Crystal Structure and Functional Characterization of an S-Formylglutathione Hydrolase (BuSFGH) from Burkholderiaceae sp.
Abstract
1. Introduction
2. Materials and Methods
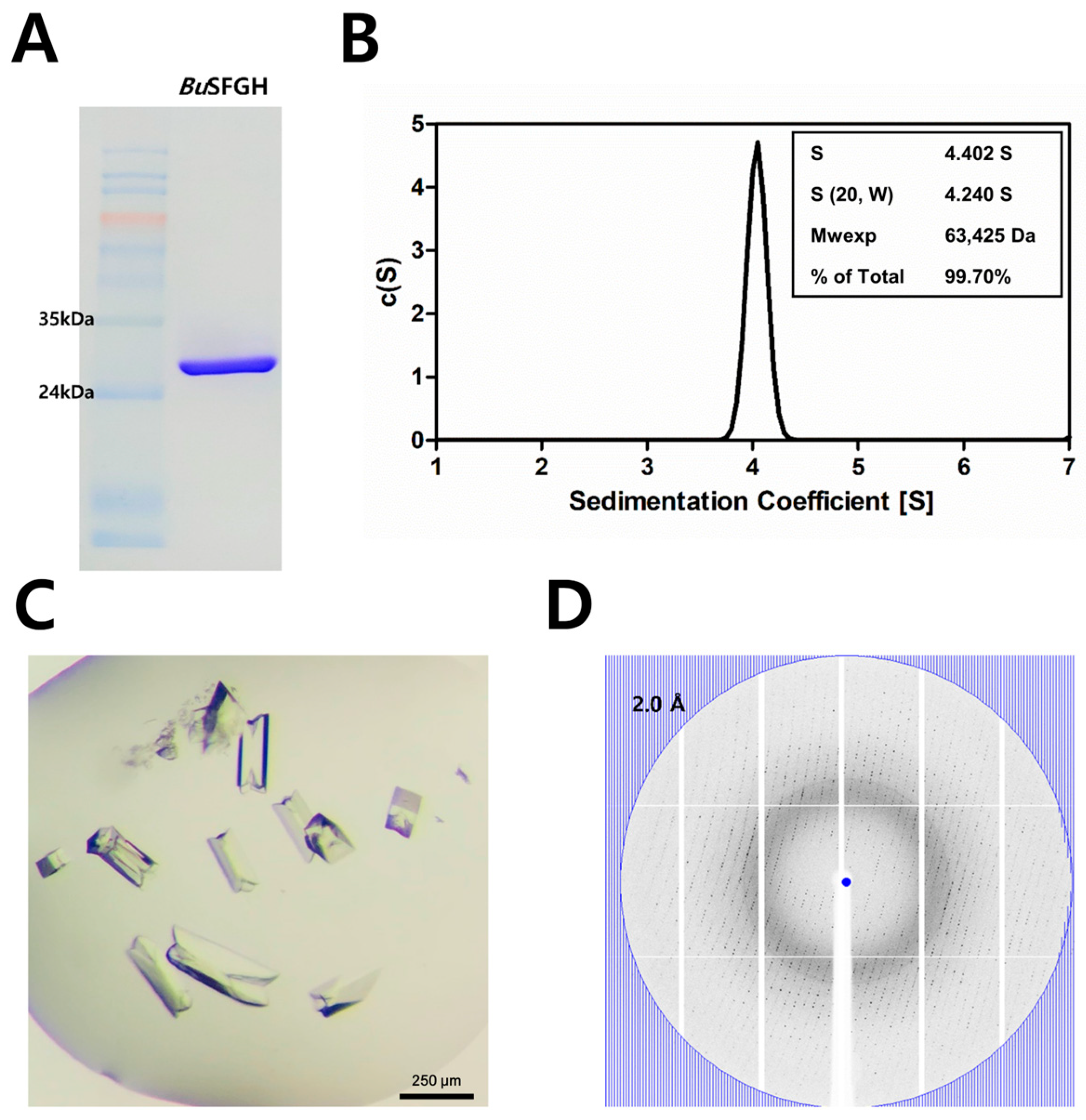
2.1. BuSFGH Cloning, Expression, and Purification
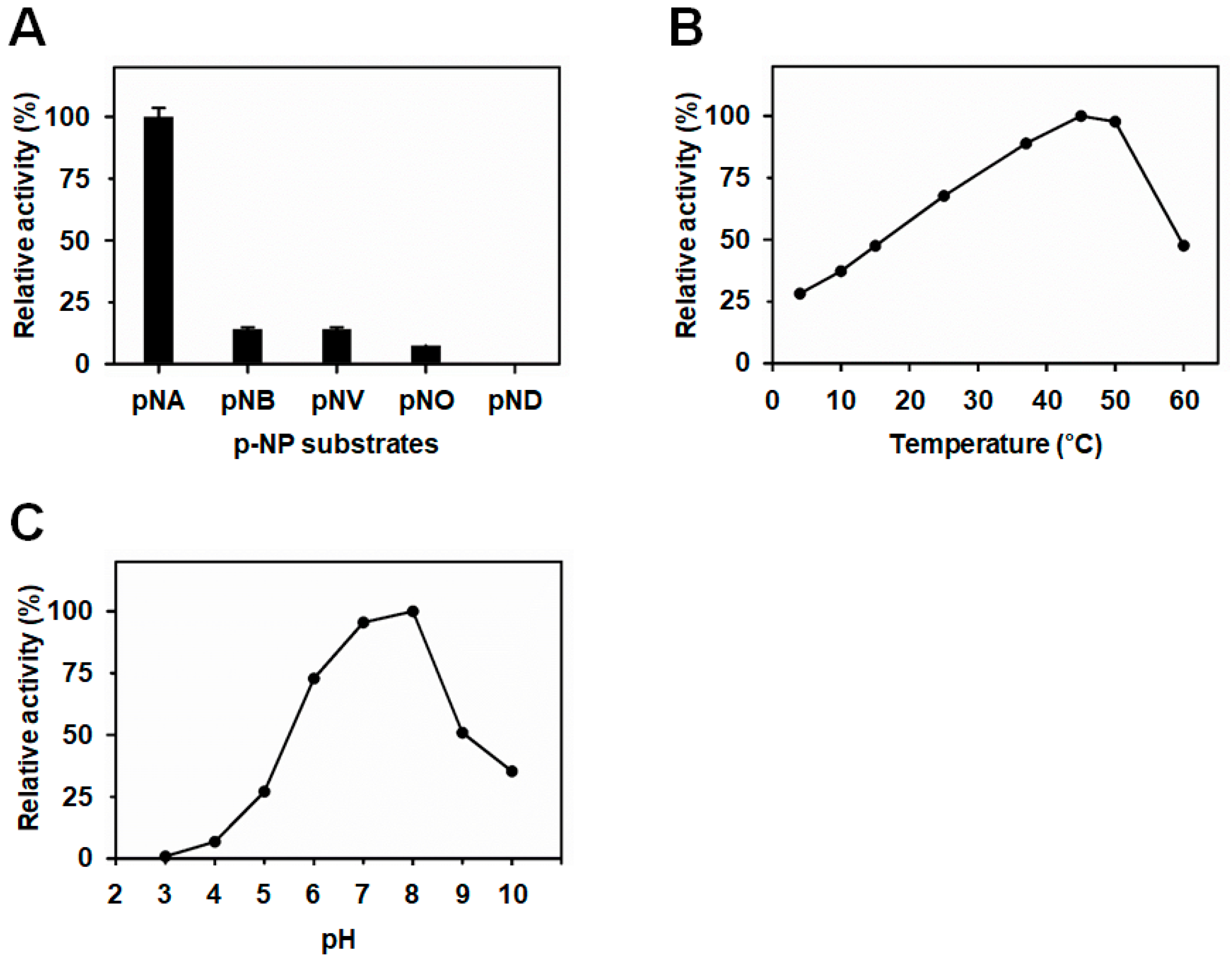
2.2. Substrate Specificity and Optimal Conditions for Enzymatic Activity
2.3. Crystallization, X-ray Diffraction, and Structure Determination
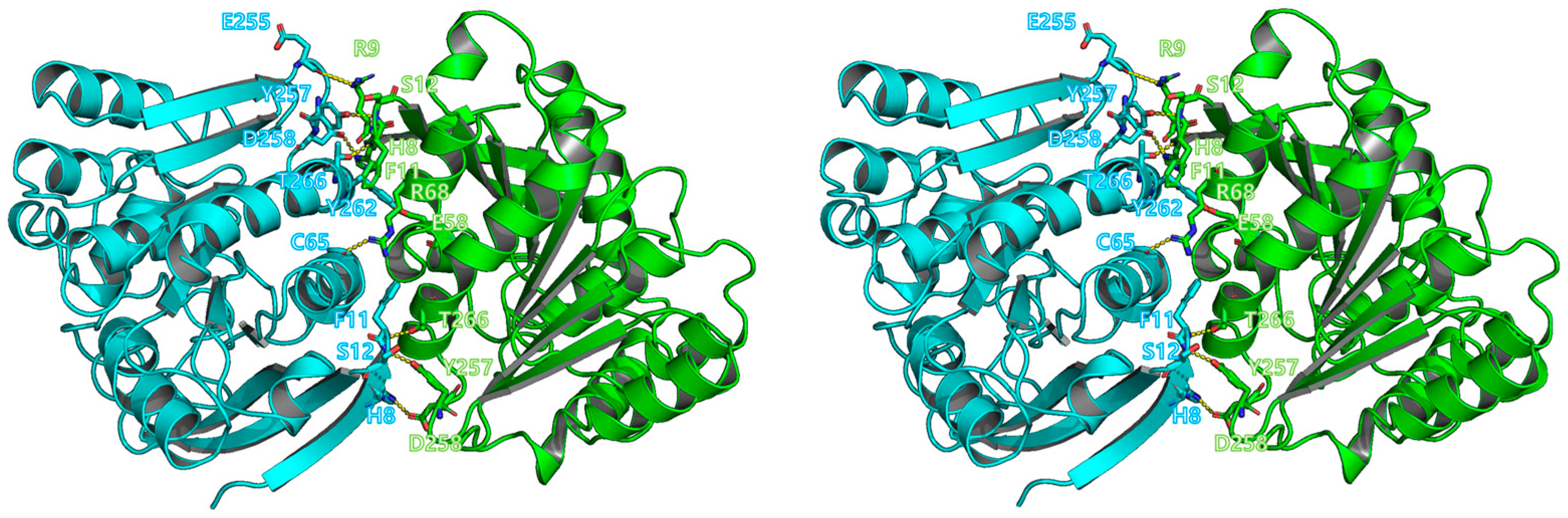
2.4. Analytical Ultracentrifugation (AUC) and Interface Analysis
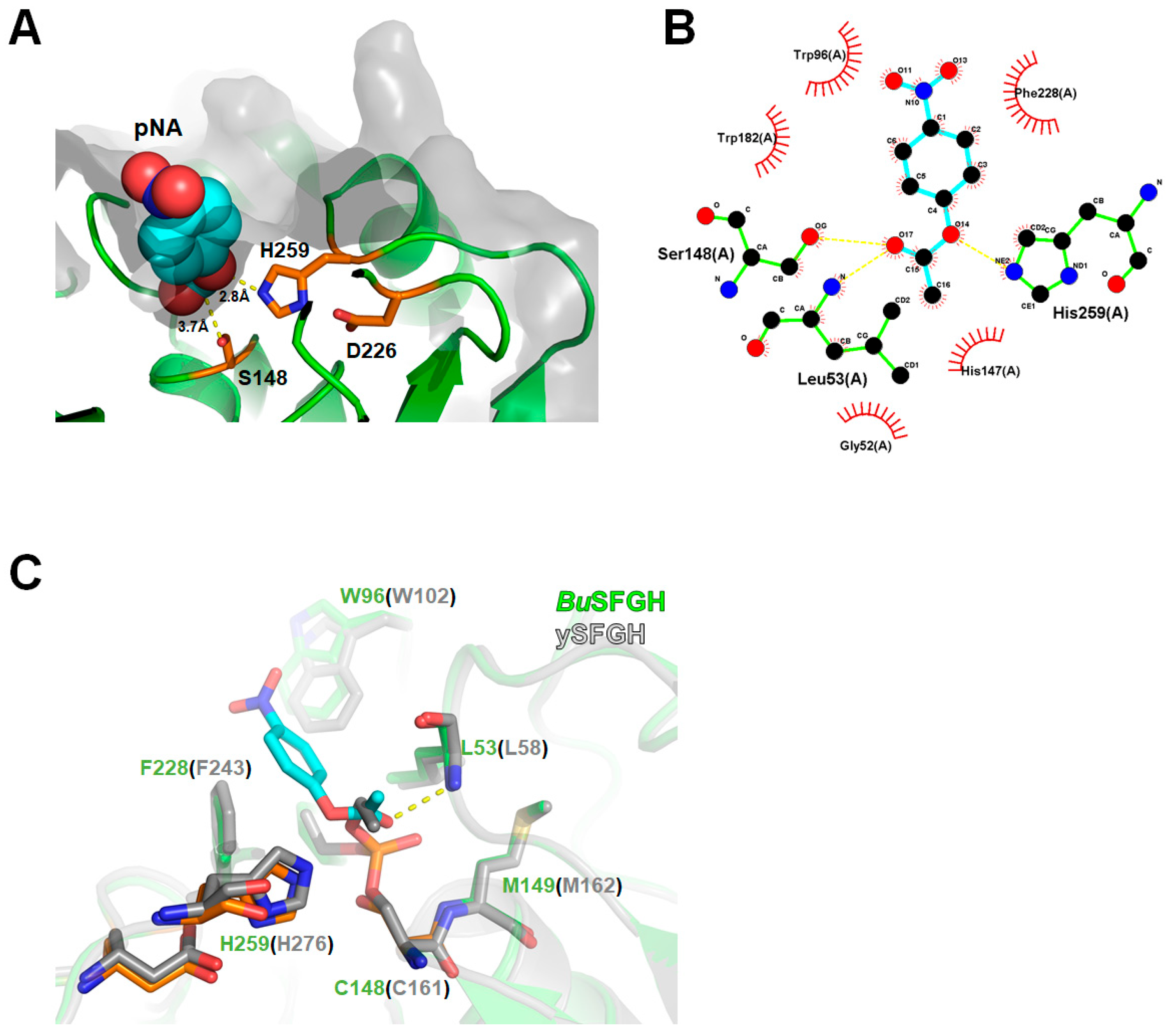
2.5. Molecular Docking Simulation
3. Results and Discussion
3.1. Recombinant Protein Expression, Crystallization, and X-ray Diffraction of BuSFGH
3.2. BuSFGH Activity Assay and Substrate Specificity
3.3. Overall Structure of BuSFGH
3.4. Computational Modeling of p-NA-Bound BuSFGH
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chen, N.H.; Couñago, R.M.; Djoko, K.Y.; Jennings, M.P.; Apicella, M.A.; Kobe, B.; McEwan, A.G. A glutathione-dependent detoxification system is required for formaldehyde resistance and optimal survival of Neisseria meningitidis in biofilms. Antioxid. Redox Signal. 2013, 18, 743–755. [Google Scholar] [CrossRef] [PubMed]
- Chen, N.H.; Djoko, K.Y.; Veyrier, F.J.; McEwan, A.G. Formaldehyde stress responses in bacterial pathogens. Front. Microbiol. 2016, 7, 257. [Google Scholar] [CrossRef] [PubMed]
- Cummins, I.; McAuley, K.; Fordham-Skelton, A.; Schwoerer, R.; Steel, P.G.; Davis, B.G.; Edwards, R. Unique regulation of the active site of the serine esterase S-formylglutathione hydrolase. J. Mol. Biol. 2006, 359, 422–432. [Google Scholar] [CrossRef] [PubMed]
- Harms, N.; Ras, J.; Reijnders, W.N.; van Spanning, R.J.; Stouthamer, A.H. S-formylglutathione hydrolase of Paracoccus denitrificans is homologous to human esterase D: A universal pathway for formaldehyde detoxification? J. Bacteriol. 1996, 178, 6296–6299. [Google Scholar] [CrossRef] [PubMed]
- Akoh, C.C.; Lee, G.C.; Liaw, Y.C.; Huang, T.H.; Shaw, J.F. GDSL family of serine esterases/lipases. Prog. Lipid Res. 2004, 43, 534–552. [Google Scholar] [CrossRef]
- Alterio, V.; Aurilia, V.; Romanelli, A.; Parracino, A.; Saviano, M.; D’Auria, S.; De Simone, G. Crystal structure of an S-formylglutathione hydrolase from Pseudoalteromonas haloplanktis TAC125. Biopolymers 2010, 93, 669–677. [Google Scholar] [CrossRef]
- Kordic, S.; Cummins, I.; Edwards, R. Cloning and characterization of an S-formylglutathione hydrolase from Arabidopsis thaliana. Arch. Biochem. Biophys. 2002, 399, 232–238. [Google Scholar] [CrossRef]
- Legler, P.M.; Leary, D.H.; Hervey, W.J., 4th; Millard, C.B. A role for His-160 in peroxide inhibition of S. cerevisiae S-formylglutathione hydrolase: Evidence for an oxidation sensitive motif. Arch. Biochem. Biophys. 2012, 528, 7–20. [Google Scholar] [CrossRef]
- Lee, C.W.; Yoo, W.; Park, S.H.; Le, L.T.H.L.; Jeong, C.S.; Ryu, B.H.; Shin, S.C.; Kim, H.W.; Park, H.; Kim, K.K.; et al. Structural and functional characterization of a novel cold-active S-formylglutathione hydrolase (SfSFGH) homolog from Shewanella frigidimarina, a psychrophilic bacterium. Microb. Cell Fact. 2019, 18, 140. [Google Scholar] [CrossRef]
- Fellner, M.; Lentz, C.S.; Jamieson, S.A.; Brewster, J.L.; Chen, L.; Bogyo, M.; Mace, P.D. Structural basis for the inhibitor and substrate specificity of the unique Fph serine hydrolases of Staphylococcus aureus. ACS Infect. Dis. 2020, 6, 2771–2782. [Google Scholar] [CrossRef]
- Miller, J.J.; Shah, I.T.; Hatten, J.; Barekatain, Y.; Mueller, E.A.; Moustafa, A.M.; Edwards, R.L.; Dowd, C.S.; Hoops, G.C.; Johnson, R.J.; et al. Structure-guided microbial targeting of antistaphylococcal prodrugs. eLife 2021, 10, e66657. [Google Scholar] [CrossRef]
- Hwang, J.; Kim, B.; Lee, M.J.; Nam, Y.; Youn, U.J.; Lee, C.S.; Oh, T.J.; Park, H.H.; Do, H.; Lee, J.H. Structural basis for the substrate specificity of an S-formylglutathione hydrolase derived from Variovorax sp. PAMC 28711. Biochem. Biophys. Res. Commun. 2022, 629, 159–164. [Google Scholar] [CrossRef]
- Otwinowski, Z.; Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997, 276, 307–326. [Google Scholar] [CrossRef]
- Matthews, B.W. Solvent content of protein crystals. J. Mol. Biol. 1968, 33, 491–497. [Google Scholar] [CrossRef]
- Read, R.J. Pushing the boundaries of molecular replacement with maximum likelihood. Acta Crystallogr. D Biol. Crystallogr. 2001, 57, 1373–1382. [Google Scholar] [CrossRef]
- Winn, M.D.; Ballard, C.C.; Cowtan, K.D.; Dodson, E.J.; Emsley, P.; Evans, P.R.; Keegan, R.M.; Krissinel, E.B.; Leslie, A.G.; McCoy, A.; et al. Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 2011, 67, 235–242. [Google Scholar] [CrossRef]
- Emsley, P.; Cowtan, K. Coot: Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004, 60, 2126–2132. [Google Scholar] [CrossRef]
- Murshudov, G.N.; Skubák, P.; Lebedev, A.A.; Pannu, N.S.; Steiner, R.A.; Nicholls, R.A.; Winn, M.D.; Long, F.; Vagin, A.A. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D Biol. Crystallogr. 2011, 67, 355–367. [Google Scholar] [CrossRef]
- Afonine, P.V.; Grosse-Kunstleve, R.W.; Echols, N.; Headd, J.J.; Moriarty, N.W.; Mustyakimov, M.; Terwilliger, T.C.; Urzhumtsev, A.; Zwart, P.H.; Adams, P.D. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. D Biol. Crystallogr. 2012, 68, 352–367. [Google Scholar] [CrossRef]
- Chen, V.B.; Arendall, W.B., 3rd; Headd, J.J.; Keedy, D.A.; Immormino, R.M.; Kapral, G.J.; Murray, L.W.; Richardson, J.S.; Richardson, D.C. MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 2010, 66, 12–21. [Google Scholar] [CrossRef]
- DeLano, W.L. The PyMOL Molecular Graphics System; DeLano Scientific LLC: San Carlos, CA, USA, 2002. [Google Scholar]
- Schuck, P. Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and Lamm equation modeling. Biophys. J. 2000, 78, 1606–1619. [Google Scholar] [CrossRef] [PubMed]
- Krissinel, E.; Henrick, K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 2007, 372, 774–797. [Google Scholar] [CrossRef] [PubMed]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef] [PubMed]
- Lebedev, A.A.; Young, P.; Isupov, M.N.; Moroz, O.V.; Vagin, A.A.; Murshudov, G.N. JLigand: A graphical tool for the CCP4 template-restraint library. Acta Crystallogr. D Biol. Crystallogr. 2012, 68, 431–440. [Google Scholar] [CrossRef] [PubMed]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef]
- Holm, L.; Rosenström, P. Dali server: Conservation mapping in 3D. Nucleic Acids Res. 2010, 38, W545–W549. [Google Scholar] [CrossRef]
- Lemak, S.; Tchigvintsev, A.; Petit, P.; Flick, R.; Singer, A.U.; Brown, G.; Evdokimova, E.; Egorova, O.; Gonzalez, C.F.; Chernikova, T.N.; et al. Structure and activity of the cold-active and anion-activated carboxyl esterase OLEI01171 from the oil-degrading marine bacterium Oleispira antarctica. Biochem. J. 2012, 445, 193–203. [Google Scholar] [CrossRef]
- Wu, D.; Li, Y.; Song, G.; Zhang, D.; Shaw, N.; Liu, Z.J. Crystal structure of human esterase D: A potential genetic marker of retinoblastoma. FASEB J. 2009, 23, 1441–1446. [Google Scholar] [CrossRef]
- van Straaten, K.E.; Gonzalez, C.F.; Valladares, R.B.; Xu, X.; Savchenko, A.V.; Sanders, D.A. The structure of a putative S-formylglutathione hydrolase from Agrobacterium tumefaciens. Protein Sci. 2009, 18, 2196–2202. [Google Scholar] [CrossRef]
- Legler, P.M.; Kumaran, D.; Swaminathan, S.; Studier, F.W.; Millard, C.B. Structural characterization and reversal of the natural organophosphate resistance of a D-type esterase, Saccharomyces cerevisiae S-formylglutathione hydrolase. Biochemistry 2008, 47, 9592–9601. [Google Scholar] [CrossRef]

| Data Set | BuSFGH |
|---|---|
| X-ray source | PAL-5C |
| Space group | P212121 |
| Unit-cell parameters (Å, °) | A = 50.37, b = 109.29, c = 112.62, α = β = γ = 90 |
| Wavelength (Å) | 0.9796 |
| Resolution (Å) | 29.52–1.73 (1.76–1.73) |
| Total reflections | 816,722 (85260) |
| Unique reflections | 65,529 (6421) |
| Average I/σ (I) | 20.63 (6.17) |
| Rmerge a | 0.081 (0.397) |
| Redundancy | 12.5 (13.3) |
| CC(1/2) | 0.999 (0.969) |
| Completeness (%) | 99.7 (98.9) |
| Refinement | |
| Resolution range (Å) | 29.52–1.73 (1.79–1.73) |
| No. of reflections of working set | 65,525 (6428) |
| No. of reflections of test set | 3277 (322) |
| No. of amino acid residues | 562 |
| No. of water molecules | 416 |
| Rcryst b | 0.195 (0.225) |
| Rfree c | 0.210 (0.237) |
| R.m.s. bond length (Å) | 0.011 |
| R.m.s. bond angle (°) | 1.15 |
| Average B value (Å2) (protein) | 20.12 |
| Average B value (Å2) (solvent) | 27.49 |
| Protein | PDB Code | DALI Z-Score | UniProtKB Code | Sequence % ID with BuSFGH (Aligned Residue Number) | Oligomerization State | Reference |
|---|---|---|---|---|---|---|
| Esterase from the oil-degrading bacterium Oleispira antarctica | 3I6Y | 47.7 | D0VWZ4 | 53 (276/278) | Dimer | [28] |
| human esterase D | 3FCX | 47.4 | P10768 | 56 (274/275) | Dimer | [29] |
| S-formylglutathione hydrolase homolog from Shewanella frigidimarina | 6JZL | 46.8 | Q07XK4 | 55 (275/278) | Dimer | [9] |
| S-formylglutathione hydrolase from Pseudoalteromonas haloplanktis | 3LS2 | 46.7 | Q3IL66 | 49 (274/278) | Dimer | [6] |
| S-Formylglutathione Hydrolase from Agrobacterium tumefaciens | 3E4D | 45.7 | A9CJ11 | 46 (277/278) | Dimer | [30] |
| Esterase D from Neisseria meningitidis | 4B6G | 45.7 | Q9JZ43 | 52 (274/275) | Monomer | [1] |
| S-formylglutathione hydrolase from Saccharomyces cerevisiae | 3C6B | 42.5 | P40363 | 42 (276/291) | Dimer | [31] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hwang, J.; Do, H.; Shim, Y.-S.; Lee, J.H. Crystal Structure and Functional Characterization of an S-Formylglutathione Hydrolase (BuSFGH) from Burkholderiaceae sp. Crystals 2023, 13, 621. https://doi.org/10.3390/cryst13040621
Hwang J, Do H, Shim Y-S, Lee JH. Crystal Structure and Functional Characterization of an S-Formylglutathione Hydrolase (BuSFGH) from Burkholderiaceae sp. Crystals. 2023; 13(4):621. https://doi.org/10.3390/cryst13040621
Chicago/Turabian StyleHwang, Jisub, Hackwon Do, Youn-Soo Shim, and Jun Hyuck Lee. 2023. "Crystal Structure and Functional Characterization of an S-Formylglutathione Hydrolase (BuSFGH) from Burkholderiaceae sp." Crystals 13, no. 4: 621. https://doi.org/10.3390/cryst13040621
APA StyleHwang, J., Do, H., Shim, Y.-S., & Lee, J. H. (2023). Crystal Structure and Functional Characterization of an S-Formylglutathione Hydrolase (BuSFGH) from Burkholderiaceae sp. Crystals, 13(4), 621. https://doi.org/10.3390/cryst13040621








