derivatization of Oligonucleotides with Selenium for X-ray Crystallography Using MAD. J. Am. Chem. Soc. 2001, 124, 24–25. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; A Hendrickson, W. Crystallographic phasing from weak anomalous signals. Curr. Opin. Struct. Biol. 2015, 34, 99–107. [Google Scholar] [CrossRef] [Green Version]
- Guo, G.; Zhu, P.; Fuchs, M.R.; Shi, W.; Andi, B.; Gao, Y.; Hendrickson, W.A.; McSweeney, S.; Liu, Q. Synchrotron microcrystal native-SAD phasing at a low energy. IUCrJ 2019, 6, 532–542. [Google Scholar] [CrossRef]
- Cianci, M.; Groves, M.; Barford, D.; Schneider, T.R. Data collection with a tailored X-ray beam size at 2.69 Å wavelength (4.6 keV): Sulfur SAD phasing of Cdc23(Nterm). Acta Crystallogr. Sect. D Struct. Biol. 2016, 72, 403–412. [Google Scholar] [CrossRef] [Green Version]
- Aurelius, O.; Duman, R.; El Omari, K.; Mykhaylyk, V.; Wagner, A. Long-wavelength macromolecular crystallography—First successful native SAD experiment close to the sulfur edge. Nucl. Instrum. Methods Phys. Res. Sect. B 2016, 411, 12–16. [Google Scholar] [CrossRef]
- Wang, W.; Yang, X.; Chang, G.; An, P.; Cha, K.; Dong, Y.; Liu, P. A method to stabilize the incident X-ray energy for anomalous diffraction measurements. J. Synchrotron Radiat. 2017, 24, 781–786. [Google Scholar] [CrossRef]
- Su, X.-D.; Zhang, H.; Terwilliger, T.C.; Liljas, A.; Xiao, J.; Dong, Y. Protein Crystallography from the Perspective of Technology Developments. Crystallogr. Rev. 2015, 21, 122–153. [Google Scholar] [CrossRef]
- Basu, S.; Olieric, V.; Leonarski, F.; Matsugaki, N.; Kawano, Y.; Takashi, T.; Huang, C.-Y.; Yamada, Y.; Vera, L.; Olieric, N.; et al. Long-wavelength native-SAD phasing: Op-portunities and challenges. IUCrJ 2019, 6, 373–386. [Google Scholar] [CrossRef] [Green Version]
- Olieric, V.; Weinert, T.; Finke, A.D.; Anders, C.; Li, D.; Olieric, N.; Borca, C.N.; Steinmetz, M.O.; Caffrey, M.; Jinek, M.; et al. Data-collection strategy for challenging native SAD phasing. Acta Crystallogr. D 2016, 72, 421–429. [Google Scholar] [CrossRef] [PubMed]
- Cianci, M.; Nanao, M.; Schneider, T.R. Long-wavelength Mesh&Collect native SAD phasing from microcrystals. Acta Crystallogr. Sect. D Struct. Biol. 2019, 75, 192–199. [Google Scholar] [CrossRef] [Green Version]
- Rose, J.P.; Wang, B.-C.; Weiss, M.S. Native SAD is maturing. IUCrJ 2015, 2, 431–440. [Google Scholar] [CrossRef] [PubMed]
- Marchal, J.; Wägner, A. Performance of PILATUS detector technology for long-wavelength macromolecular crystallography. Nucl. Instruments Methods Phys. Res. Sect. A 2011, 633, S121–S124. [Google Scholar] [CrossRef]
- Wagner, A.; Duman, R.; Henderson, K.; Mykhaylyk, V. In-vacuum long-wavelength macromolecular crystallography. Acta Crystallogr. Sect. D 2016, 72, 430–439. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, C.-Y.; Olieric, V.; Ma, P.; Panepucci, E.; Diederichs, K.; Wang, M.; Caffrey, M. In meso in situ serial X-ray crystallography of soluble and membrane proteins. Acta Crystallogr. Sect. D Biol. Crystallogr. 2015, 71, 1238–1256. [Google Scholar] [CrossRef] [Green Version]
- Walden, H. Selenium incorporation using recombinant techniques. Acta Crystallogr. Sect. D 2010, 66, 352–357. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gubellini, F.; Verdon, G.; Karpowich, N.K.; Luff, J.D.; Boel, G.; Gauthier, N.; Handelman, S.; Ades, S.E.; Hunt, J.F. Physiological Response to Membrane Protein Overexpression in E. coli. Mol. Cell. Proteom. 2011, 10, 1–17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gialama, D.; Kostelidou, K.; Michou, M.; Delivoria, D.C.; Kolisis, F.N.; Skretas, G. Development of Escherichia coli Strains That Withstand Membrane Protein-Induced Toxicity and Achieve High-Level Recombinant Membrane Protein Production. ACS Synth. Biol. 2016, 6, 284–300. [Google Scholar] [CrossRef]
- Wagner, S.; Klepsch, M.M.; Schlegel, S.; Appel, A.; Draheim, R.; Tarry, M.; Högbom, M.; van Wijk, K.J.; Slotboom, D.J.; Persson, J.O.; et al. Tuning Escherichia coli for mem-brane protein overexpression. Proc. Natl. Acad. Sci. USA 2008, 105, 14371–14376. [Google Scholar] [CrossRef] [Green Version]
- Wagner, S.; Bader, M.L.; Drew, D.; de Gier, J.-W. Rationalizing membrane protein overexpression. Trends Biotechnol. 2006, 24, 364–371. [Google Scholar] [CrossRef] [PubMed]
- Pike, A.C.; Garman, E.F.; Krojer, T.; Von Delft, F.; Carpenter, E.P. An overview of heavy-atom derivatization of protein crystals. Acta Crystallogr. Sect. D Struct. Biol. 2016, 72, 303–318. [Google Scholar] [CrossRef] [Green Version]
- Garman, E.; Murray, J.W. Heavy-atom derivatization. Acta Crystallogr. Sect. D 2003, 59, 1903–1913. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boggon, T.J.; Shapiro, L. Screening for phasing atoms in protein crystallography. Structure 2000, 8, R143–R149. [Google Scholar] [CrossRef] [Green Version]
- Lu, J.; Sun, P.D. A rapid and rational approach to generating isomorphous heavy-atom phasing derivatives. FEBS J. 2014, 281, 4021–4028. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, D.; Pye, V.E.; Caffrey, M. Experimental phasing for structure determination using membrane-protein crystals grown by the lipid cubic phase method. Acta Crystallogr. D 2015, 71, 104–122. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, Y.; Kalathur, R.C.; Liu, Q.; Kloss, B.; Bruni, R.; Ginter, C.; Kloppmann, E.; Rost, B.; Hendrickson, W.A. Structure and activity of tryptophan-rich TSPO proteins. Science 2015, 347, 551–555. [Google Scholar] [CrossRef] [Green Version]
- Nakane, T.; Hanashima, S.; Suzuki, M.; Saiki, H.; Hayashi, T.; Kakinouchi, K.; Sugiyama, S.; Kawatake, S.; Matsuoka, S.; Matsumori, N.; et al. Membrane protein structure determination by SAD, SIR, or SIRAS phasing in serial femtosecond crystallography using an iododetergent. Proc. Natl. Acad. Sci. USA 2016, 113, 13039–13044. [Google Scholar] [CrossRef] [Green Version]
- Luo, Z. Selenourea: A convenient phasing vehicle for macromolecular X-ray crystal structures. Sci. Rep. 2016, 6, 37123. [Google Scholar] [CrossRef] [Green Version]
- Caffrey, M. A comprehensive review of the lipid cubic phase or in meso method for crystallizing membrane and soluble proteins and complexes. Acta Crystallogr. F 2015, 71, 3–18. [Google Scholar] [CrossRef] [Green Version]
- Li, Z.; Tang, Y.; Wu, Y.; Zhao, S.; Bao, J.; Luo, Y.; Li, D. Structural insights into the committed step of bacterial phospholipid biosynthesis. Nat. Commun. 2017, 8, 1691. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ren, S.; Caforio, A.; Yang, Q.; Sun, B.; Yu, F.; Zhu, X.; Wang, J.; Dou, C.; Fu, Q.; Huang, N.; et al. Structural and mechanistic insights into the biosynthesis of CDP-archaeol in membranes. Cell Res. 2017, 27, 1378–1391. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scott, A.J.; Niitsu, A.; Kratochvil, H.T.; Lang, E.J.M.; Sengel, J.T.; Dawson, W.M.; Mahendran, K.R.; Mravic, M.; Thomson, A.R.; Brady, R.L.; et al. Constructing ion channels from water-soluble α-helical barrels. Nat. Chem. 2021, 13, 643–650. [Google Scholar] [CrossRef] [PubMed]
- Yang, B.; Yao, H.; Li, D.; Liu, Z. The phosphatidylglycerol phosphate synthase PgsA utilizes a trifurcated amphipathic cavity for catalysis at the membrane-cytosol interface. Curr. Res. Struct. Biol. 2021, 3, 312–323. [Google Scholar] [CrossRef]
- Sammito, M.; Millán, C.; Frieske, D.; Rodríguez-Freire, E.; Borges, R.J.; Usón, I. ARCIMBOLDO_LITE: Single-workstation implementation and use. Acta Crystallogr. D Biol. Crystallogr. 2015, 71, 1921–1930. [Google Scholar] [CrossRef]
- Sjodt, M.; Brock, K.; Dobihal, G.; Rohs, P.; Green, A.; Hopf, T.A.; Meeske, A.; Srisuknimit, V.; Kahne, D.; Walker, S.; et al. Structure of the peptidoglycan polymerase RodA resolved by evolutionary coupling analysis. Nature 2018, 556, 118–121. [Google Scholar] [CrossRef]
- Xiao, Q.; Wang, L.; Supekar, S.; Shen, T.; Liu, H.; Ye, F.; Huang, J.; Fan, H.; Wei, Z.; Zhang, C. Structure of human steroid 5α-reductase 2 with the anti-androgen drug finasteride. Nat. Commun. 2020, 11, 5430. [Google Scholar] [CrossRef]
- Drew, D.; Lerch, M.; Kunji, E.; Slotboom, D.J.; de Gier, J.W. Optimization of membrane protein overexpression and purification using GFP fusions. Nat. Methods 2006, 3, 303–313. [Google Scholar] [CrossRef]
- Aherne, M.; Lyons, J.; Caffrey, M. A fast, simple and robust protocol for growing crystals in the lipidic cubic phase. J. Appl. Crystallogr. 2012, 45, 1330–1333. [Google Scholar] [CrossRef] [Green Version]
- Cheng, A.; Hummel, B.; Qiu, H.; Caffrey, M. A simple mechanical mixer for small viscous lipid-containing samples. Chem. Phys. Lipids 1998, 95, 11–21. [Google Scholar] [CrossRef]
- Caffrey, M.; Porter, C. Crystallizing Membrane Proteins for Structure Determination using Lipidic Mesophases. J. Vis. Exp. 2010, 45, e1712. [Google Scholar] [CrossRef] [Green Version]
- Caffrey, M.; Eifert, R.; Li, D.; Howe, N. The lipid cubic phase or in meso method for crystallizing proteins. Bushings for better manual dispensing. J. Appl. Crystallogr. 2014, 47, 1804–1806. [Google Scholar] [CrossRef] [PubMed]
- Caffrey, M.; Cherezov, V. Crystallizing membrane proteins using lipidic mesophases. Nat. Protoc. 2009, 4, 706–731. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Z.; Tang, Y.; Li, D. In Meso Crystallization of the Integral Membrane Glycerol 3-Phosphate Acyltransferase with Substrates. Cryst. Growth Des. 2018, 18, 2243–2258. [Google Scholar] [CrossRef]
- Li, D.; Boland, C.; Walsh, K.; Caffrey, M. Use of a Robot for High-throughput Crystallization of Membrane Proteins in Lipidic Mesophases. J. Vis. Exp. 2012, 67, e4000. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Boland, C.; Aragao, D.; Walsh, K.; Caffrey, M. Harvesting and Cryo-cooling Crystals of Membrane Proteins Grown in Lipidic Mesophases for Structure Determination by Macromolecular Crystallography. J. Vis. Exp. 2012, 67, e4001. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Pan, Q.; Yang, L.; Zhou, H.; Xu, C.; Yu, F.; Wang, Q.; Huang, S.; He, J. Automatic crystal centring procedure at the SSRF macromolecular crystallography beamline. J. Synchrotron Radiat. 2016, 23, 1323–1332. [Google Scholar] [CrossRef]
- Zhang, W.-Z.; Tang, J.-C.; Wang, S.-S.; Wang, Z.-J.; Qin, W.-M.; He, J.-H. The protein complex crystallography beamline (BL19U1) at the Shanghai Synchrotron Radiation Facility. Nucl. Sci. Technol. 2019, 30, 170. [Google Scholar] [CrossRef]
- Otwinowski, Z.; Minor, W. Processing of X-ray Diffraction Data Collected in Oscillation Mode. In Methods Enzymol; Academic Press: Cambridge, MA, USA, 1997; Volume 276, pp. 307–326. [Google Scholar]
- Sheldrick, G.M. Experimental phasing with SHELXC/D/E: Combining chain tracing with density modification. Acta Crystallogr. D 2010, 66, 479–485. [Google Scholar] [CrossRef] [Green Version]
- Cowtan, K. The Buccaneer software for automated model building. 1. Tracing protein chains. Acta Crystallogr. D 2006, 62, 1002–1011. [Google Scholar] [CrossRef] [Green Version]
- Potterton, L.; Agirre, J.; Ballard, C.; Cowtan, K.; Dodson, E.; Evans, P.R.; Jenkins, H.T.; Keegan, R.; Krissinel, E.; Stevenson, K.; et al. CCP4i2: The new graphical user interface to the CCP4 program suite. Acta Crystallogr. D 2018, 74, 68–84. [Google Scholar] [CrossRef] [Green Version]
- Emsley, P.; Lohkamp, B.; Scott, W.G.; Cowtan, K. Features and development of Coot. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010, 66, 486–501. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- PyMOL. The PyMOL Molecular Graphics System; DeLano Scientific: San Carlos, CA, USA, 2009. [Google Scholar]
- Cherezov, V.; Fersi, H.; Caffrey, M. Crystallization Screens: Compatibility with the Lipidic Cubic Phase for in Meso Crystal-lization of Membrane Proteins. Biophys. J. 2001, 81, 225–242. [Google Scholar] [CrossRef] [Green Version]
- Misquitta, Y.; Caffrey, M. Detergents destabilize the cubic phase of monoolein: Implications for membrane protein crystalli-zation. Biophys. J. 2003, 85, 3084–3096. [Google Scholar] [CrossRef] [Green Version]
- Ai, X.; Caffrey, M. Membrane Protein Crystallization in Lipidic Mesophases: Detergent Effects. Biophys. J. 2000, 79, 394–405. [Google Scholar] [CrossRef] [Green Version]
- Cherezov, V.; Clogston, J.; Misquitta, Y.; Abdel-Gawad, W.; Caffrey, M. Membrane Protein Crystallization in Meso: Lipid Type-Tailoring of the Cubic Phase. Biophys. J. 2002, 83, 3393–3407. [Google Scholar] [CrossRef] [Green Version]
- Cherezov, V.; Clogston, J.; Papiz, M.Z.; Caffrey, M. Room to Move: Crystallizing Membrane Proteins in Swollen Lipidic Mesophases. J. Mol. Biol. 2006, 357, 1605–1618. [Google Scholar] [CrossRef]
- Levin, E.J.; Cao, Y.; Enkavi, G.; Quick, M.; Pan, Y.; Tajkhorshid, E.; Zhou, M. Structure and permeation mechanism of a mammalian urea transporter. Proc. Natl. Acad. Sci. USA 2012, 109, 11194–11199. [Google Scholar] [CrossRef] [Green Version]
- Strub, M.-P.; Hoh, F.; Sanchez, J.-F.; Strub, J.-M.; Böck, A.; Aumelas, A.; Dumas, C. Selenomethionine and Selenocysteine Double Labeling Strategy for Crystallographic Phasing. Structure 2003, 11, 1359–1367. [Google Scholar] [CrossRef] [Green Version]
- Li, D.; Lee, J.; Caffrey, M. Crystallizing Membrane Proteins in Lipidic Mesophases. A Host Lipid Screen. Cryst. Growth Des. 2011, 11, 530–537. [Google Scholar] [CrossRef] [Green Version]
- Li, D.; Howe, N.; Dukkipati, A.; Shah, S.T.; Bax, B.D.; Edge, C.; Bridges, A.; Hardwicke, P.; Singh, O.M.; Giblin, G.; et al. Crystallizing Membrane Proteins in the Lipidic Mesophase. Experience with Human Prostaglandin E2 Synthase 1 and an Evolving Strategy. Cryst. Growth Des. 2014, 14, 2034–2047. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Shah, S.T.; Caffrey, M. Host Lipid and Temperature as Important Screening Variables for Crystallizing Integral Membrane Proteins in Lipidic Mesophases. Trials with Diacylglycerol Kinase. Cryst. Growth Des. 2013, 13, 2846–2857. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, W.; Hanson, M.A.; Stevens, R.C.; Cherezov, V. LCP-Tm: An assay to measure and understand stability of membrane proteins in a membrane environment. Biophys. J. 2010, 98, 1539–1548. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cherezov, V.; Liu, J.; Griffith, M.; Hanson, M.A.; Stevens, R.C. LCP-FRAP Assay for Pre-Screening Membrane Proteins for in Meso Crystallization. Cryst. Growth Des. 2008, 8, 4307–4315. [Google Scholar] [CrossRef] [Green Version]
- Cherezov, V.; Abola, E.; Stevens, R.C. Recent progress in the structure determination of GPCRs, a membrane protein family with high potential as pharmaceutical targets. Methods Mol. Biol. 2010, 654, 141–168. [Google Scholar] [CrossRef] [Green Version]
- Magnani, F.; Serrano-Vega, M.J.; Shibata, Y.; Abdul-Hussein, S.; Lebon, G.; Miller-Gallacher, J.; Singhal, A.; Strege, A.; Thomas, J.A.; Tate, C.G. A mutagenesis and screening strategy to generate optimally thermostabilized membrane proteins for structural studies. Nat. Protoc. 2016, 11, 1554–1571. [Google Scholar] [CrossRef] [Green Version]
- Yamamoto, M.; Hirata, K.; Yamashita, K.; Hasegawa, K.; Ueno, G.; Ago, H.; Kumasaka, T. Protein microcrystallography using synchrotron radiation. IUCrJ 2017, 4, 529–539. [Google Scholar] [CrossRef]
- Mehrabi, P.; Bücker, R.; Bourenkov, G.; Ginn, H.M.; von Stetten, D.; Müller-Werkmeister, H.M.; Kuo, A.; Morizumi, T.; Eger, B.T.; Ou, W.L.; et al. Oghbaey S, Sarracini A, Besaw JE, Pare-Labrosse O, Meier S, Schikora H, Tellkamp F, Marx A. Serial femtosecond and serial synchrotron crystallography can yield data of equivalent quality: A systematic comparison. Sci. Adv. 2021, 7, eabf1380. [Google Scholar] [CrossRef]
- Li, D.; Stansfeld, P.J.; Sansom, M.S.P.; Keogh, A.; Vogeley, L.; Howe, N.; Lyons, J.A.; Aragao, D.; Fromme, P.; Fromme, R.; et al. Ternary structure reveals mechanism of a membrane diacylglycerol kinase. Nat. Commun. 2015, 6, 10140. [Google Scholar] [CrossRef] [Green Version]
- Zhu, P.; Yu, X.; Wang, C.; Zhang, Q.; Liu, W.; McSweeney, S.; Shanklin, J.; Lam, E.; Liu, Q. Structural basis for Ca2+-dependent activation of a plant metacaspase. Nat. Commun. 2020, 11, 2249. [Google Scholar] [CrossRef]
- McCoy, A.J.; Read, R.J. Experimental phasing: Best practice and pitfalls. Acta Crystallogr. D 2010, 66, 458–469. [Google Scholar] [CrossRef] [PubMed] [Green Version]




| Lysozyme + Se-Urea | PlsY + Se-Urea | |
|---|---|---|
| Data collection | ||
| Space group | P43212 | P21 |
| Cell dimensions | ||
| a, b, c (Å) | 78.73, 78.73, 37.24 | 57.88, 89.99, 53.98 |
| α, β, γ (°) | 90, 90, 90 | 90, 95.19, 90 |
| Wavelength (Å) | 0.97915 | 0.97907 |
| Resolution (Å) | 50.00–1.71 (1.77–1.71) | 50.00–1.80 (1.86–1.80) a |
| Rmerge | 0.072 (0.487) | 0.094 (0.973) |
| Rpim | 0.014 (0.097) | 0.039 (0.400) |
| <I/σ(I)> | 51.3 (8.0) | 18.6 (1.7) |
| Completeness (%) | 99.5 (100.0) | 99.6 (100.0) |
| Multiplicity | 25.5 (25.7) | 6.7 (6.8) |
| CC1/2 | 0.999 (0.974) | 1.000 (0.687) |
| Refinement | ||
| Resolution (Å) | 33.66–1.71 | 36.37–1.80 |
| No. reflections | 12,454 | 42,088 |
| Rwork/Rfree | 0.1704/0.1988 | 0.1564/0.1902 |
| No. atoms | 1156 | 3687 |
| Protein | 1009 | 3070 |
| Ligand/ion | 30 | 497 |
| Water | 117 | 120 |
| No. residues | 129 | 400 |
| B-factors (Å2) | 22.83 | 26.93 |
| Protein | 21.41 | 22.98 |
| Ligand/ion | 35.89 | 50.39 |
| Water | 31.68 | 30.80 |
| R.m.s deviations | ||
| Bond lengths (Å) | 0.010 | 0.010 |
| Bond angles (°) | 1.616 | 1.452 |
| Ramachandran | ||
| Favoured (%) | 98.43 | 98.74 |
| Allowed (%) | 1.57 | 1.26 |
| Outlier (%) | 0 | 0 |
| PDB ID | 7CQO | 7CQM |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Luo, Z.; Gu, W.; Wang, Y.; Tang, Y.; Li, D. Selenourea for Experimental Phasing of Membrane Protein Crystals Grown in Lipid Cubic Phase. Crystals 2022, 12, 976. https://doi.org/10.3390/cryst12070976
Luo Z, Gu W, Wang Y, Tang Y, Li D. Selenourea for Experimental Phasing of Membrane Protein Crystals Grown in Lipid Cubic Phase. Crystals. 2022; 12(7):976. https://doi.org/10.3390/cryst12070976
Chicago/Turabian StyleLuo, Zhipu, Weijie Gu, Yichao Wang, Yannan Tang, and Dianfan Li. 2022. "Selenourea for Experimental Phasing of Membrane Protein Crystals Grown in Lipid Cubic Phase" Crystals 12, no. 7: 976. https://doi.org/10.3390/cryst12070976






