Structural Basis for the Influence of A1, 5A, and W51W57 Mutations on the Conductivity of the Geobacter sulfurreducens Pili
Abstract
:1. Introduction
2. Materials and Methods
2.1. Molecular Dynamics Simulation
2.2. Cross-Correlation Analysis
2.3. Electrostatic Properties Analysis
2.4. Constructing a Pilus Structure from Subunit Data with Sparse Constraints
2.5. Residue Interaction Network
2.6. Algorithm for Exploring Potential Electrically Conductive Pathway
3. Results
3.1. Structures of Pilins
3.2. Structures of Wild-Type Pilus and Its Mutant Pili for G. Sulfurreducens
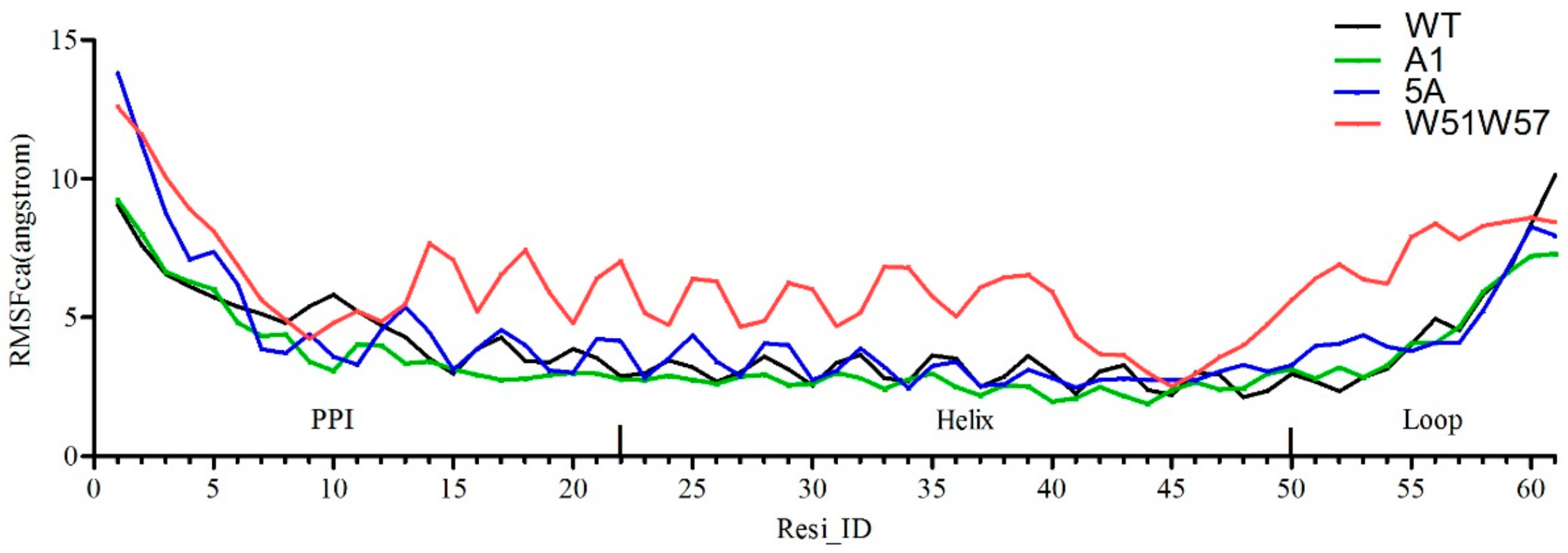
3.3. Structural Changes in Geobacter Pili Were Partly Attributed to More Unstable and More Flexible Pilins
3.4. Structural Basis for Diverse Values of Conductivity
3.5. Potential Electron Transfer Pathway in Geobacter Pili
4. Discussion
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Reguera, G.; McCarthy, K.D.; Mehta, T.; Nicoll, J.S.; Tuominen, M.T.; Lovley, D.R. Extracellular electron transfer via microbial nanowires. Nature 2005, 435, 1098–1101. [Google Scholar] [CrossRef] [PubMed]
- Adhikari, R.Y.; Malvankar, N.S.; Tuominen, M.T.; Lovley, D.R. Conductivity of individual Geobacter pili. RSC Adv. 2016, 6, 8363–8366. [Google Scholar] [CrossRef]
- Malvankar, N.S.; Vargas, M.; Nevin, K.; Tremblay, P.L.; Evans-Lutterodt, K.; Nykypanchuk, D.; Martz, E.; Tuominen, M.T.; Lovley, D.R. Structural basis for metallic-like conductivity in microbial nanowires. mBio 2015, 6, e00084-15. [Google Scholar] [CrossRef] [PubMed]
- Bond, D.R.; Lovley, D.R. Electricity production by Geobacter sulfurreducens attached to electrodes. Appl. Environ. Microbiol. 2003, 69, 1548–1555. [Google Scholar] [CrossRef] [PubMed]
- Leang, C.; Qian, X.; Mester, T.; Lovley, D.R. Alignment of the c-type cytochrome OmcS along pili of Geobacter sulfurreducens. Appl. Environ. Microb. 2010, 76, 4080–4084. [Google Scholar] [CrossRef] [PubMed]
- Reardon, P.N.; Mueller, K.T. Structure of the type IVa major pilin from the electrically conductive bacterial nanowires of Geobacter sulfurreducens. J. Biol. Chem. 2013, 288, 29260–29266. [Google Scholar] [CrossRef] [PubMed]
- Craig, L.; Pique, M.E.; Tainer, J.A. Type IV pilus structure and bacterial pathogenicity. Nat. Rev. Microbiol. 2004, 2, 363–378. [Google Scholar] [CrossRef] [PubMed]
- Xiao, K.; Malvankar, N.S.; Shu, C.; Martz, E.; Lovley, D.R.; Sun, X. Low energy atomic models suggesting a pilus structure that could account for electrical conductivity of Geobacter sulfurreducens pili. Sci. Rep. UK 2016, 6, 23385. [Google Scholar] [CrossRef] [PubMed]
- Qin, N.; Shen, Y.; Yang, X.; Su, L.; Tang, R.; Li, W.; Wang, M. Site-directed mutagenesis under the direction of in silico protein docking modeling reveals the active site residues of 3-ketosteroid-Delta1-dehydrogenase from Mycobacterium neoaurum. World. J. Microb. Biotechnol. 2017, 33, 146. [Google Scholar] [CrossRef] [PubMed]
- Shih, C.; Museth, A.K.; Abrahamsson, M.; Blanco-Rodriguez, A.M.; Di Bilio, A.J.; Sudhamsu, J.; Crane, B.R.; Ronayne, K.L.; Towrie, M., Jr.; Vlcek, A.; et al. Tryptophan-accelerated electron flow through proteins. Science 2008, 320, 1760–1762. [Google Scholar] [CrossRef] [PubMed]
- Vargas, M.; Malvankar, N.S.; Tremblay, P.L.; Leang, C.; Smith, J.A.; Patel, P.; Snoeyenbos-West, O.; Nevin, K.P.; Lovley, D.R. Aromatic amino acids required for pili conductivity and long-range extracellular electron transport in Geobacter sulfurreducens. mBio 2013, 4, e105–e113. [Google Scholar] [CrossRef]
- Tan, Y.; Adhikari, R.Y.; Malvankar, N.S.; Pi, S.; Ward, J.E.; Woodard, T.L.; Nevin, K.P.; Xia, Q.; Tuominen, M.T.; Lovley, D.R. Synthetic biological protein nanowires with high conductivity. Small 2016, 12, 4481–4485. [Google Scholar] [CrossRef] [PubMed]
- Richter, L.V.; Sandler, S.J.; Weis, R.M. Two isoforms of Geobacter sulfurreducens PilA have distinct roles in pilus biogenesis, cytochrome localization, extracellular electron transfer, and biofilm formation. J. Bacteriol. 2012, 194, 2551–2563. [Google Scholar] [CrossRef] [PubMed]
- Shu, C.; Xiao, K.; Yan, Q.; Sun, X. Comparative analysis of Type IV pilin in Desulfuromonadales. Front. Microbiol. 2016, 7, 2080. [Google Scholar] [CrossRef] [PubMed]
- Shu, C.; Xiao, K.; Cao, C.; Ding, D.; Sun, X. Predicting and interpreting the structure of type IV pilus of electricigens by molecular dynamics simulations. Molecules 2017, 8, 1342. [Google Scholar] [CrossRef] [PubMed]
- Yan, H.; Chuang, C.; Zhugayevych, A.; Tretiak, S.; Dahlquist, F.W.; Bazan, G.C. Inter-aromatic distances in Geobacter sulfurreducens pili relevant to biofilm charge transport. Adv. Mater. 2015, 27, 1908–1911. [Google Scholar] [CrossRef] [PubMed]
- Xiao, K.; Shu, C.; Yan, Q.; Sun, X. Predicting homogeneous pilus structure from monomeric data and sparse constraints. BioMed Res. Int. 2015, 2015, 817134. [Google Scholar] [CrossRef] [PubMed]
- Case, D.A.; Darden, T.A.; Cheatham, T.E., III; Simmerling, C.L.; Wang, J.; Duke, R.E.; Luo, R.; Walker, R.C.; Zhang, W.; Merz, K.M.; et al. Amber 12; University of California: San Francisco, CA, USA, 2012. [Google Scholar]
- Tan, Z.; Spasic, A.; Mathews, D.H. 96 Benchmark of Amber ff12SB force field by comparison of estimated hairpin loop folding stabilities to experiments. J. Biomol. Struct. Dyn. 2015, 33 (Suppl. 1), 61–62. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, S.; Phu, M.; de Morais, T.P.; Nascimento, R.; Goulart, L.R.; Rao, B.J.; Asgeirsson, B.; Dandekar, A.M. The PDB database is a rich source of alpha-helical anti-microbial peptides to combat disease causing pathogens. F1000Research 2014, 3, 295. [Google Scholar] [CrossRef] [PubMed]
- McKay, T.; Hart, K.; Bernstein, H.; Tedla-Boyd, W.; Craig, P. Protein function prediction using ProMOL and PyMOL. FASEB J. 2014, 1, 722–725. [Google Scholar]
- Choudhury, C.; Deva, P.U.; Sastry, G.N. Molecular dynamics investigation of the active site dynamics of mycobacterial cyclopropane synthase during various stages of the cyclopropanation process. J. Struct. Biol. 2014, 187, 38–48. [Google Scholar] [CrossRef] [PubMed]
- Needham, P.J.; Bhuiyan, A.; Walker, R.C. Extension of the AMBER molecular dynamics software to Intel’s Many Integrated Core (MIC) architecture. Comput. Phys. Commun. 2016, 201, 95–105. [Google Scholar] [CrossRef]
- Feliciano, G.T.; Steidl, R.J.; Reguera, G. Structural and functional insights into the conductive pili of Geobacter sulfurreducens revealed in molecular dynamics simulations. Phys. Chem. Chem. Phys. 2015, 17, 22217–22226. [Google Scholar] [CrossRef] [PubMed]
- Roe, D.R.; Cheatham, T.R. PTRAJ and CPPTRAJ: Software for processing and analysis of molecular dynamics trajectory data. J. Chem. Theory. Comput. 2013, 9, 3084–3095. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.M.; Xia, L.Q.; Yang, X.P.; Peng, X.Y. Flexibility analysis of bacillus thuringiensis Cry1Aa. Biomed. Environ. Sci. 2015, 28, 634–641. [Google Scholar] [PubMed]
- Liu, M.; Wang, L.; Sun, X.; Zhao, X. Investigating the impact of Asp181 point mutations on interactions between PTP1B and phosphotyrosine substrate. Sci. Rep. 2014, 4, 5095. [Google Scholar] [CrossRef] [PubMed]
- Unni, S.; Huang, Y.; Hanson, R.M.; Tobias, M.; Krishnan, S.; Li, W.W.; Nielsen, J.E.; Baker, N.A. Web servers and services for electrostatics calculations with APBS and PDB2PQR. J. Comput. Chem. 2011, 32, 1488–1491. [Google Scholar] [CrossRef] [PubMed]
- Craig, L.; Volkmann, N.; Arvai, A.S.; Pique, M.E.; Yeager, M.; Egelman, E.H.; Tainer, J.A. Type IV pilus structure by cryo-electron microscopy and crystallography: Implications for pilus assembly and functions. Mol. Cell 2006, 23, 651–662. [Google Scholar] [CrossRef] [PubMed]
- Dolinsky, T.J.; Czodrowski, P.; Li, H.; Nielsen, J.E.; Jensen, J.H.; Klebe, G.; Baker, N.A. PDB2PQR: Expanding and upgrading automated preparation of biomolecular structures for molecular simulations. Nucleic Acids Res. 2007, 35, W522–W525. [Google Scholar] [CrossRef] [PubMed]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996, 14, 27–28, 33–38. [Google Scholar] [CrossRef]
- Piovesan, D.; Minervini, G.; Tosatto, S.C.E. The RING 2.0 web server for high quality residue interaction networks. Nucleic Acids Res. 2016, 44, W367–W374. [Google Scholar] [CrossRef] [PubMed]
- Ihlen, E.A.F. Introduction to multifractal detrended fluctuation analysis in Matlab. Front. Physiol. 2012, 3, 141. [Google Scholar] [CrossRef] [PubMed]
- Dijkstra, E.W. A note on two problems in connexion with graphs. Numer. Math. 1959, 1, 269–271. [Google Scholar] [CrossRef]
- DeLano, W.L. Pymol: An open-source molecular graphics tool. CCP4 Newsl. Protein Crystallogr. 2002, 40, 82–92. [Google Scholar]
- Liu, C. Modifications of steepest descent method and conjugate gradient method against noise for ill-posed linear systems. Commun. Numer. Anal. 2012, 2012, cna-00115. [Google Scholar] [CrossRef]
- Mandell, A.J.; Selz, K.A.; Shlesinger, M.F. Transformational homologies in amino acid sequences suggest memberships in protein families. J. Stat. Phys. 1998, 93, 673–697. [Google Scholar] [CrossRef]
- Yiqing, H.; Qiang, L.X.; Jia, Z. Using evolutionary couplings to optimize dimer prediction with Rosetta SymDock. In Proceedings of the 2013 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), Shanghai, China, 18–21 December 2013; IEEE: Piscataway, NJ, USA, 2013; pp. 65–68. [Google Scholar]
- Leaver-Fay, A.; Tyka, M.; Lewis, S.M.; Lange, O.F.; Thompson, J.; Jacak, R.; Kaufman, K.; Renfrew, P.D.; Smith, C.A.; Sheffler, W.; et al. ROSETTA3: An object-oriented software suite for the simulation and design of macromolecules. Methods Enzymol. 2011, 487, 545–574. [Google Scholar] [PubMed]
- Das, R.; Baker, D. Macromolecular modeling with Rosetta. Annu. Rev. Biochem. 2008, 77, 363–382. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Nussinov, R. Salt bridge stability in monomeric proteins. J. Mol. Biol. 1999, 293, 1241–1255. [Google Scholar] [CrossRef] [PubMed]
- Bosshard, H.R.; Marti, D.N.; Jelesarov, I. Protein stabilization by salt bridges: Concepts, experimental approaches and clarification of some misunderstandings. J. Mol. Recognit. 2004, 17. [Google Scholar] [CrossRef] [PubMed]
- Min, B.K.; Cheng, S.A.; Logan, B.E. Electricity generation using membrane and salt bridge microbial fuel cells. Water Res. 2005, 39, 1675–1686. [Google Scholar] [CrossRef] [PubMed]
- Campos, M.; Cisneros, D.A.; Nivaskumar, M.; Francetic, O. The type II secretion system - a dynamic fiber assembly nanomachine. Res. Microbiol. 2013, 164, 545–555. [Google Scholar] [CrossRef] [PubMed]
- Dobbins, S.E.; Lesk, V.I.; Sternberg, M.J.E. Insights into protein flexibility: The relationship between normal modes and conformational change upon protein-protein docking. Proc. Natl. Acad. Sci. USA 2008, 105, 10390–10395. [Google Scholar] [CrossRef] [PubMed]
- Giltner, C.L.; Nguyen, Y.; Burrows, L.L. Type IV pilin proteins: Versatile molecular modules. Microbiol. Mol. Biol. Rev. 2012, 76, 740–772. [Google Scholar] [CrossRef] [PubMed]
- Fuglestad, B.; Gasper, P.M.; McCammon, J.A.; Markwick, P.R.L.; Komives, E.A. Correlated motions and residual frustration in thrombin. J. Phys. Chem. B 2013, 117, 12857–12863. [Google Scholar] [CrossRef] [PubMed]
- Malvankar, N.S.; Vargas, M.; Nevin, K.P.; Franks, A.E.; Leang, C.; Kim, B.C.; Inoue, K.; Mester, T.; Covalla, S.F.; Johnson, J.P.; et al. Tunable metallic-like conductivity in microbial nanowire networks. Nat. Nanotechnol. 2011, 6, 573–579. [Google Scholar] [CrossRef] [PubMed]
- Donald, J.E.; Kulp, D.W.; DeGrado, W.F. Salt bridges: Geometrically specific, designable interactions. Proteins 2011, 79, 898–915. [Google Scholar] [CrossRef] [PubMed]
- Hunter, C.A.; Sanders, J. The nature of pi-pi interactions. J. Am. Chem. Soc. 1990, 112, 5525–5534. [Google Scholar] [CrossRef]
- Sinnokrot, M.O.; Sherrill, C.D. Highly accurate coupled cluster potential energy curves for the benzene dimer: Sandwich, T-shaped, and parallel-displaced configurations. J. Phys. Chem. A 2004, 108, 10200–10207. [Google Scholar] [CrossRef]
- Page, C.C.; Moser, C.C.; Chen, X.; Dutton, P.L. Natural engineering principles of electron tunnelling in biological oxidation-reduction. Nature 1999, 402, 47–52. [Google Scholar] [CrossRef] [PubMed]
- Aas, F.E.; Winther-Larsen, H.C.; Wolfgang, M.; Frye, S.; Lovold, C.; Roos, N.; van Putten, J.P.M.; Koomey, M. Substitutions in the N-terminal alpha helical spine of Neisseria gonorrhoeae pilin affect type IV pilus assembly, dynamics and associated functions. Mol. Microbiol. 2007, 63, 69–85. [Google Scholar] [CrossRef] [PubMed]
- Stoodley, C.J.; Limperopoulos, C. Structure-function relationships in the developing cerebellum: Evidence from early-life cerebellar injury and neurodevelopmental disorders. Semin. Fetal Neonatal Med. 2016, 21, 356–364. [Google Scholar] [CrossRef] [PubMed]
- Tan, Y.; Adhikari, R.Y.; Malvankar, N.S.; Ward, J.E.; Woodard, T.L.; Nevin, K.P.; Lovley, D.R. Expressing the Geobacter metallireducens PilA in Geobacter sulfurreducens yields pili with exceptional conductivity. mBio 2017, 8, e2203–e2216. [Google Scholar] [CrossRef] [PubMed]
- Walker, D.J.F.; Adhikari, R.Y.; Holmes, D.E.; Ward, J.E.; Woodard, T.L.; Nevin, K.P.; Lovley, D.R. Electrically conductive Pili from pilin genes of phylogenetically diverse microorganisms. ISME J. 2017, 2017, 141. [Google Scholar]
- Kumar, S.; Nussinov, R. Close-range electrostatic interactions in proteins. ChemBioChem. 2002, 3, 604–617. [Google Scholar] [CrossRef]
- Martinez, C.R.; Iverson, B.L. Rethinking the term “pi-stacking”. Chem. Sci. 2012, 3, 2191–2201. [Google Scholar] [CrossRef]









| Pilus | Before ✶ | After-Assembly | |||
|---|---|---|---|---|---|
| Intra ✶✶ | Inter ⌑-Common ⌑⌑ | Inter-Different ⌑⌑ | Number ✦ | ||
| WT | 0 | 48E-44K | 0 | 28R-60E1, 60E-28R2, 5E-41R3, 41R-5E4 | 21 |
| A1 | 0 | 48E-44K | 30K-53D1, 53D-30K2 | 0 | 39 |
| 5A | 0 | 48E-44K | 28R-39D1, 39D-28R2 | 53D-30K3, 30K-53D4 | 38 |
| W51W57 | 0 | 48E-44K | 39D-41R1, 41R-39D2, 44K-60E3, 60E-44K4 | 5E-41R5, 41R-5E6 | 41 |
| N ⟡ | D (Å) | R (Å) | ρ (1021 cm−3) | Conductivity (μs·cm−1) | |
|---|---|---|---|---|---|
| WT | 6 | 31.6 | 10.7 | 4.29 | 5 [48] |
| A1 | 5 | 30.3 | 10.9 | 1.91 | UN ✦ |
| 5A | 1 | 32.6 | 7.2 | 0.57 | WT*1/100 [11] |
| W51W57 | 8 | 23.6 | 10.5 | 12.02 | 500*WT [12] |
| Pathway | Distance (Å) | |
|---|---|---|
| WT | (F1p, Y27p+4, F24p+4, F1p+1, Y27p+5, F24p+5, F1p+2, Y27p+6, F24p+6, F1p+3, Y27p+7, F24p+7, F1p+4, Y27p+8, F24p+8, F1p+5, Y27p+9, F24p+9, F1p+6) | (5.3, 6.2, 4.3, 5.3, 6.2, 4.3, 5.3, 6.2, 4.3, 5.3, 6.2, 4.3, 5.3, 6.2, 4.3, 5.3, 6.2, 4.3) |
| W51W57 | (F1p, Y27p+4, F24p+4, F1p+1, Y27p+5, F24p+5, F1p+2, Y27p+6, F24p+6, F1p+3, Y27p+7, F24p+7, F1p+4, Y27p+8, F24p+8, F1p+5, Y27p+9, F24p+9, F1p+6) | (5.1, 6.0, 3.7, 5.1, 6.0, 3.7, 5.1, 6.0, 3.7, 5.1, 6.0, 3.7, 5.1, 6.0, 3.7, 5.1, 6.0, 3.7) |
| A1 | (Y27p, F24p, F51p+4, Y27p+1, F24p+1, F51p+5, Y27p+2, F24p+2, F51p+6, Y27p+3) | (6.7, 10.3, 4.6, 6.7, 10.3, 4.6, 6.7, 10.3, 4.6) |
| 5A | (F1p, F1p+1, F1p+2, F1p+3) | (11.3, 11.3, 11.3) |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shu, C.; Xiao, K.; Sun, X. Structural Basis for the Influence of A1, 5A, and W51W57 Mutations on the Conductivity of the Geobacter sulfurreducens Pili. Crystals 2018, 8, 10. https://doi.org/10.3390/cryst8010010
Shu C, Xiao K, Sun X. Structural Basis for the Influence of A1, 5A, and W51W57 Mutations on the Conductivity of the Geobacter sulfurreducens Pili. Crystals. 2018; 8(1):10. https://doi.org/10.3390/cryst8010010
Chicago/Turabian StyleShu, Chuanjun, Ke Xiao, and Xiao Sun. 2018. "Structural Basis for the Influence of A1, 5A, and W51W57 Mutations on the Conductivity of the Geobacter sulfurreducens Pili" Crystals 8, no. 1: 10. https://doi.org/10.3390/cryst8010010
APA StyleShu, C., Xiao, K., & Sun, X. (2018). Structural Basis for the Influence of A1, 5A, and W51W57 Mutations on the Conductivity of the Geobacter sulfurreducens Pili. Crystals, 8(1), 10. https://doi.org/10.3390/cryst8010010





