Effects of Radiation Damage on Metal-Binding Sites in Thermolysin
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Preparation
2.2. X-ray Diffraction and Data Processing
2.3. Structure Determination
3. Results
3.1. Data Collection
3.2. Structure Analysis
3.3. Metal Ion–Binding Sites
4. Discussion
Supplementary Materials
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Smyth, M.S.; Martin, J.H. X-ray crystallography. Mol. Pathol. 2000, 53, 8–14. [Google Scholar] [CrossRef] [PubMed]
- Su, X.D.; Zhang, H.; Terwilliger, T.C.; Liljas, A.; Xiao, J.; Dong, Y. Protein Crystallography from the Perspective of Technology Developments. Crystallogr. Rev. 2015, 21, 122–153. [Google Scholar] [CrossRef] [PubMed]
- Maveyraud, L.; Mourey, L. Protein X-ray Crystallography and Drug Discovery. Molecules 2020, 25, 1030. [Google Scholar] [CrossRef] [PubMed]
- Kim, I.J.; Kim, S.R.; Bornscheuer, U.T.; Nam, K.H. Engineering of GH11 Xylanases for Optimal pH Shifting for Industrial Applications. Catalysts 2023, 13, 1405. [Google Scholar] [CrossRef]
- Aitipamula, S.; Vangala, V.R. X-ray Crystallography and its Role in Understanding the Physicochemical Properties of Pharmaceutical Cocrystals. J. Indian Inst. Sci. 2017, 97, 227–243. [Google Scholar] [CrossRef]
- Batool, M.; Ahmad, B.; Choi, S. A Structure-Based Drug Discovery Paradigm. Int. J. Mol. Sci. 2019, 20, 2783. [Google Scholar] [CrossRef]
- Anderson, A.C. The Process of Structure-Based Drug Design. Biol. Chem. 2003, 10, 787–797. [Google Scholar] [CrossRef]
- Xu, Y.; Nam, K.H. Xylitol binding to the M1 site of glucose isomerase induces a conformational change in the substrate binding channel. Biochem. Biophys. Res. Commun. 2023, 682, 21–26. [Google Scholar] [CrossRef]
- Jelsch, C.; Teeter, M.M.; Lamzin, V.; Pichon-Pesme, V.; Blessing, R.H.; Lecomte, C. Accurate protein crystallography at ultra-high resolution: Valence electron distribution in crambin. Proc. Natl. Acad. Sci. USA 2000, 97, 3171–3176. [Google Scholar] [CrossRef]
- Laulumaa, S.; Kursula, P. Sub-Atomic Resolution Crystal Structures Reveal Conserved Geometric Outliers at Functional Sites. Molecules 2019, 24, 3044. [Google Scholar] [CrossRef]
- Tan, L.; Tian, S.; Liu, X.; Zhang, W.; Wu, X.; Gong, Y. Emittance and energy spread compensation in future synchrotron light sources. Nucl. Instrum. Methods Phys. Res. A 2023, 1052, 168278. [Google Scholar] [CrossRef]
- Kim, Y.; Cho, B.G.; Koo, T.Y. Design and simulation of a mirror-mitigated thermal load on Korea-4GSR hard X-ray undulator beamline optics. J. Korean Phys. Soc. 2022, 81, 273–277. [Google Scholar] [CrossRef]
- Cho, B.-G.; Kim, Y.; Shin, S.; Koo, T.-Y. Comparative study of hard X-ray undulator beamline performance in the Korean 4GSR and the PLS-II. J. Korean Phys. Soc. 2021, 78, 467–475. [Google Scholar] [CrossRef]
- Schroer, C.G.; Falkenberg, G. Hard X-ray nanofocusing at low-emittance synchrotron radiation sources. J. Synchrotron Radiat. 2014, 21, 996–1005. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Kim, H.-Y.; Park, J.; Kim, S.; Kim, S.; Rah, S.; Lim, J.; Nam, K.H. Focusing X-ray free-electron laser pulses using Kirkpatrick–Baez mirrors at the NCI hutch of the PAL-XFEL. J. Synchrotron Radiat. 2018, 25, 289–292. [Google Scholar] [CrossRef]
- Gu, D.H.; Eo, C.; Hwangbo, S.A.; Ha, S.C.; Kim, J.H.; Kim, H.; Lee, C.S.; Seo, I.D.; Yun, Y.D.; Lee, W.; et al. BL-11C Micro-MX: A high-flux microfocus macromolecular-crystallography beamline for micrometre-sized protein crystals at Pohang Light Source II. J. Synchrotron Radiat. 2021, 28, 1210–1215. [Google Scholar] [CrossRef]
- Kim, J.; Nam, K.H. X-ray-Induced Heating in the Vicinity of the X-ray Interaction Point. Appl. Sci. 2023, 13, 717. [Google Scholar] [CrossRef]
- Shelley, K.L.; Garman, E.F. Quantifying and comparing radiation damage in the Protein Data Bank. Nat. Commun. 2022, 13, 1314. [Google Scholar] [CrossRef]
- Gebicki, J.M.; Nauser, T. Initiation and Prevention of Biological Damage by Radiation-Generated Protein Radicals. Int. J. Mol. Sci. 2021, 23, 396. [Google Scholar] [CrossRef]
- Sliz, P.; Harrison, S.C.; Rosenbaum, G. How does Radiation Damage in Protein Crystals Depend on X-Ray Dose? Structure 2003, 11, 13–19. [Google Scholar] [CrossRef]
- Ravelli, R.B.G.; McSweeney, S.M. The ‘fingerprint’ that X-rays can leave on structures. Structure 2000, 8, 315–328. [Google Scholar] [CrossRef] [PubMed]
- Murray, J.; Garman, E. Investigation of possible free-radical scavengers and metrics for radiation damage in protein cryocrystallography. J. Synchrotron Radiat. 2002, 9, 347–354. [Google Scholar] [CrossRef] [PubMed]
- Carugo, O.; Carugo, K.D. When X-rays modify the protein structure: Radiation damage at work. Trends Biochem. Sci. 2005, 30, 213–219. [Google Scholar] [CrossRef] [PubMed]
- Weiss, M.S.; Panjikar, S.; Mueller-Dieckmann, C.; Tucker, P.A. On the influence of the incident photon energy on the radiation damage in crystalline biological samples. J. Synchrotron Radiat. 2005, 12, 304–309. [Google Scholar] [CrossRef] [PubMed]
- Corbett, M.C.; Latimer, M.J.; Poulos, T.L.; Sevrioukova, I.F.; Hodgson, K.O.; Hedman, B. Photoreduction of the active site of the metalloprotein putidaredoxin by synchrotron radiation. Acta Crystallogr. Sect. D Biol. Crystallogr. 2007, 63, 951–960. [Google Scholar] [CrossRef]
- Garman, E.F. Radiation damage in macromolecular crystallography: What is it and why should we care? Acta Crystallogr. Sect. D Biol. Crystallogr. 2010, 66, 339–351. [Google Scholar] [CrossRef]
- Burmeister, W.P. Structural changes in a cryo-cooled protein crystal owing to radiation damage. Acta Crystallogr. Sect. D Biol. Crystallogr. 2000, 56, 328–341. [Google Scholar] [CrossRef]
- Nam, K.H. Radiation Damage on Selenomethionine-Substituted Single-Domain Substrate-Binding Protein. Crystals 2023, 13, 1620. [Google Scholar] [CrossRef]
- McCoy, A.J.; Read, R.J. Experimental phasing: Best practice and pitfalls. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010, 66, 458–469. [Google Scholar] [CrossRef]
- Schmidt, M.; Šrajer, V.; Purwar, N.; Tripathi, S. The kinetic dose limit in room-temperature time-resolved macromolecular crystallography. J. Synchrotron Radiat. 2012, 19, 264–273. [Google Scholar] [CrossRef]
- Matsui, Y.; Sakai, K.; Murakami, M.; Shiro, Y.; Adachi, S.-I.; Okumura, H.; Kouyama, T. Specific Damage Induced by X-ray Radiation and Structural Changes in the Primary Photoreaction of Bacteriorhodopsin. J. Mol. Biol. 2002, 324, 469–481. [Google Scholar] [CrossRef] [PubMed]
- Taberman, H. Radiation Damage in Macromolecular Crystallography—An Experimentalist’s View. Crystals 2018, 8, 157. [Google Scholar] [CrossRef]
- Harding, M.M.; Nowicki, M.W.; Walkinshaw, M.D. Metals in protein structures: A review of their principal features. Crystallogr. Rev. 2010, 16, 247–302. [Google Scholar] [CrossRef]
- Harding, M.M. The architecture of metal coordination groups in proteins. Acta Crystallogr. Sect. D Biol. Crystallogr. 2004, 60, 849–859. [Google Scholar] [CrossRef] [PubMed]
- Zheng, H.; Chruszcz, M.; Lasota, P.; Lebioda, L.; Minor, W. Data mining of metal ion environments present in protein structures. J. Inorg. Biochem. 2008, 102, 1765–1776. [Google Scholar] [CrossRef] [PubMed]
- Dokmanić, I.; Šikić, M.; Tomić, S. Metals in proteins: Correlation between the metal-ion type, coordination number and the amino-acid residues involved in the coordination. Acta Crystallogr. Sect. D Biol. Crystallogr. 2008, 64, 257–263. [Google Scholar] [CrossRef]
- Neufeld, M.J.; Lutzke, A.; Pratx, G.; Sun, C. High-Z Metal–Organic Frameworks for X-ray Radiation-Based Cancer Theranostics. Chem. Eur. J. 2020, 27, 3229–3237. [Google Scholar] [CrossRef]
- Yano, J.; Kern, J.; Irrgang, K.-D.; Latimer, M.J.; Bergmann, U.; Glatzel, P.; Pushkar, Y.; Biesiadka, J.; Loll, B.; Sauer, K.; et al. X-ray damage to the Mn4Ca complex in single crystals of photosystem II: A case study for metalloprotein crystallography. Proc. Natl. Acad. Sci. USA 2005, 102, 12047–12052. [Google Scholar] [CrossRef]
- Pfanzagl, V.; Beale, J.H.; Michlits, H.; Schmidt, D.; Gabler, T.; Obinger, C.; Djinović-Carugo, K.; Hofbauer, S. X-ray–induced photoreduction of heme metal centers rapidly induces active-site perturbations in a protein-independent manner. J. Biol. Chem. 2020, 295, 13488–13501. [Google Scholar] [CrossRef]
- Wuerges, J.; Lee, J.-W.; Yim, Y.-I.; Yim, H.-S.; Kang, S.-O.; Carugo, K.D. Crystal structure of nickel-containing superoxide dismutase reveals another type of active site. Proc. Natl. Acad. Sci. USA 2004, 101, 8569–8574. [Google Scholar] [CrossRef]
- Bury, C.S.; Brooks-Bartlett, J.C.; Walsh, S.P.; Garman, E.F. Estimate your dose: RADDOSE-3D. Protein Sci. 2018, 27, 217–228. [Google Scholar] [CrossRef] [PubMed]
- Otwinowski, Z.; Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997, 276, 307–326. [Google Scholar] [CrossRef]
- Vagin, A.; Teplyakov, A. Molecular replacement with MOLREP. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010, 66, 22–25. [Google Scholar] [CrossRef]
- Agirre, J.; Atanasova, M.; Bagdonas, H.; Ballard, C.B.; Baslé, A.; Beilsten-Edmands, J.; Borges, R.J.; Brown, D.G.; Burgos-Mármol, J.J.; Berrisford, J.M.; et al. The CCP4 suite: Integrative software for macromolecular crystallography. Acta Crystallogr. Sect. D Struct. Biol. 2023, 79, 449–461. [Google Scholar] [CrossRef] [PubMed]
- Nam, K.H. Structural analysis of metal chelation of the metalloproteinase thermolysin by 1,10-phenanthroline. J. Inorg. Biochem. 2021, 215, 111319. [Google Scholar] [CrossRef] [PubMed]
- Emsley, P.; Cowtan, K. Coot: Model-building tools for molecular graphics. Acta Crystallogr. Sect. D Biol. Crystallogr. 2004, D60, 2126–2132. [Google Scholar] [CrossRef]
- Murshudov, G.N.; Skubak, P.; Lebedev, A.A.; Pannu, N.S.; Steiner, R.A.; Nicholls, R.A.; Winn, M.D.; Long, F.; Vagin, A.A. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. Sect. D Biol. Crystallogr. 2011, 67, 355–367. [Google Scholar] [CrossRef]
- Williams, C.J.; Headd, J.J.; Moriarty, N.W.; Prisant, M.G.; Videau, L.L.; Deis, L.N.; Verma, V.; Keedy, D.A.; Hintze, B.J.; Chen, V.B.; et al. MolProbity: More and better reference data for improved all-atom structure validation. Protein Sci. 2018, 27, 293–315. [Google Scholar] [CrossRef]
- Zheng, H.; Cooper, D.R.; Porebski, P.J.; Shabalin, I.G.; Handing, K.B.; Minor, W. CheckMyMetal: A macromolecular metal-binding validation tool. Acta Crystallogr. Sect. D Struct. Biol. 2017, 73, 223–233. [Google Scholar] [CrossRef]
- Roche, R.S.; Voordouw, G. The structural and functional roles of metal ions in thermolysin. CRC Crit. Rev. Biochem. 1978, 5, 1–23. [Google Scholar] [CrossRef]
- Park, J.; Park, S.; Kim, J.; Park, G.; Cho, Y.; Nam, K.H. Polyacrylamide injection matrix for serial femtosecond crystallography. Sci. Rep. 2019, 9, 2525. [Google Scholar] [CrossRef] [PubMed]
- Nam, K.H. Radiation Damage on Thaumatin: A Case Study of Crystals That Are Larger Than the Microfocusing X-ray Beam. Appl. Sci. 2023, 13, 1876. [Google Scholar] [CrossRef]
- Wherland, S.; Pecht, I. Radiation chemists look at damage in redox proteins induced by X-rays. Proteins 2018, 86, 817–826. [Google Scholar] [CrossRef]
- Nass, K. Radiation damage in protein crystallography at X-ray free-electron lasers. Acta Crystallogr. Sect. D Struct. Biol. 2019, 75, 211–218. [Google Scholar] [CrossRef]
- Koppenol, W.H.; Hider, R.H. Iron and redox cycling. Do’s and don’ts. Free Radic. Biol. Med. 2019, 133, 3–10. [Google Scholar] [CrossRef]
- Leclercq, B.; Kabbour, H.; Arevalo-Lopez, A.M.; Daviero-Minaud, S.; Minaud, C.; David, R.; Mentré, O. Synthesis, structure and magnetic behavior of iron arsenites with hierarchical magnetic units. Inorg. Chem. Front. 2020, 7, 3987–3999. [Google Scholar] [CrossRef]
- Hu, N.; Li, X.-Y.; Liu, S.-M.; Wang, Z.; He, X.-K.; Hou, Y.-X.; Wang, Y.-X.; Deng, Z.; Chen, L.-H.; Su, B.-L. Enhanced stability of highly-dispersed copper catalyst supported by hierarchically porous carbon for long term selective hydrogenation. Chin. J. Catal. 2020, 41, 1081–1090. [Google Scholar] [CrossRef]
- Bhattacharyya, R.; Dhar, J.; Ghosh Dastidar, S.; Chakrabarti, P.; Weiss, M.S. The susceptibility of disulfide bonds towards radiation damage may be explained by S...O interactions. IUCrJ 2020, 7, 825–834. [Google Scholar] [CrossRef]
- Chevion, M.; Korbashi, P.; Katzhandler, J.; Saltman, P. Zinc—A Redox-Inactive Metal Provides a Novel Approach for Protection Against Metal-Mediated Free Radical Induced Injury: Study of Paraquat Toxicity in E. coli. In Antioxidants in Therapy and Preventive Medicine; Advances in Experimental Medicine and Biology; Springer: Berlin/Heidelberg, Germany, 1990; pp. 217–222. [Google Scholar] [CrossRef]
- Saito, K.; Nakagawa, M.; Mandal, M.; Ishikita, H. Role of redox-inactive metals in controlling the redox potential of heterometallic manganese–oxido clusters. Photosynth. Res. 2021, 148, 153–159. [Google Scholar] [CrossRef]
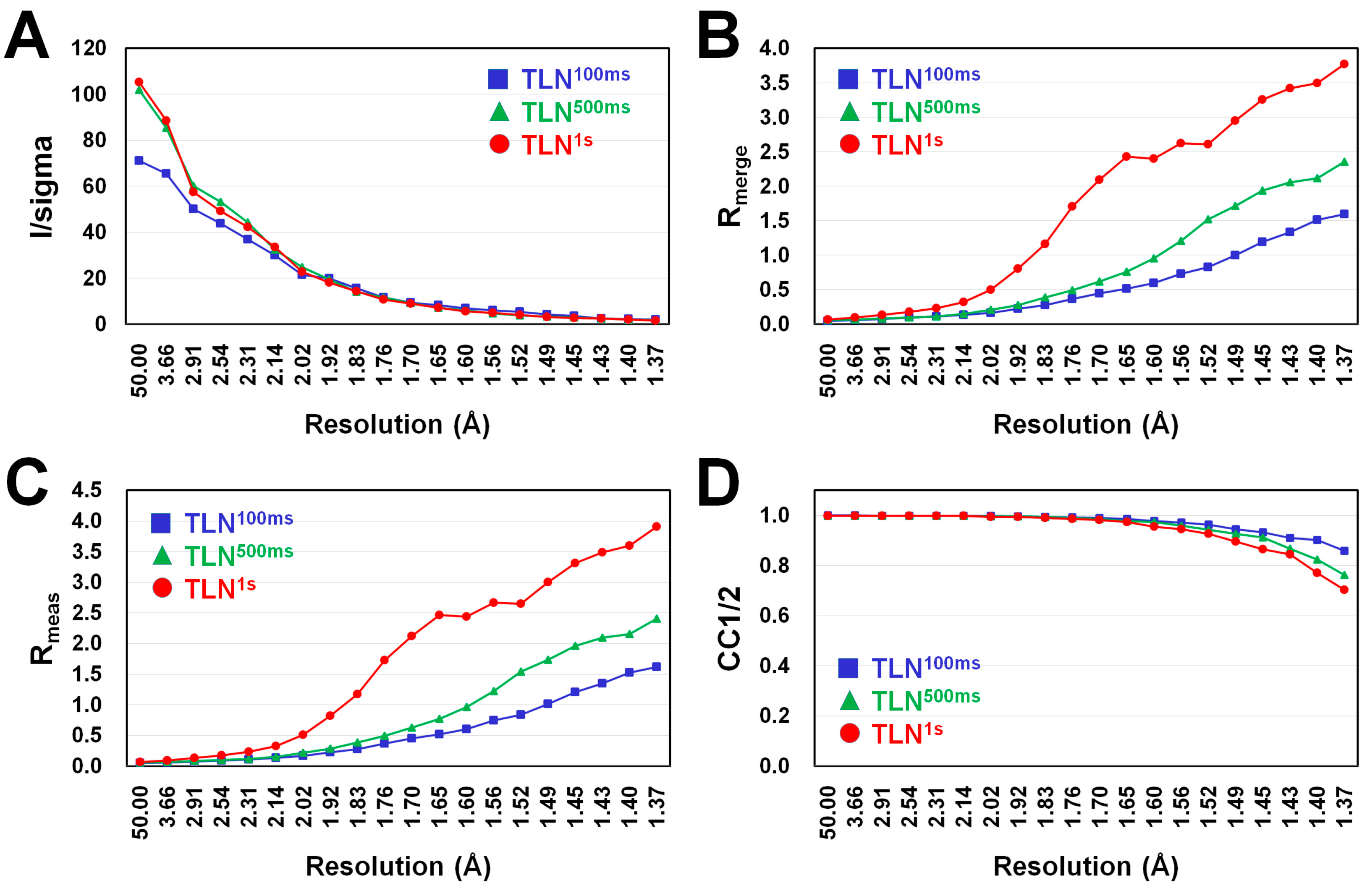
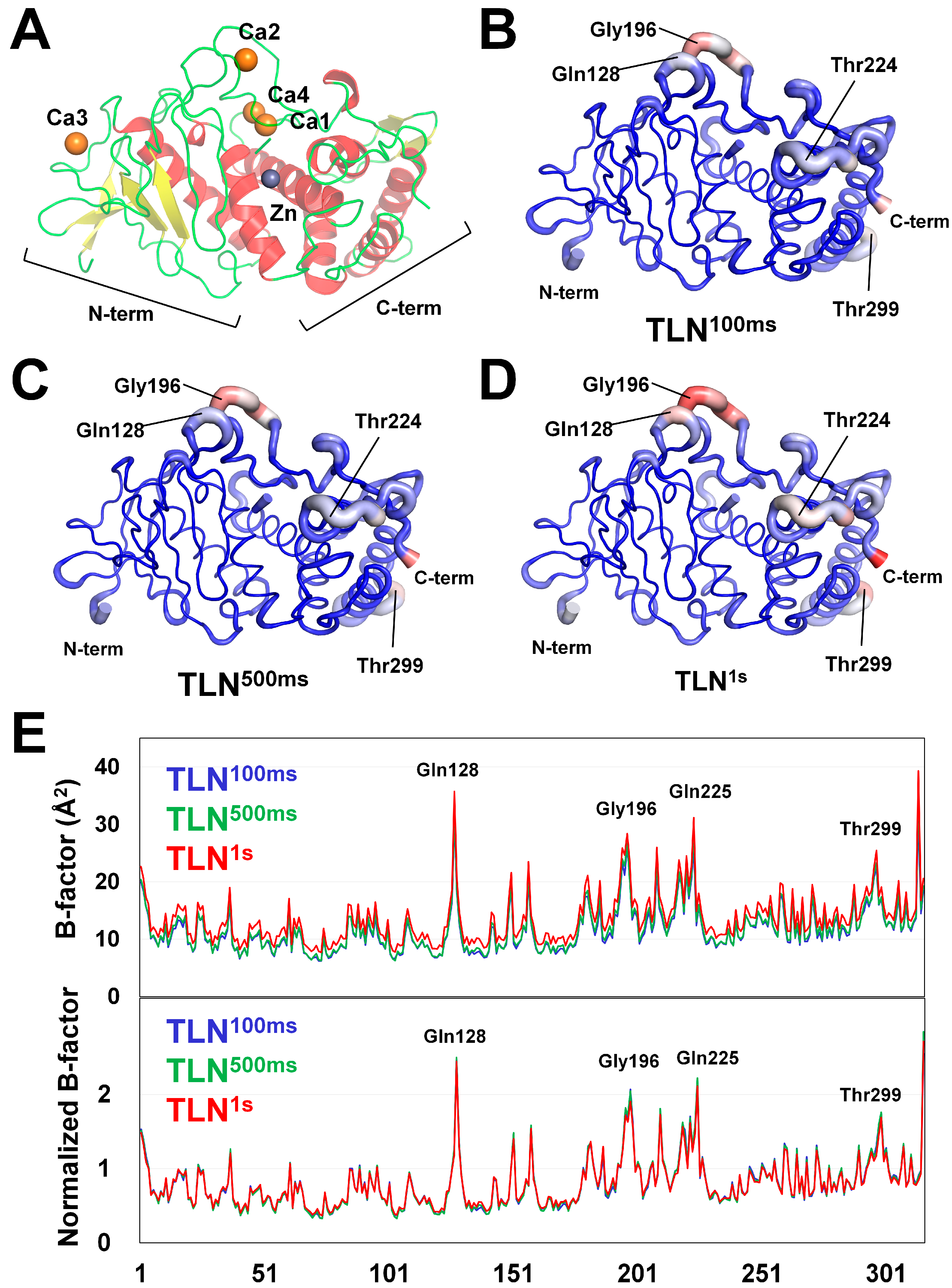

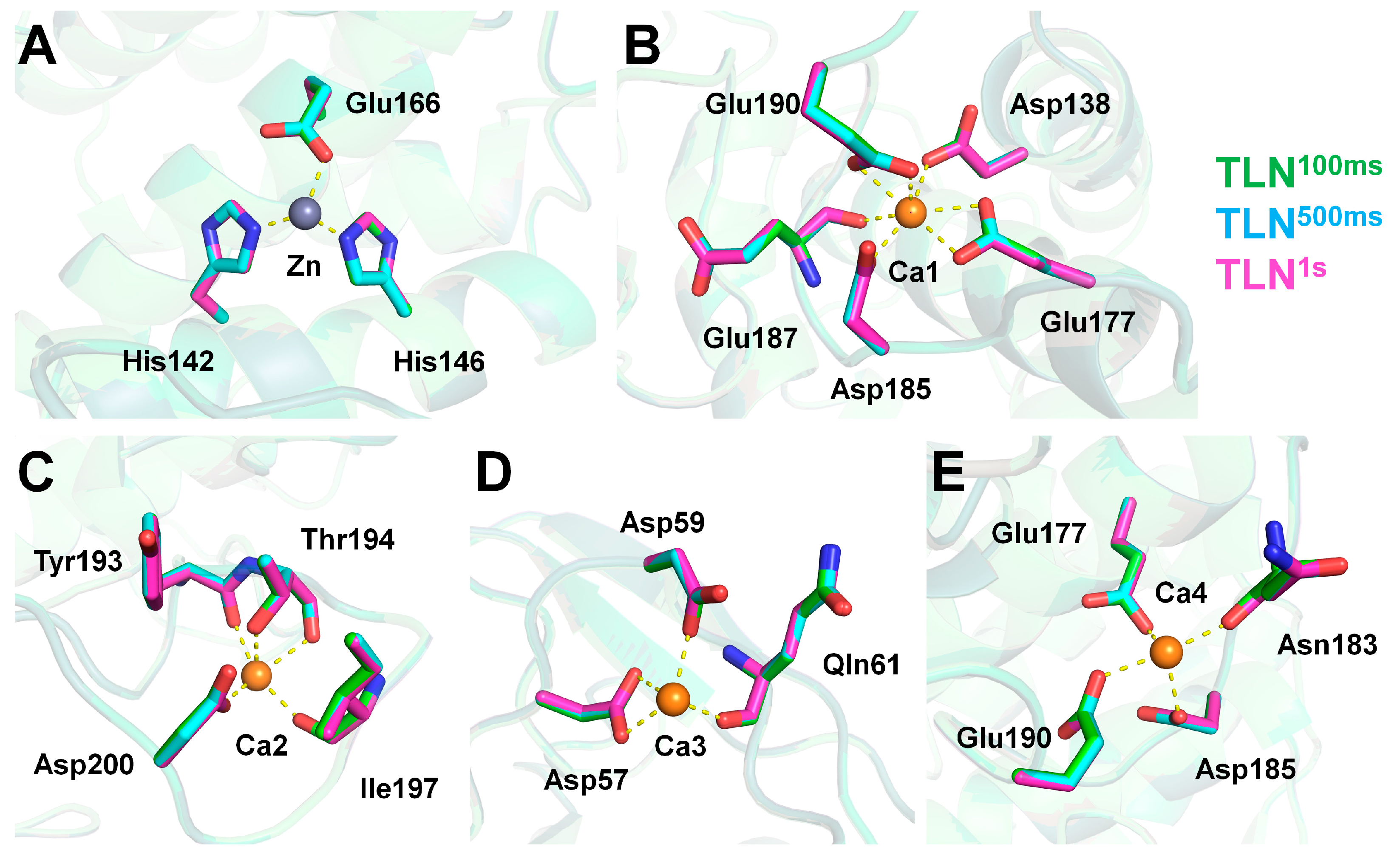
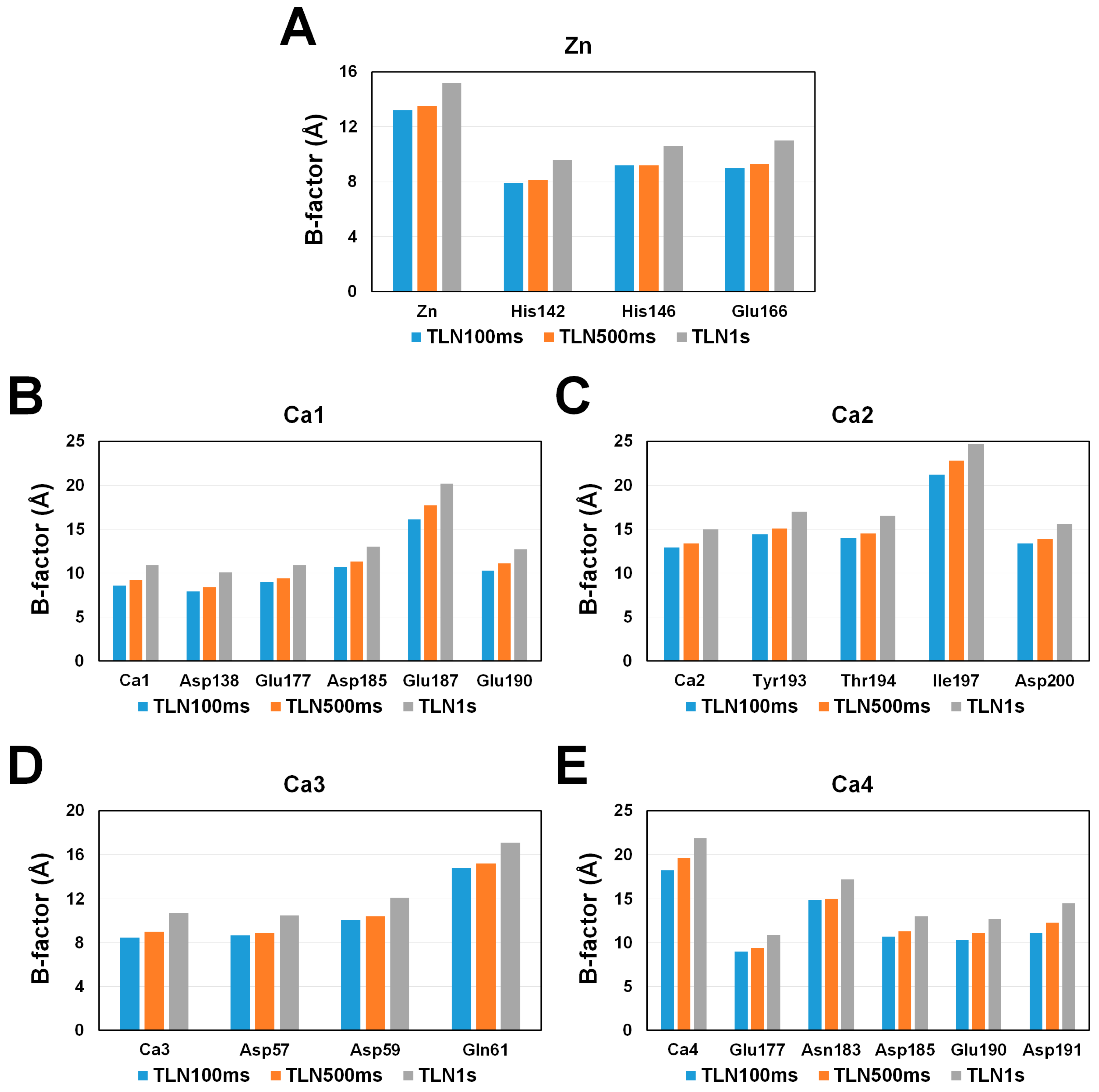
| Data Collection | TLN100ms | TLN500ms | TLN1s |
|---|---|---|---|
| Beamline | Beamline 11C, PLS-II | Beamline 11C, PLS-II | Beamline 11C, PLS-II |
| Temperature (K) | 100 | 100 | 100 |
| Exposure time (ms/°) | 100 | 500 | 1000 |
| Space group | P6122 | P6122 | P6122 |
| Unit cell dimension | |||
| a, b, c (Å) | 92.919, 92.919, 128.378 | 93.081, 93.081, 128.694 | 93.141, 93.141,128.777 |
| α, β, γ (°) | 90.00, 90.00, 120.00 | 90.00, 90.00, 120.00 | 90.00, 90.00, 120.00 |
| Resolution range (Å) | 50.0–1.35 (1.37–1.35) | 50.0–1.30 (1.32–1.30) | 50.0–1.35 (1.37–1.35) |
| No. of unique reflections | 72,111 (3536) | 81,578 (3991) | 74,793 (3665) |
| Completeness (%) | 100.0 (100.0) | 100.0 (100.0) | 100.0 (100.0) |
| Redundancy | 38.4 (33.3) | 35.7 (22.5) | 32.6 (14.0) |
| I/sigma(I) | 28.25 (2.00) | 31.94 (1.76) | 32.89 (1.60) |
| Rmerge | 0.144 (0.422) | 0.169 (0.472) | 0.317 (3.771) |
| Rmeas | 0.146 (1.617) | 0.172 (2.406) | 0.322 (3.907) |
| CC1/2 | 0.998 (0.859) | 0.998 (0.762) | 0.996 (0.703) |
| Refinement | TLN100ms | TLN500ms | TLN1s |
|---|---|---|---|
| Resolution range (Å) | 34.09–1.40 | 34.16–1.40 | 34.18–1.40 |
| Rworka | 0.14822 | 0.14948 | 0.15182 |
| Rfreeb | 0.16730 | 0.16843 | 0.17244 |
| R.m.s. deviations | |||
| Bonds (Å) | 0.012 | 0.012 | 0.012 |
| Angles (°) | 1.780 | 1.801 | 1.758 |
| Average B factors (Å2) | |||
| Protein | 11.642 | 12.001 | 13.603 |
| Water | 26.294 | 26.688 | 27.514 |
| Ramachandran plot | |||
| Most favored (%) | 96.2 | 96.2 | 96.5 |
| Allowed (%) | 3.8 | 3.8 | 3.5 |
| PDB code | 8ZM4 | 8ZM5 | 8ZM6 |
| Metal Ion | Residues (atom) | TLN100ms | TLN500ms | TLN1s |
|---|---|---|---|---|
| Zn | His142 (NE2) | 2.10 | 2.09 | 2.09 |
| His146 (NE2) | 2.03 | 2.03 | 2.05 | |
| Glu166 (OE2) | 2.08 | 2.10 | 2.11 | |
| Ca1 | Asp138 (OD2) | 2.40 | 2.38 | 2.38 |
| Glu177 (OE1/OE2) | 2.53/2.58 | 2.53/2.60 | 2.54/2.59 | |
| Asp185 (OD1) | 2.42 | 2.45 | 2.45 | |
| Glu187 (O) | 2.38 | 2.36 | 2.36 | |
| Glu190 (OE1/OE2) | 2.56/2.45 | 2.57/2.47 | 2.58/2.45 | |
| Ca2 | Tyr193 (O) | 2.33 | 2.33 | 2.33 |
| Thr194 (O/OG1) | 2.40/2.44 | 2.42/2.43 | 2.42/2.44 | |
| Ile197 (O) | 2.24 | 2.24 | 2.27 | |
| Asp200 (OD1) | 2.34 | 2.37 | 2.35 | |
| Ca3 | Asp57 (OD1/OD2) | 2.57/2.38 | 2.57/2.38 | 2.57/2.38 |
| Asp59 (OD1) | 2.39 | 2.38 | 2.39 | |
| Gln61 (O) | 2.30 | 2.28 | 2.28 | |
| Ca4 | Glu177 (OE2) | 2.75 | 2.77 | 2.79 |
| Asn183 (O) | 2.28 | 2.29 | 2.27 | |
| Asp185 (OD2) | 2.39 | 2.39 | 2.40 | |
| Glu190 (OE2) | 2.34 | 2.35 | 2.37 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nam, K.H. Effects of Radiation Damage on Metal-Binding Sites in Thermolysin. Crystals 2024, 14, 876. https://doi.org/10.3390/cryst14100876
Nam KH. Effects of Radiation Damage on Metal-Binding Sites in Thermolysin. Crystals. 2024; 14(10):876. https://doi.org/10.3390/cryst14100876
Chicago/Turabian StyleNam, Ki Hyun. 2024. "Effects of Radiation Damage on Metal-Binding Sites in Thermolysin" Crystals 14, no. 10: 876. https://doi.org/10.3390/cryst14100876
APA StyleNam, K. H. (2024). Effects of Radiation Damage on Metal-Binding Sites in Thermolysin. Crystals, 14(10), 876. https://doi.org/10.3390/cryst14100876






