The Promise and Challenge of High Pressure Macromolecular Crystallography
Abstract
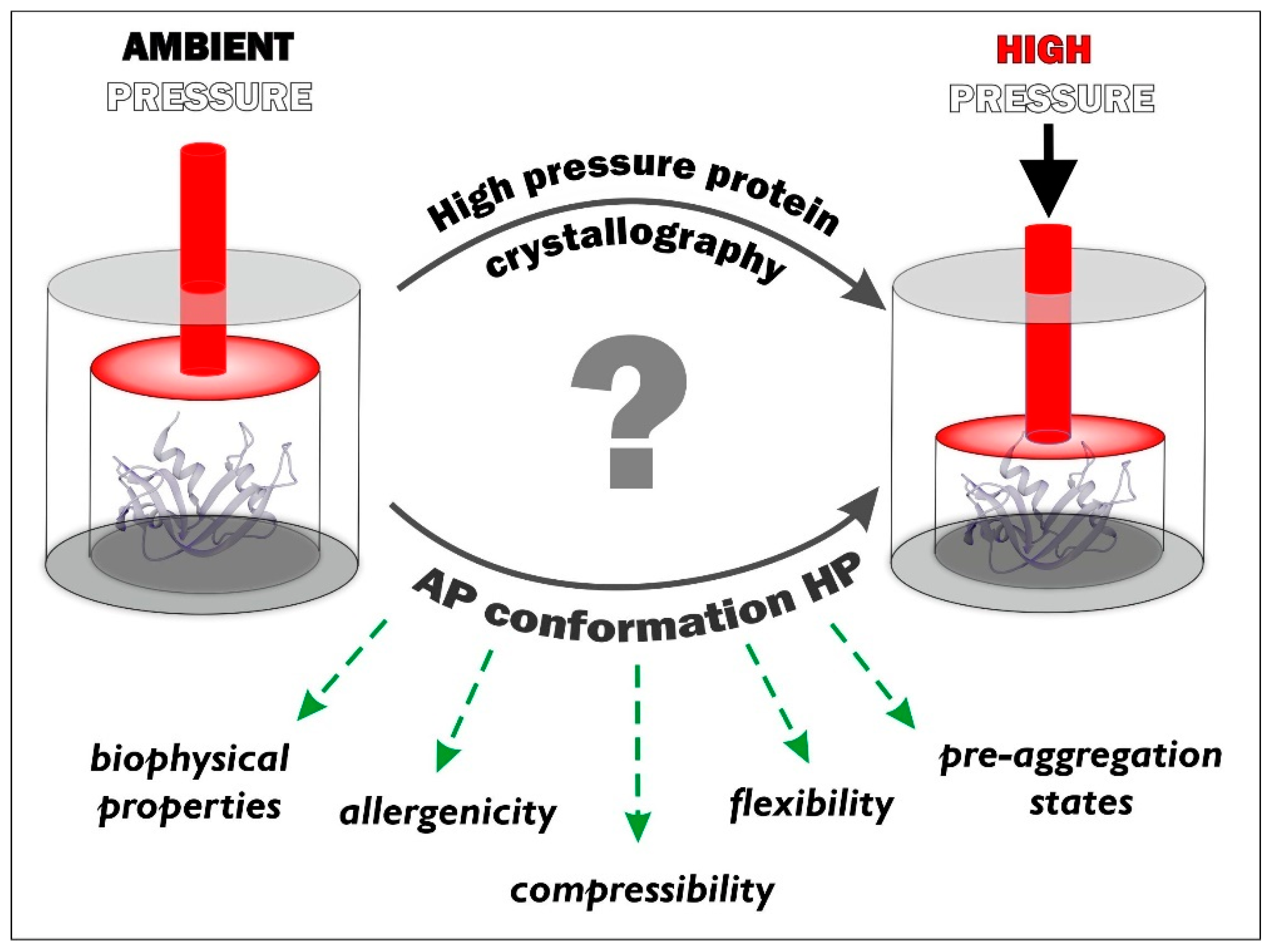
:1. Introduction
2. High Pressure in Structural Studies
3. High Pressure Macromolecular Crystallography Instrumentation
4. Examples of HPMX Achievements and Applications
5. Perspectives of HPMX
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Hermann, A.; Ashcroft, N.W.; Hoffmann, R. High Pressure Ices. Proc. Natl. Acad. Sci. USA 2012, 109, 745–750. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Collins, M.D.; Kim, C.U.; Gruner, S.M. High-Pressure Protein Crystallography and NMR to Explore Protein Conformations. Annu. Rev. Biophys. 2011, 40, 81–98. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Somkuti, J.; Houska, M.; Smeller, L. Pressure and Temperature Stability of the Main Apple Allergen Mal D1. Eur. Biophys. J. 2011, 40, 143–151. [Google Scholar] [CrossRef] [PubMed]
- Tölgyesi, E.; Böde, C.S.; Smelleri, L.; Kim, D.R.; Kim, K.K.; Heremans, K.; Fidy, J. Pressure Activation of the Chaperone Function of Small Heat Shock Proteins. Cell. Mol. Biol. 2004, 50, 361–369. [Google Scholar] [CrossRef]
- Picart, L.; Thiebaud, M.; René, M.; Guiraud, J.P.; Cheftel, J.C.; Dumay, E. Effects of High Pressure Homogenisation of Raw Bovine Milk on Alkaline Phosphatase and Microbial Inactivation. A Comparison with Continuous Short-Time Thermal Treatments. J. Dairy Res. 2006, 73, 454–463. [Google Scholar] [CrossRef]
- Chien, S.H.; Teixeira, L.A.; Cantarella, H.; Rehm, G.W.; Grant, C.A.; Gearhart, M.M. Agronomic Effectiveness of Granular Nitrogen/Phosphorus Fertilizers Containing Elemental Sulfur with and without Ammonium Sulfate: A Review. Agron. J. 2016, 108, 1203–1213. [Google Scholar] [CrossRef] [Green Version]
- de Castro Leite Júnior, B.R.; Tribst, A.A.L.; Cristianini, M. Effect of High-Pressure Technologies on Enzymes Applied in Food Processing. In Enzyme Inhibitors and Activators; IntechOpen: London, UK, 2017; pp. 49–72. [Google Scholar] [CrossRef] [Green Version]
- Eisenmenger, M.J.; Reyes-De-Corcuera, J.I. High Pressure Enhancement of Enzymes: A Review. Enzym. Microb. Technol. 2009, 45, 331–347. [Google Scholar] [CrossRef]
- Arnold, F.H. Directed Evolution: Bringing New Chemistry to Life. Angew. Chem.-Int. Ed. 2018, 57, 4143–4148. [Google Scholar] [CrossRef] [Green Version]
- Cao, Y.; Li, X.; Ge, J. Enzyme Catalyst Engineering toward the Integration of Biocatalysis and Chemocatalysis. Trends Biotechnol. 2021, 39, 1173–1183. [Google Scholar] [CrossRef]
- Torrent, J.; Marchal, S.; Tortora, P.; Lange, R.; Balny, C. High Pressure, an Alternative Approach to Understand Protein Misfolding Diseases. Mol. Cell. Biol. 2004, 50, 377–385. [Google Scholar]
- de Oliveira, G.A.P.; Marques, M.A.; Pedrote, M.M.; Silva, J.L. High Pressure Studies on the Misfolding and Aggregation of P53 in Cancer and of α-Synuclein in Parkinson’s Disease. High Press. Res. 2019, 39, 193–201. [Google Scholar] [CrossRef]
- Randolph, T.W.; Seefeldt, M.; Carpenter, J.F. High Hydrostatic Pressure as a Tool to Study Protein Aggregation and Amyloidosis. Biochim. Biophys. Acta-Protein Struct. Mol. Enzymol. 2002, 1595, 224–234. [Google Scholar] [CrossRef]
- Munir, M.; Nadeem, M.; Qureshi, T.M.; Leong, T.S.H.; Gamlath, C.J.; Martin, G.J.O.; Ashokkumar, M. Effects of High Pressure, Microwave and Ultrasound Processing on Proteins and Enzyme Activity in Dairy Systems—A Review. Innov. Food Sci. Emerg. Technol. 2019, 57, 102192. [Google Scholar] [CrossRef]
- Winter, R. Synchrotron X-Ray and Neutron Small-Angle Scattering of Lyotropic Lipid Mesophases, Model Biomembranes and Proteins in Solution at High Pressure. Biochim. Biophys. Acta 2002, 1595, 160–184. [Google Scholar] [CrossRef]
- Reich, J.A.; Aßmann, M.; Hölting, K.; Bubenheim, P.; Kuballa, J.; Liese, A. Shift of the Reaction Equilibrium at High Pressure in the Continuous Synthesis of Neuraminic Acid. Beilstein J. Org. Chem. 2022, 18, 567–579. [Google Scholar] [CrossRef]
- Ishimaru, D.; Sá-Carvalho, D.; Silva, J.L. Pressure-Inactivated FMDV: A Potential Vaccine. Vaccine 2004, 22, 2334–2339. [Google Scholar] [CrossRef]
- Yan, S.; Liu, K.; Mu, L.; Liu, J.; Tang, W.; Liu, B. Research and Application of Hydrostatic High Pressure in Tumor Vaccines (Review). Oncol. Rep. 2021, 45, 75. [Google Scholar] [CrossRef]
- Govaris, A.; Pexara, A. Inactivation of Foodborne Viruses by High-Pressure Processing (Hpp). Foods 2021, 10, 215. [Google Scholar] [CrossRef]
- Abera, G. Review on High-Pressure Processing of Foods. Cogent Food Agric. 2019, 5, 1568725. [Google Scholar] [CrossRef]
- Simonin, H.; Duranton, F.; de Lamballerie, M. New Insights into the High-Pressure Processing of Meat and Meat Products. Compr. Rev. Food Sci. Food Saf. 2012, 11, 285–306. [Google Scholar] [CrossRef]
- Diels, A.M.J.; Michiels, C.W. High-Pressure Homogenization as a Non-Thermal Technique for the Inactivation of Microorganisms. Crit. Rev. Microbiol. 2006, 32, 201–216. [Google Scholar] [CrossRef]
- Wang, Q.; Liu, Y.; Zhang, C.; Guo, F.; Feng, C.; Li, X.; Shi, H.; Su, Z. High Hydrostatic Pressure Enables Almost 100% Refolding of Recombinant Human Ciliary Neurotrophic Factor from Inclusion Bodies at High Concentration. Protein Expr. Purif. 2017, 133, 152–159. [Google Scholar] [CrossRef] [PubMed]
- Goto, M.; Kazama, A.; Fukuhara, K.; Sato, H.; Tamai, N.; Ito, H.-O.; Matsuki, H. Membrane Fusion of Phospholipid Bilayers under High Pressure: Spherical and Irreversible Growth of Giant Vesicles. Biophys. Chem. 2021, 277, 106639. [Google Scholar] [CrossRef] [PubMed]
- Caliò, A.; Dubois, C.; Fontanay, S.; Koza, M.M.; Hoh, F.; Roumestand, C.; Oger, P.; Peters, J. Unravelling the Adaptation Mechanisms to High Pressure in Proteins. Int. J. Mol. Sci. 2022, 23, 8649. [Google Scholar] [CrossRef] [PubMed]
- Lerch, M.T.; Horwitz, J.; McCoy, J.; Hubbell, W.L. Circular Dichroism and Site-Directed Spin Labeling Reveal Structural and Dynamical Features of High-Pressure States of Myoglobin. Proc. Natl. Acad. Sci. USA 2013, 110, E4714–E4722. [Google Scholar] [CrossRef] [Green Version]
- Maeno, A.; Akasaka, K. High-Pressure Fluorescence Spectroscopy. Subcell Biochem. 2015, 72, 687–705. [Google Scholar] [CrossRef]
- Dzwolak, W.; Kato, M.; Taniguchi, Y. Fourier Transform Infrared Spectroscopy in High-Pressure Studies on Proteins. Biochim. Biophys. Acta-Protein Struct. Mol. Enzymol. 2002, 1595, 131–144. [Google Scholar] [CrossRef]
- Rai, D.K.; Gillilan, R.E.; Huang, Q.; Miller, R.; Ting, E.; Lazarev, A.; Tate, M.W.; Gruner, S.M. High-Pressure Small-Angle X-Ray Scattering Cell for Biological Solutions and Soft Materials. J. Appl. Crystallogr. 2021, 54, 111–122. [Google Scholar] [CrossRef]
- Teixeira, S.C.M. High-Pressure Small-Angle Neutron Scattering for Food Studies. Curr. Opin. Colloid Interface Sci. 2019, 42, 99–109. [Google Scholar] [CrossRef]
- Roche, J.; Royer, C.A.; Roumestand, C. Monitoring Protein Folding through High Pressure NMR Spectroscopy. Prog. Nucl. Magn. Reson. Spectrosc. 2017, 102–103, 15–31. [Google Scholar] [CrossRef]
- Fourme, R.; Kahn, R.; Mezouar, M.; Girard, E.; Hoerentrup, C.; Prangé, T.; Ascone, I. High-Pressure Protein Crystallography (HPPX): Instrumentation, Methodology and Results on Lysozyme Crystals. J. Synchrotron Radiat. 2001, 8, 1149–1156. [Google Scholar] [CrossRef] [Green Version]
- Shen, G.; Mao, H.K. High-Pressure Studies with x-Rays Using Diamond Anvil Cells. Rep. Prog. Phys. 2017, 80, 16101. [Google Scholar] [CrossRef] [Green Version]
- Rahm, M.; Cammi, R.; Ashcroft, N.W.; Hoffmann, R. Squeezing All Elements in the Periodic Table: Electron Configuration and Electronegativity of the Atoms under Compression. J. Am. Chem. Soc. 2019, 141, 10253–10271. [Google Scholar] [CrossRef]
- Burley, S.K.; Bhikadiya, C.; Bi, C.; Bittrich, S.; Chen, L.; Crichlow, G.V.; Christie, C.H.; Dalenberg, K.; Di Costanzo, L.; Duarte, J.M.; et al. RCSB Protein Data Bank: Powerful New Tools for Exploring 3D Structures of Biological Macromolecules for Basic and Applied Research and Education in Fundamental Biology, Biomedicine, Biotechnology, Bioengineering and Energy Sciences. Nucleic Acids Res. 2021, 49, D437–D451. [Google Scholar] [CrossRef]
- Girard, E.; Kahn, R.; Mezouar, M.; Dhaussy, A.C.; Lin, T.; Johnson, J.E.; Fourme, R. The First Crystal Structure of a Macromolecular Assembly under High Pressure: CpMV at 330 MPa. Biophys. J. 2005, 88, 3562–3571. [Google Scholar] [CrossRef] [Green Version]
- Lin, T.; Schildkamp, W.; Brister, K.; Doerschuk, P.C.; Somayazulu, M.; Mao, H.K.; Johnson, J.E. The Mechanism of High-Pressure-Induced Ordering in a Macromolecular Crystal. Acta Crystallogr. Sect. D Biol. Crystallogr. 2005, 61, 737–743. [Google Scholar] [CrossRef]
- Yamada, H.; Nagae, T.; Watanabe, N. High-Pressure Protein Crystallography of Hen Egg-White Lysozyme. Acta Crystallogr. Sect. D Biol. Crystallogr. 2015, 71, 742–753. [Google Scholar] [CrossRef] [Green Version]
- Kurpiewska, K.; Miłaczewska, A.; Lewiński, K. Insulin Conformational Changes under High Pressure in Structural Studies and Molecular Dynamics Simulations. J. Mol. Struct. 2020, 1202, 127251. [Google Scholar] [CrossRef]
- Collins, M.D.; Hummer, G.; Quillin, M.L.; Matthews, B.W.; Gruner, S.M. Cooperative Water Filling of a Nonpolar Protein Cavity Observed by High-Pressure Crystallography and Simulation. Proc. Natl. Acad. Sci. USA 2005, 102, 16668–16671. [Google Scholar] [CrossRef] [Green Version]
- Collins, M.D.; Quillin, M.L.; Hummer, G.; Matthews, B.W.; Gruner, S.M. Structural Rigidity of a Large Cavity-Containing Protein Revealed by High-Pressure Crystallography. J. Mol. Biol. 2007, 367, 752–763. [Google Scholar] [CrossRef] [Green Version]
- Prangé, T.; Girard, E.; Fourme, R.; Dhaussy, A.C.; Edwards, B.; Vaishnav, A.; Patel, C.; Guy-Evans, H.; Hervé, G.; Evans, D.R. Pressure-Induced Activation of Latent Dihydroorotase from Aquifex Aeolicus as Revealed by High Pressure Protein Crystallography. FEBS J. 2019, 286, 1204–1213. [Google Scholar] [CrossRef] [PubMed]
- Ascone, I.; Savino, C.; Kahn, R.; Fourme, R. Flexibility of the Cu,Zn Superoxide Dismutase Structure Investigated at 0.57 GPa. Acta Crystallogr. Sect. D 2010, 66, 654–663. [Google Scholar] [CrossRef] [PubMed]
- Girard, E.; Dhaussy, A.C.; Couzinet, B.; Chervin, J.C.; Mezouar, M.; Kahn, R.; Ascone, I.; Fourme, R. Toward Fully Fledged High-Pressure Macromolecular Crystallography. J. Appl. Crystallogr. 2007, 40, 912–918. [Google Scholar] [CrossRef]
- Kundrot, C.E.; Richards, F.M. Crystal Structure of Hen Egg-White Lysozyme at a Hydrostatic Pressure of 1000 Atmospheres. J. Mol. Biol. 1987, 193, 157–170. [Google Scholar] [CrossRef]
- Nagae, T.; Kawamura, T.; Chavas, L.M.G.; Niwa, K.; Hasegawa, M.; Kato, C.; Watanabe, N. High-Pressure-Induced Water Penetration into 3-Isopropylmalate Dehydrogenase. Acta Crystallogr. Sect. D Biol. Crystallogr. 2012, 68, 300–309. [Google Scholar] [CrossRef] [Green Version]
- Ascone, I.; Kahn, R.; Girard, E.; Prangé, T.; Dhaussy, A.C.; Mezouar, M.; Ponikwicki, N.; Fourme, R. Isothermal Compressibility of Macromolecular Crystals and Macromolecules Derived from High-Pressure X-Ray Crystallography. J. Appl. Crystallogr. 2010, 43, 407–416. [Google Scholar] [CrossRef]
- Colloc’h, N.; Sacquin-Mora, S.; Avella, G.; Dhaussy, A.C.; Prangé, T.; Vallone, B.; Girard, E. Determinants of Neuroglobin Plasticity Highlighted by Joint Coarse-Grained Simulations and High Pressure Crystallography. Sci. Rep. 2017, 7, 1858. [Google Scholar] [CrossRef] [Green Version]
- Kurpiewska, K.; Dziubek, K.; Katrusiak, A.; Font, J.; Ribò, M.; Vilanova, M.; Lewiński, K. Structural Investigation of Ribonuclease A Conformational Preferences Using High Pressure Protein Crystallography. Chem. Phys. 2016, 468, 53–62. [Google Scholar] [CrossRef]
- Hamajima, Y.; Nagae, T.; Watanabe, N.; Ohmae, E.; Kato-Yamada, Y.; Kato, C. Pressure Adaptation of 3-Isopropylmalate Dehydrogenase from an Extremely Piezophilic Bacterium Is Attributed to a Single Amino Acid Substitution. Extremophiles 2016, 20, 177–186. [Google Scholar] [CrossRef]
- Zhang, Z.; Yang, Y.; Zhou, P.; Zhang, X.; Wang, J. Effects of High Pressure Modification on Conformation and Gelation Properties of Myofibrillar Protein. Food Chem. 2017, 217, 678–686. [Google Scholar] [CrossRef]
- Prangé, T.; Carpentier, P.; Dhaussy, A.-C.; van der Linden, P.; Girard, E.; Colloc’h, N. Comparative Study of the Effects of High Hydrostatic Pressure per Se and High Argon Pressure on Urate Oxidase Ligand Stabilization. Acta Crystallogr. Sect. D 2022, 78, 162–173. [Google Scholar] [CrossRef]
- Girard, E.; Marchal, S.; Perez, J.; Finet, S.; Kahn, R.; Fourme, R.; Marassio, G.; Dhaussy, A.-C.; Prangé, T.; Giffard, M.; et al. Structure-Function Perturbation and Dissociation of Tetrameric Urate Oxidase by High Hydrostatic Pressure. Biophys. J. 2010, 98, 2365–2373. [Google Scholar] [CrossRef] [Green Version]
- Cao, Y.; Xia, T.; Zhou, G.; Xu, X. The Mechanism of High Pressure-Induced Gels of Rabbit Myosin. Innov. Food Sci. Emerg. Technol. 2012, 16, 41–46. [Google Scholar] [CrossRef]
- Guo, Z.; Huang, Z.; Guo, Y.; Li, B.; Yu, W.; Zhou, L.; Jiang, L.; Teng, F.; Wang, Z. Effects of High-Pressure Homogenization on Structural and Emulsifying Properties of Thermally Soluble Aggregated Kidney Bean (Phaseolus Vulgaris L.) Proteins. Food Hydrocoll. 2021, 119, 106835. [Google Scholar] [CrossRef]
- Chen, Y.; Xu, A.; Yang, R.; Jia, R.; Zhang, J.; Xu, D.; Yang, W. Myofibrillar Protein Structure and Gel Properties of Trichiurus Haumela Surimi Subjected to High Pressure or High Pressure Synergistic Heat. Food Bioprocess Technol. 2020, 13, 589–598. [Google Scholar] [CrossRef]
- Li, Z.; Liu, H.; Ma, R.; Tang, B.; Pan, D.; Peng, Y.; Ling, X.; Wang, Y.; Wu, X.; Che, L.; et al. Changes to the Tropomyosin Structure Alter the Angiotensin-Converting Enzyme Inhibitory Activity and Texture Profiles of Eel Balls under High Hydrostatic Pressure. Food Funct. 2018, 9, 6535–6543. [Google Scholar] [CrossRef]
- Cepero-Betancourt, Y.; Opazo-Navarrete, M.; Janssen, A.E.M.; Tabilo-Munizaga, G.; Pérez-Won, M. Effects of High Hydrostatic Pressure (HHP) on Protein Structure and Digestibility of Red Abalone (Haliotis Rufescens) Muscle. Innov. Food Sci. Emerg. Technol. 2020, 60, 102282. [Google Scholar] [CrossRef]
- Sun, Y.S.; Zhao, Z.; Yang, Z.N.; Xu, F.; Lu, H.J.; Zhu, Z.Y.; Shi, W.; Jiang, J.; Yao, P.P.; Zhu, H.P. Risk Factors and Preventions of Breast Cancer. Int. J. Biol. Sci. 2017, 13, 1387–1397. [Google Scholar] [CrossRef] [Green Version]
- Piermarini, G.J.; Weir, C.E. A Diamond Cell for X-Ray Diffraction Studies at High Pressures. J. Res. Natl. Bur. Stand. Sect. A Phys. Chem. 1962, 66, 325. [Google Scholar] [CrossRef]
- Block, S.; Weir, C.W.; Piermarini, G.J. High-Pressure Single-Crystal Studies of Ice VI. Science 1965, 148, 947–948. [Google Scholar] [CrossRef]
- Takele, S.; Hearne, G.R. Magnetic-Electronic Properties of FeS and Fe7S8 Studied by 57Fe Mössbauer and Electrical Measurements at High Pressure and Variable Temperatures. J. Phys. Condens. Matter 2001, 13, 10077–10088. [Google Scholar] [CrossRef]
- O’Bannon, E.F.; Jenei, Z.; Cynn, H.; Lipp, M.J.; Jeffries, J.R. Contributed Review: Culet Diameter and the Achievable Pressure of a Diamond Anvil Cell: Implications for the Upper Pressure Limit of a Diamond Anvil Cell. Rev. Sci. Instrum. 2018, 89, 111501. [Google Scholar] [CrossRef] [PubMed]
- Anzellini, S.; Boccato, S. A Practical Review of the Laser-Heated Diamond Anvil Cell for University Laboratories and Synchrotron Applications. Crystals 2020, 10, 459. [Google Scholar] [CrossRef]
- Forman, R.A.; Piermarini, G.J.; Dean Barnett, J.; Block, S. Pressure Measurement Made by the Utilization of Ruby Sharp-Line Luminescence. Science 1972, 176, 284–285. [Google Scholar] [CrossRef] [PubMed]
- Prangé, T.; Colloc’h, N.; Dhaussy, A.C.; Lecouvey, M.; Migianu-Griffoni, E.; Girard, E. Behavior of B-and Z-DNA Crystals under High Hydrostatic Pressure. Crystals 2022, 12, 871. [Google Scholar] [CrossRef]
- Katrusiak, A.; Dauter, Z. Compressibility of Lysozyme Protein Crystals by X-Ray Diffraction. Acta Crystallogr. Sect. D Biol. Crystallogr. 1996, 52, 607–608. [Google Scholar] [CrossRef] [Green Version]
- Fourme, R.; Girard, E.; Dhaussy, A.C.; Medjoubi, K.; Prangé, T.; Ascone, I.; Mezouar, M.; Kahn, R. A New Paradigm for Macromolecular Crystallography Beamlines Derived from High-Pressure Methodology and Results. J. Synchrotron Radiat. 2011, 18, 31–36. [Google Scholar] [CrossRef] [Green Version]
- Fourme, R.; Ascone, I.; Kahn, R.; Mezouar, M.; Bouvier, P.; Girard, E.; Lin, T.; Johnson, J.E. Opening the High-Pressure Domain beyond 2 Kbar to Protein and Virus Crystallography: Technical Advance. Structure 2002, 10, 1409–1414. [Google Scholar] [CrossRef]
- Girard, E.; Prangé, T.; Dhaussy, A.C.; Migianu-Griffoni, E.; Lecouvey, M.; Chervin, J.C.; Mezouar, M.; Kahn, R.; Fourme, R. Adaptation of the Base-Paired Double-Helix Molecular Architecture to Extreme Pressure. Nucleic Acids Res. 2007, 35, 4800–4808. [Google Scholar] [CrossRef] [Green Version]
- Colloc’h, N.; Girard, E.; Dhaussy, A.C.; Kahn, R.; Ascone, I.; Mezouar, M.; Fourme, R. High Pressure Macromolecular Crystallography: The 140-MPa Crystal Structure at 2.3 Å Resolution of Urate Oxidase, a 135-KDa Tetrameric Assembly. Biochim. Biophys. Acta-Proteins Proteom. 2006, 1764, 391–397. [Google Scholar] [CrossRef]
- Meents, A.; Wiedorn, M.O.; Srajer, V.; Henning, R.; Sarrou, I.; Bergtholdt, J.; Barthelmess, M.; Reinke, P.Y.A.; Dierksmeyer, D.; Tolstikova, A.; et al. Pink-Beam Serial Crystallography. Nat. Commun. 2017, 8, 1281. [Google Scholar] [CrossRef] [Green Version]
- Quirnheim Pais, D.; Rathmann, B.; Koepke, J.; Tomova, C.; Wurzinger, P.; Thielmann, Y. A Standardized Technique for High-Pressure Cooling of Protein Crystals. Acta Crystallogr. Sect. D Struct. Biol. 2017, 73, 997–1006. [Google Scholar] [CrossRef]
- Kurpiewska, K.; Lewiński, K. High Pressure Macromolecular Crystallography for Structural Biology: A Review. Cent. Eur. J. Biol. 2010, 5, 531–542. [Google Scholar] [CrossRef]
- Kaneshina, S. High Pressure Bioscience. Rev. High Press. Sci. Technol. 1999, 9, 182. [Google Scholar] [CrossRef]
- Marassio, G.; Prange, T.; David, H.N.; Santos, J.S.O.; Gabison, L.; Delcroix, N.; Abraini, J.H.; Colloc’h, N. Pressure-response Analysis of Anesthetic Gases Xenon and Nitrous Oxide on Urate Oxidase: A Crystallographic Study. FASEB J. 2011, 25, 2266–2275. [Google Scholar] [CrossRef]
- Lafumat, B.; Mueller-Dieckmann, C.; Leonard, G.; Colloc’H, N.; Prangé, T.; Giraud, T.; Dobias, F.; Royant, A.; Van Der Linden, P.; Carpentier, P. Gas-Sensitive Biological Crystals Processed in Pressurized Oxygen and Krypton Atmospheres: Deciphering Gas Channels in Proteins Using a Novel “soak-and-Freeze” Methodology. J. Appl. Crystallogr. 2016, 49, 1478–1487. [Google Scholar] [CrossRef]
- Abraini, J.H.; Marassio, G.; David, H.N.; Vallone, B.; Prangé, T.; Colloc’h, N. Crystallographic Studies with Xenon and Nitrous Oxide Provide Evidence for Protein-Dependent Processes in the Mechanisms of General Anesthesia. Anesthesiology 2014, 121, 1018–1027. [Google Scholar] [CrossRef] [Green Version]
- Ardiccioni, C.; Arcovito, A.; Della Longa, S.; Van Der Linden, P.; Bourgeois, D.; Weik, M.; Montemiglio, L.C.; Savino, C.; Avell, G.; Exertier, C.; et al. Ligand Pathways in Neuroglobin Revealed by Low-Temperature Photodissociation and Docking Experiments. IUCrJ 2019, 6, 832–842. [Google Scholar] [CrossRef]
- Colloc’h, N.; Carpentier, P.; Montemiglio, L.C.; Vallone, B.; Prangé, T. Mapping Hydrophobic Tunnels and Cavities in Neuroglobin with Noble Gas under Pressure. Biophys. J. 2017, 113, 2199–2206. [Google Scholar] [CrossRef] [Green Version]
- Moschetti, T.; Mueller, U.; Schulze, J.; Brunori, M.; Vallone, B. The Structure of Neuroglobin at High Xe and Kr Pressure Reveals Partial Conservation of Globin Internal Cavities. Biophys. J. 2009, 97, 1700–1708. [Google Scholar] [CrossRef] [Green Version]
- Morrison, C.N.; Hoy, J.A.; Zhang, L.; Einsle, O.; Rees, D.C. Substrate Pathways in the Nitrogenase MoFe Protein by Experimental Identification of Small Molecule Binding Sites. Biochemistry 2015, 54, 2052–2060. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nagae, T.; Yamada, H.; Watanabe, N. High-Pressure Protein Crystal Structure Analysis of Escherichia Coli Dihydrofolate Reductase Complexed with Folate and NADP+. Acta Crystallogr. Sect. D Struct. Biol. 2018, 74, 895–905. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kurpiewska, K.; Biela, A.; Loch, J.I.; Świątek, S.; Jachimska, B.; Lewiński, K. Investigation of High Pressure Effect on the Structure and Adsorption of β-Lactoglobulin. Colloids Surf. B Biointerfaces 2018, 161, 387–393. [Google Scholar] [CrossRef] [PubMed]
- Saleem-Batcha, R.; Stull, F.; Sanders, J.N.; Moore, B.S.; Palfey, B.A.; Houk, K.N.; Teufel, R. Enzymatic Control of Dioxygen Binding and Functionalization of the Flavin Cofactor. Proc. Natl. Acad. Sci. USA 2018, 115, 4909–4919. [Google Scholar] [CrossRef] [Green Version]
- Matthews, A.; Saleem-Batcha, R.; Sanders, J.N.; Stull, F.; Houk, K.N.; Teufel, R. Aminoperoxide Adducts Expand the Catalytic Repertoire of Flavin Monooxygenases. Nat. Chem. Biol. 2020, 16, 556–563. [Google Scholar] [CrossRef]
- Markova, K.; Chmelova, K.; Marques, S.M.; Carpentier, P.; Bednar, D.; Damborsky, J.; Marek, M. Decoding the Intricate Network of Molecular Interactions of a Hyperstable Engineered Biocatalyst. Chem. Sci. 2020, 11, 11162–11178. [Google Scholar] [CrossRef]
- Zacarias, S.; Temporão, A.; Carpentier, P.; van der Linden, P.; Pereira, I.A.C.; Matias, P.M. Exploring the Gas Access Routes in a [NiFeSe] Hydrogenase Using Crystals Pressurized with Krypton and Oxygen. J. Biol. Inorg. Chem. 2020, 25, 863–874. [Google Scholar] [CrossRef]
- Carpentier, P.; Leprêtre, C.; Basset, C.; Douki, T.; Torelli, S.; Duarte, V.; Hamdane, D.; Fontecave, M.; Atta, M. Structural, Biochemical and Functional Analyses of TRNA-Monooxygenase Enzyme MiaE from Pseudomonas Putida Provide Insights into TRNA/MiaE Interaction. Nucleic Acids Res. 2020, 48, 9918–9930. [Google Scholar] [CrossRef]
- Girard, E.; Lopes, P.; Spoerner, M.; Dhaussy, A.C.; Prangé, T.; Kalbitzer, H.R.; Colloc’h, N. Equilibria between Conformational States of the Ras Oncogene Protein Revealed by High Pressure Crystallography. Chem. Sci. 2022, 13, 2001–2010. [Google Scholar] [CrossRef]
- Melnikov, I.; Orekhov, P.; Rulev, M.; Kovalev, K.; Astashkin, R.; Bratanov, D.; Ryzhykau, Y.; Balandin, T.; Bukhdruker, S.; Okhrimenko, I.; et al. High-Pressure Crystallography Shows Noble Gas Intervention into Protein-Lipid Interaction and Suggests a Model for Anaesthetic Action. Commun. Biol. 2022, 5, 360. [Google Scholar] [CrossRef]
- Fourme, R.; Girard, E.; Akasaka, K. High-Pressure Macromolecular Crystallography and NMR: Status, Achievements and Prospects. Curr. Opin. Struct. Biol. 2012, 22, 636–642. [Google Scholar] [CrossRef]
- Kurpiewska, K.; Biela, A.; Loch, J.I.; Lipowska, J.; Siuda, M.; Lewiński, K. Towards Understanding the Effect of High Pressure on Food Protein Allergenicity: β-Lactoglobulin Structural Studies. Food Chem. 2019, 270, 315–321. [Google Scholar] [CrossRef]
- Shima, F.; Ijiri, Y.; Muraoka, S.; Liao, J.; Ye, M.; Araki, M.; Matsumoto, K.; Yamamoto, N.; Sugimoto, T.; Yoshikawa, Y.; et al. Structural Basis for Conformational Dynamics of GTP-Bound Ras Protein. J. Biol. Chem. 2010, 285, 22696–22705. [Google Scholar] [CrossRef] [Green Version]
- Kundrot, C.E.; Richards, F.M. Effect of Hydrostatic Pressure on the Solvent in Crystals of Hen Egg-White Lysozyme. J. Mol. Biol. 1988, 200, 401–410. [Google Scholar] [CrossRef]
- Mertens, H.D.T.; Svergun, D.I. Structural Characterization of Proteins and Complexes Using Small-Angle X-Ray Solution Scattering. J. Struct. Biol. 2010, 172, 128–141. [Google Scholar] [CrossRef]
- Krywka, C.; Sternemann, C.; Paulus, M.; Tolan, M.; Royer, C.; Winter, R. Effect of Osmolytes on Pressure-Induced Unfolding of Proteins: A High-Pressure SAXS Study. ChemPhysChem 2008, 9, 2809–2815. [Google Scholar] [CrossRef]
- Jaworek, M.W.; Möbitz, S.; Gao, M.; Winter, R. Stability of the Chaperonin System GroEL-GroES under Extreme Environmental Conditions. Phys. Chem. Chem. Phys. 2020, 22, 3734–3743. [Google Scholar] [CrossRef]
- König, N.; Paulus, M.; Julius, K.; Schulze, J.; Voetz, M.; Tolan, M. Antibodies under Pressure: A Small-Angle X-Ray Scattering Study of Immunoglobulin G under High Hydrostatic Pressure. Biophys. Chem. 2017, 231, 45–49. [Google Scholar] [CrossRef]
- Miller, R.C.; Cummings, C.; Huang, Q.; Ando, N.; Gillilan, R.E. Inline Small-Angle X-Ray Scattering-Coupled Chromatography under Extreme Hydrostatic Pressure. Protein Sci. 2022, 31, e4489. [Google Scholar] [CrossRef]
- Boldyreva, E.V. High-Pressure Studies of Crystalline Amino Acids and Simple Peptides. High Press. Biosci. Biotechnol. 2006, 1, 28–46. [Google Scholar]





| Sample | Space Group | Pressure [MPa] | Resolution [A] | PDB id | Reference | Structure of Monomer |
|---|---|---|---|---|---|---|
| Cu,Zn Superoxide dismutase | P212121 | 570 | 2.00 | 3HW7 | [43] |  |
| Urate oxidase | I222 | 0.1; 0.2; 0.5; 0.8; 1.0; 1.5; 1.8; 2.0; 3.0; 3.2; 3.5; 4.2; 6.5; 9.0 | 1.10–1.85 | 3PK5, PK6, 3PKF, 3PK8, 3PLE, 3PKK, 3PKL, 3PKU, 3PLG, 3PJK, 3PKH, 3PKS, 3PLH, 3PLI, 3PKG, 3PKT, 3PLM, 3PLJ, 3PK3, 3PK4, 5FRC, 6I9X, 6I9Z, 6IC1 | [52,76,77] |  |
| 12; 22 | 1.69–2.36 | 6IA1, 6IA1, 6I19 | [52] | |||
| 200; 210; 310 | 1.50–2.40 | 6RGM, 7P0D, 7P0C | [52] | |||
| P21212 | 60; 100; 150; 200 | 1.64–2.19 | 6IA9, 7PUF, 7PWN, 7Q09 | [52] | ||
| 3-Isopropylmalate dehydrogenase from Shewanella oneidensis MR-1 | C121 | 0.1 | 1.90 | 3WZV, 3WZX | [50] |  |
| 160, 340, 410, 580, 650 | 1.55–2.20 | 3VL2, 3VL3, 3VL4, 3VL6, 3VL7, 3WZY, 3WZW | [46,50] | |||
| Lysozyme | P43212 | 0.1; 3; 4.5; 10.0 | 1.20–1.65 | 4NWE, 4NWH, 4WLD, 5FSJ, 5FST | [38,77,78] |  |
| 190, 280, 380, 440, 500, 600, 710, 800, 890, 920 | 1.55–1.70 | 4WLT, 4WLX, 4WLY, 4WM1, 4WM2, 4WM3, 4WM4, 4WM5, 4XEN, 6Q8R *, 8F2G * | [38] | |||
| P43 | 950 | 1.85 | 4WM6 | [38] | ||
| Myoglobin | P1211 | 3 | 1.55–1.60 | 4NXA, 4NXC | [78] |  |
| Murine neuroglobine | H32 | 1; 2; 3; 5; 8; 10; 15 | 1.60–2.31 | 4O4T, 4O4Z, 5MJD, 5MJC, 5NVI, 5NW6, 5O1K, 5O17, 5O27, 3GK9, 3GKT, 3GKN, 6R1Q * | [78,79,80,81] |  |
| 150, 240, 270, 280, 310 | 1.90–2.40 | 5EV5, 5EYJ, 5EOH, 5F0B, 5EQM, 5F2A | [48] | |||
| Nitrogenase MoFe Protein from Clostridium | P1 | 24.5 | 1.90 | 4WN9 | [82] |  |
| Nitrogenase MoFe Protein from Azotobacter vinelandii | P1211 | 24.5 | 2.00 | 4WNA | [81] |  |
| Dihydrofolate reductase from Escherichia coli complexed with folate and NADP+ | P1211 | 0.1 | 1.70 | 4X5F | [83] |  |
| P1211 | 270 | 1.90 | 4X5G | [83] | ||
| C121 | 500, 660, 750 | 1.80–1.90 | 4X5H, 4X5I, 4X5J | [83] | ||
| Dihydrofolate reductase from Escherichia coli | P212121 | 0.1 | 1.80 | 5Z6F, 5Z6L, 5Z6M, 5Z6J, 5Z6K | [83] |  |
| Thermolysin | P6122 | 4; 10; 45 | 1.20–1.70 | 5FSP, 5FSJ, 5FSS | [77] |  |
| Bovine beta-lactoglobulin complex with dodecane | P3221 | 430-550 | 2.85 | 5IO7, 5LKF | [84] |  |
| Uncharacterized Protein Lpg1496 from Legionella pneumophila | P42212 | 300; 350 | 1.48 | 5T8B *, 5T8C * | - |  |
| Vacuolar-sorting protein Snf7 | P1211 | 200; 350 | 2.20 | 5T8N *, 5T8L * | - |  |
| Putative FAD-dependent oxygenase EncM complexed with dioxygen | C222 | 0.5; 1; 1.5 | 1.39–2.04 | 6FOW, 6FOQ, 6FP3, 6FYC | [85] |  |
| Putative FAD-dependent oxygenase EncM complex with xenon | I222 | 1.5 | 3.65 | 6FY9 | [85] |  |
| Dihydroorotase from Aquifex aeolicus | C2221 | 60; 120 | 2.50–2.60 | 6GDF, 6GDD | [42] |  |
| Bovine insulin | I213 | 60; 100; 200 | 2.00–2.15 | 6Q8Q *, 6QQG, 6QRK, 6QRH | [39] |  |
| Elastase (PPE) | P212121 | 200 | 1.80 | 6Q8S * | - |  |
| Thermolysine | P6122 | 200 | 1.85 | 6QAR * | - |  |
| Horse spleen apoferritin | F432 | 200 | 2.00 | 6RA8 * | - |  |
| Monooxygenase RutA complexed with dioxygen/uracil and dioxygen | P3121 | 0.5; 15 | 1.80 | 6SGG, 6TEF, 6TEEG | [86] |  |
| Hyperstable haloalkane dehalogenase variant DhaA115 | P212121 | 15 | 1.55 | 6SP8 | [87] |  |
| [NiFeSe] hydrogenase from Desulfovibrio vulgaris hildenborough | P1211 | 7.5; 10 | 1.09–2.20 | 6Z7R, 6Z8J, 6Z8O, 6Z9G | [88] |  |
| C121 | 7.5; 10 | 1.02–1.53 | 6Z8M, 6Z9O | [88] | ||
| tRNA-monooxygenase | C121 | 200 | 1.80–2.50 | 6ZMC, 6ZMA | [89] |  |
| GTPase HRAS | P3221 | 200; 500; 650; 900 | 1.70–2.10 | 7OGE, 7OGF, 7OGA, 7OGC, 7OGB | [90] |  |
| Proton pump MAR rhodopsin | P121 | 13.0 | 2.25 | 7Q37 | [91] |  |
| Mutant bacterio-rhodopsin | C222 | 13.3; 13.2 | 2.00 | 7Q35, 7Q38 | [91] |  |
| KR2 sodium pump rhodopsin | I222 | 13.1 | 2.60 | 7Q36 | [91] |  |
| RNase A | P3221 | 22; 50; 72; 100; 120 | 1.48–1.60 | 7Q7A *, 7Q77 *, 7Q76 *, 7Q79 *, 7Q78 * | - |  |
| Human Serine/Threonine kinase | P4122 | 111 | 2.60 | 7Q7D * | - |  |
| Rhodobacter sphaeroides Photosynthetic Reaction Center | P4122 | 75; 100 | 2.55–2.87 | 7Q7M *, 7Q7J *, 7Q7G *, 7Q7N * | - |  |
| d(CGCGCG)2 Z-DNA (Rich’s DNA) | P212121 | 300; 540; 715 | 1.41–1.44 | 7ZQO, 7ZQN, 7ZQM | [66] |  |
| d(CGCGTTAACGCG)2 B-DNA (Dickerson’s DNA) | P212121 | 310 | 2.55 | 7ZQL | [66] |  |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kurpiewska, K.; Sławek, J.; Klonecka, A.; Kozak, M. The Promise and Challenge of High Pressure Macromolecular Crystallography. Crystals 2023, 13, 560. https://doi.org/10.3390/cryst13040560
Kurpiewska K, Sławek J, Klonecka A, Kozak M. The Promise and Challenge of High Pressure Macromolecular Crystallography. Crystals. 2023; 13(4):560. https://doi.org/10.3390/cryst13040560
Chicago/Turabian StyleKurpiewska, Katarzyna, Joanna Sławek, Agnieszka Klonecka, and Maciej Kozak. 2023. "The Promise and Challenge of High Pressure Macromolecular Crystallography" Crystals 13, no. 4: 560. https://doi.org/10.3390/cryst13040560
APA StyleKurpiewska, K., Sławek, J., Klonecka, A., & Kozak, M. (2023). The Promise and Challenge of High Pressure Macromolecular Crystallography. Crystals, 13(4), 560. https://doi.org/10.3390/cryst13040560






