Textile Functionalization by Porous Protein Crystal Conjugation and Guest Molecule Loading
Abstract
1. Introduction
2. Materials and Methods
3. Results
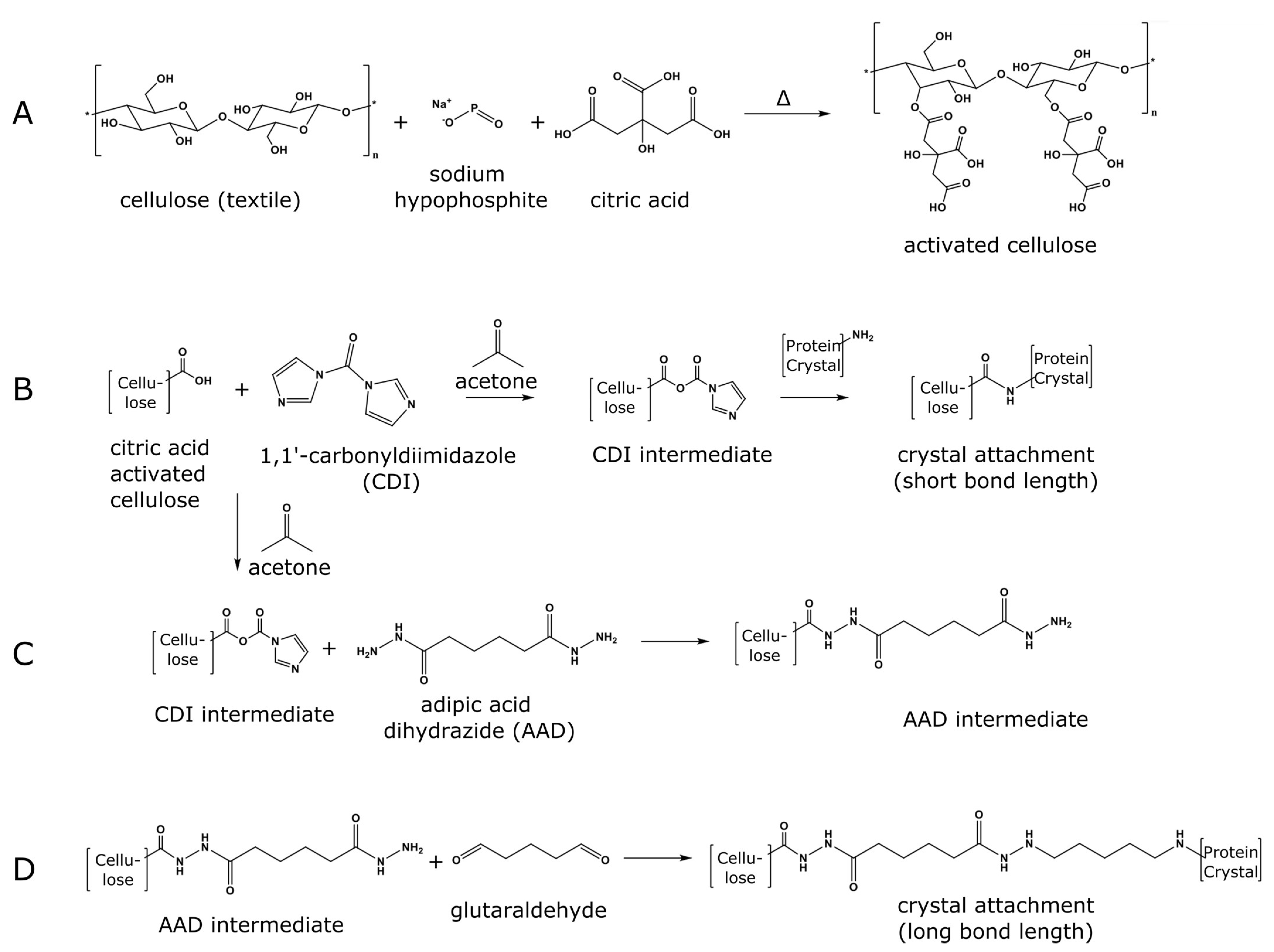
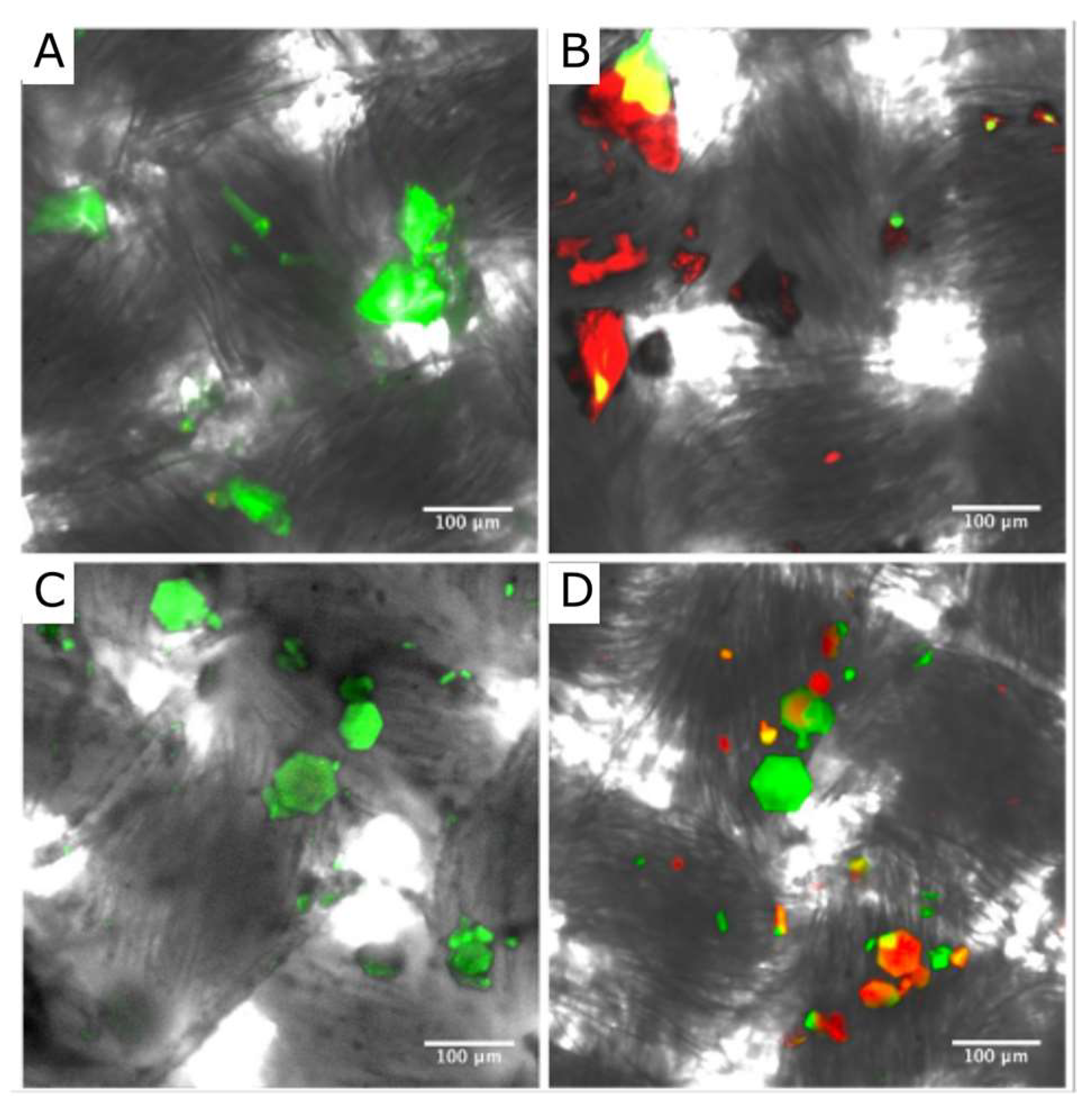
3.1. Attaching the HEWL Protein Crystals to Cotton Fabric
3.2. Assessing HEWL Crystal Attachment Strength
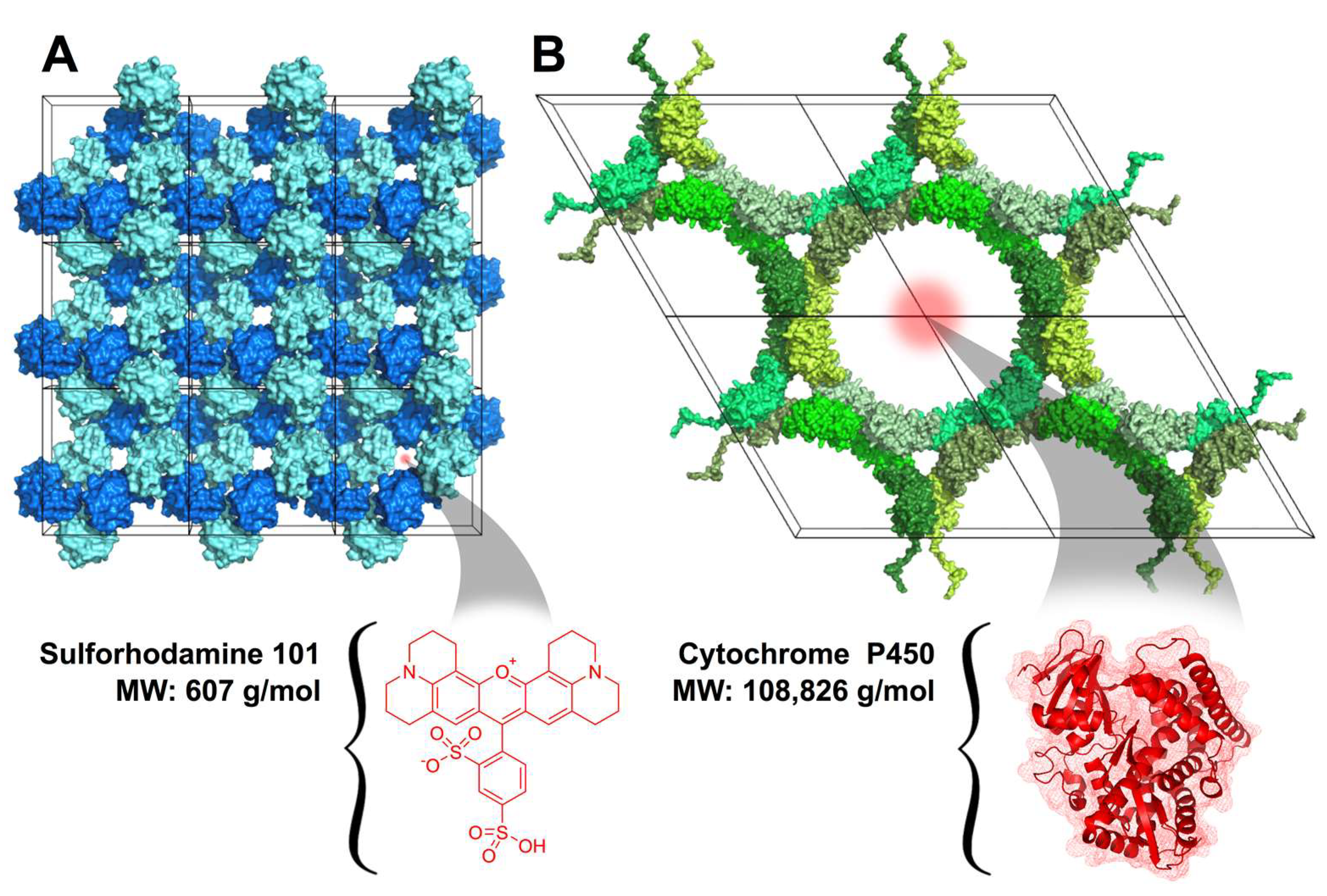
3.3. Attaching Porous Protein Crystals to Cotton Fabric
3.4. Loading Guest Molecules into Textile-Attached Protein Crystals
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tabe, H.; Shimoi, T.; Boudes, M.; Abe, S.; Coulibaly, F.; Kitagawa, S.; Mori, H.; Ueno, T. Photoactivatable CO release from engineered protein crystals to modulate NF-κB activation. Chem. Commun. 2016, 52, 4545–4548. [Google Scholar] [CrossRef] [PubMed]
- Hartje, L.F.; Bui, H.T.; Andales, D.A.; James, S.P.; Huber, T.R.; Snow, C.D. Characterizing the Cytocompatibility of Various Cross-Linking Chemistries for the Production of Biostable Large-Pore Protein Crystal Materials. ACS Biomater. Sci. Eng. 2018, 4, 826–831. [Google Scholar] [CrossRef] [PubMed]
- Noritomi, H.; Koyama, K.; Kato, S.; Nagahama, K. Increased Thermostability of Cross-Linked Enzyme Crystals of Subtilisin in Organic Solvents. Biotechnol. Tech. 1998, 12, 467–469. [Google Scholar] [CrossRef]
- Sobolov, S.B.; Bartoszko-Malik, A.; Oeschger, T.R.; Montelbano, M.M. Cross-linked enzyme crystals of fructose diphosphate aldolase: Development as a biocatalyst for synthesis. Tetrahedron Lett. 1994, 35, 7751–7754. [Google Scholar] [CrossRef]
- Sobolov, S.B.; Leonida, M.D.; Bartoszko-Malik, A.; Voivodov, K.I.; McKinney, F.; Kim, J.; Fry, A.J. Cross-Linked LDH Crystals for Lactate Synthesis Coupled to Electroenzymatic Regeneration of NADH. J. Org. Chem. 1996, 61, 2125–2128. [Google Scholar] [CrossRef]
- Khalaf, N.; Govardhan, C.P.; Lalonde, J.J.; Persichetti, R.A.; Wang, Y.-F.; Margolin, A.L. Cross-Linked Enzyme Crystals as Highly Active Catalysts in Organic Solvents. J. Am. Chem. Soc. 1996, 118, 5494–5495. [Google Scholar] [CrossRef]
- Xu, K.; Klibanov, A.M. pH Control of the Catalytic Activity of Cross-Linked Enzyme Crystals in Organic Solvents. J. Am. Chem. Soc. 1996, 118, 9815–9819. [Google Scholar] [CrossRef]
- Wang, Y.-F.; Yakovlevsky, K.; Zhang, B.; Margolin, A.L. Cross-Linked Crystals of Subtilisin: Versatile Catalyst for Organic Synthesis. J. Org. Chem. 1997, 62, 3488–3495. [Google Scholar] [CrossRef]
- Visuri, K.; Pastinen, O.; Wu, X.; Mäkinen, K.; Leisola, M. Stability of native and cross-linked crystalline glucose isomerase. Biotechnol. Bioeng. 1999, 64, 377–380. [Google Scholar] [CrossRef]
- St. Clair, N.; Wang, Y.-F.; Margolin, A.L. Cofactor-Bound Cross-Linked Enzyme Crystals (CLEC) of Alcohol Dehydrogenase. Angew. Chem. Int. Ed. 2000, 39, 380–383. [Google Scholar] [CrossRef]
- Roy, J.J.; Abraham, T.E. Continuous biotransformation of pyrogallol to purpurogallin using cross-linked enzyme crystals of laccase as catalyst in a packed-bed reactor. J. Chem. Technol. Biotechnol. 2006, 81, 1836–1839. [Google Scholar] [CrossRef]
- Lopez, S.; Rondot, L.; Leprêtre, C.; Marchi-Delapierre, C.; Ménage, S.; Cavazza, C. Cross-Linked Artificial Enzyme Crystals as Heterogeneous Catalysts for Oxidation Reactions. J. Am. Chem. Soc. 2017, 139, 17994–18002. [Google Scholar] [CrossRef] [PubMed]
- Vilenchik, L.Z.; Griffith, J.P.; St. Clair, N.; Navia, M.A.; Margolin, A.L. Protein Crystals as Novel Microporous Materials. J. Am. Chem. Soc. 1998, 120, 4290–4294. [Google Scholar] [CrossRef]
- Pastinen, O.; Visuri, K.; Leisola, M. Xylitol purification by cross-linked glucose isomerase crystals. Biotechnol. Tech. 1998, 12, 557–560. [Google Scholar] [CrossRef]
- Pastinen, O.; Jokela, J.; Eerikäinen, T.; Schwabe, T.; Leisola, M. Cross-linked glucose isomerase crystals as a liquid chromatographic separation material. Enzyme Microb. Technol. 2000, 26, 550–558. [Google Scholar] [CrossRef]
- Leisola, M.; Jokela, J.; Finell, J.; Pastinen, O. Simultaneous catalysis and product separation by cross-linked enzyme crystals. Biotechnol. Bioeng. 2001, 72, 501–505. [Google Scholar] [CrossRef]
- Vuolanto, A.; Kiviharju, K.; Nevanen, T.K.; Leisola, M.; Jokela, J. Development of Cross-Linked Antibody Fab Fragment Crystals for Enantioselective Separation of a Drug Enantiomer. Cryst. Growth Des. 2003, 3, 777–782. [Google Scholar] [CrossRef]
- Vuolanto, A.; Leisola, M.; Jokela, J. Enantioselective Affinity Chromatography of a Chiral Drug by Crystalline and Carrier-Bound Antibody Fab Fragment. Biotechnol. Prog. 2004, 20, 771–776. [Google Scholar] [CrossRef]
- Basu, S.K.; Govardhan, C.P.; Jung, C.W.; Margolin, A.L. Protein crystals for the delivery of biopharmaceuticals. Expert Opin. Biol. Ther. 2004, 4, 301–317. [Google Scholar] [CrossRef]
- Puhl, S.; Li, L.; Meinel, L.; Germershaus, O. Controlled Protein Delivery from Electrospun Non-Wovens: Novel Combination of Protein Crystals and a Biodegradable Release Matrix. Mol. Pharm. 2014, 11, 2372–2380. [Google Scholar] [CrossRef]
- Tabe, H.; Shimoi, T.; Fujita, K.; Abe, S.; Ijiri, H.; Tsujimoto, M.; Kuchimaru, T.; Kizaka-Kondo, S.; Mori, H.; Kitagawa, S.; et al. Design of a CO-releasing Extracellular Scaffold Using in Vivo Protein Crystals. Chem. Lett. 2014, 44, 342–344. [Google Scholar] [CrossRef]
- Tabe, H.; Fujita, K.; Abe, S.; Tsujimoto, M.; Kuchimaru, T.; Kizaka-Kondoh, S.; Takano, M.; Kitagawa, S.; Ueno, T. Preparation of a Cross-Linked Porous Protein Crystal Containing Ru Carbonyl Complexes as a CO-Releasing Extracellular Scaffold. Inorg. Chem. 2015, 54, 215–220. [Google Scholar] [CrossRef]
- Luiz de Mattos, I.; Lukachova, L.V.; Gorton, L.; Laurell, T.; Karyakin, A.A. Evaluation of glucose biosensors based on Prussian Blue and lyophilised, crystalline and cross-linked glucose oxidases (CLEC(R)). Talanta 2001, 54, 963–974. [Google Scholar] [CrossRef]
- Roy, J.J.; Abraham, T.E.; Abhijith, K.S.; Kumar, P.V.S.; Thakur, M.S. Biosensor for the determination of phenols based on cross-linked enzyme crystals (CLEC) of laccase. Biosens. Bioelectron. 2005, 21, 206–211. [Google Scholar] [CrossRef] [PubMed]
- Laothanachareon, T.; Champreda, V.; Sritongkham, P.; Somasundrum, M.; Surareungchai, W. Cross-linked enzyme crystals of organophosphate hydrolase for electrochemical detection of organophosphorus compounds. World J. Microbiol. Biotechnol. 2008, 24, 3049–3055. [Google Scholar] [CrossRef]
- Conejero-Muriel, M.; Rodríguez-Ruiz, I.; Verdugo-Escamilla, C.; Llobera, A.; Gavira, J.A. Continuous Sensing Photonic Lab-on-a-Chip Platform Based on Cross-Linked Enzyme Crystals. Anal. Chem. 2016, 88, 11919–11923. [Google Scholar] [CrossRef]
- Matthews, B.W. Solvent content of protein crystals. J. Mol. Biol. 1968, 33, 491–497. [Google Scholar] [CrossRef] [PubMed]
- Cvetkovic, A.; Straathof, A.J.J.; Hanlon, D.N.; van der Zwaag, S.; Krishna, R.; van der Wielen, L.A.M. Quantifying anisotropic solute transport in protein crystals using 3-D laser scanning confocal microscopy visualization. Biotechnol. Bioeng. 2004, 86, 389–398. [Google Scholar] [CrossRef] [PubMed]
- Cvetkovic, A.; Picioreanu, C.; Straathof, A.J.J.; Krishna, R.; van der Wielen, L.A.M. Quantification of binary diffusion in protein crystals. J. Phys. Chem. B 2005, 109, 10561–10566. [Google Scholar] [CrossRef]
- Cvetkovic, A.; Picioreanu, C.; Straathof, A.J.J.; Krishna, R.; Wielen, L.A.M. Relation between pore sizes of protein crystals and anisotropic solute diffusivities. J. Am. Chem. Soc. 2005, 127, 875–879. [Google Scholar] [CrossRef] [PubMed]
- Kowalski, A.E.; Huber, T.R.; Ni, T.W.; Hartje, L.F.; Appel, K.L.; Yost, J.W.; Ackerson, C.J.; Snow, C.D. Gold nanoparticle capture within protein crystal scaffolds. Nanoscale 2016, 8, 12693–12696. [Google Scholar] [CrossRef]
- Hartje, L.F.; Munsky, B.; Ni, T.W.; Ackerson, C.J.; Snow, C.D. Adsorption-Coupled Diffusion of Gold Nanoclusters within a Large-Pore Protein Crystal Scaffold. J. Phys. Chem. B 2017, 121, 7652–7659. [Google Scholar] [CrossRef]
- Huber Thaddaus, R.; Hartje Luke, F.; McPherson Eli, C.; Kowalski Ann, E.; Snow Christopher, D. Programmed Assembly of Host–Guest Protein Crystals. Small 2017, 13, 1602703. [Google Scholar] [CrossRef] [PubMed]
- Huber, T.R.; McPherson, E.C.; Keating, C.E.; Snow, C.D. Installing Guest Molecules at Specific Sites within Scaffold Protein Crystals. Bioconjug. Chem. 2018, 29, 17–22. [Google Scholar] [CrossRef] [PubMed]
- AATCC Test Method (61-2013), Color Fastness to Laundering: Accelerated, Technical Manual Method American Association of Textile Chemists and Colorists. 2017; 108.
- Hekmat, D.; Hebel, D.; Schmid, H.; Weuster-Botz, D. Crystallization of lysozyme: From vapor diffusion experiments to batch crystallization in agitated ml-scale vessels. Process Biochem. 2007, 42, 1649–1654. [Google Scholar] [CrossRef]
- Vincent Edwards, J.; Prevost, N.; Condon, B.; Sethumadhavan, K.; Ullah, J. Immobilization of Lysozyme on Cotton Fabrics: Synthesis, Characterization, and Activity. AATCC Rev. 2011, 11, 73–79. [Google Scholar]
- Edwards, J.V.; Prevost, N.T.; Condon, B.; French, A. Covalent attachment of lysozyme to cotton/cellulose materials: Protein verses solid support activation. Cellulose 2011, 18, 1239–1249. [Google Scholar] [CrossRef]
- Hermanson, G.T. Bioconjugate Techniques, 3rd ed.; Elsevier/AP: London, UK; Waltham, MA, USA, 2013; ISBN 978-0-12-382239-0. [Google Scholar]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef]
- Migneault, I.; Dartiguenave, C.; Bertrand, M.J.; Waldron, K.C. Glutaraldehyde: Behavior in aqueous solution, reaction with proteins, and application to enzyme crosslinking. BioTechniques 2004, 37, 790–796, 798–802. [Google Scholar] [CrossRef]
- Otey, C.R.; Landwehr, M.; Endelman, J.B.; Hiraga, K.; Bloom, J.D.; Arnold, F.H. Structure-guided recombination creates an artificial family of cytochromes P450. PLoS Biol. 2006, 4, e112. [Google Scholar] [CrossRef]
- Kowalski, A.E.; Johnson, L.B.; Dierl, H.K.; Park, S.; Huber, T.R.; Snow, C.D. Porous protein crystals as scaffolds for enzyme immobilization. Biomater. Sci. 2019, 7, 1898–1904. [Google Scholar] [CrossRef] [PubMed]
- Bondancia, T.J.; de Aguiar, J.; Batista, G.; Cruz, A.J.G.; Marconcini, J.M.; Mattoso, L.H.C.; Farinas, C.S. Production of Nanocellulose Using Citric Acid in a Biorefinery Concept: Effect of the Hydrolysis Reaction Time and Techno-Economic Analysis. Ind. Eng. Chem. Res. 2020, 59, 11505–11516. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hartje, L.F.; Andales, D.A.; Gintner, L.P.; Johnson, L.B.; Li, Y.V.; Snow, C.D. Textile Functionalization by Porous Protein Crystal Conjugation and Guest Molecule Loading. Crystals 2023, 13, 352. https://doi.org/10.3390/cryst13020352
Hartje LF, Andales DA, Gintner LP, Johnson LB, Li YV, Snow CD. Textile Functionalization by Porous Protein Crystal Conjugation and Guest Molecule Loading. Crystals. 2023; 13(2):352. https://doi.org/10.3390/cryst13020352
Chicago/Turabian StyleHartje, Luke F., David A. Andales, Lucas P. Gintner, Lucas B. Johnson, Yan V. Li, and Christopher D. Snow. 2023. "Textile Functionalization by Porous Protein Crystal Conjugation and Guest Molecule Loading" Crystals 13, no. 2: 352. https://doi.org/10.3390/cryst13020352
APA StyleHartje, L. F., Andales, D. A., Gintner, L. P., Johnson, L. B., Li, Y. V., & Snow, C. D. (2023). Textile Functionalization by Porous Protein Crystal Conjugation and Guest Molecule Loading. Crystals, 13(2), 352. https://doi.org/10.3390/cryst13020352








