Identification and Crystallographic Analysis of a New Carbohydrate Acetylesterase (SmAcE1) from Sinorhizobium meliloti
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Sequence Alignment of SmAcE1 with Other CEs
2.3. Cloning and Site-Directed Mutagenesis
2.4. Protein Expression and Crystallization
2.5. Data Collection
2.6. Analytical Size-Exclusion Chromatography
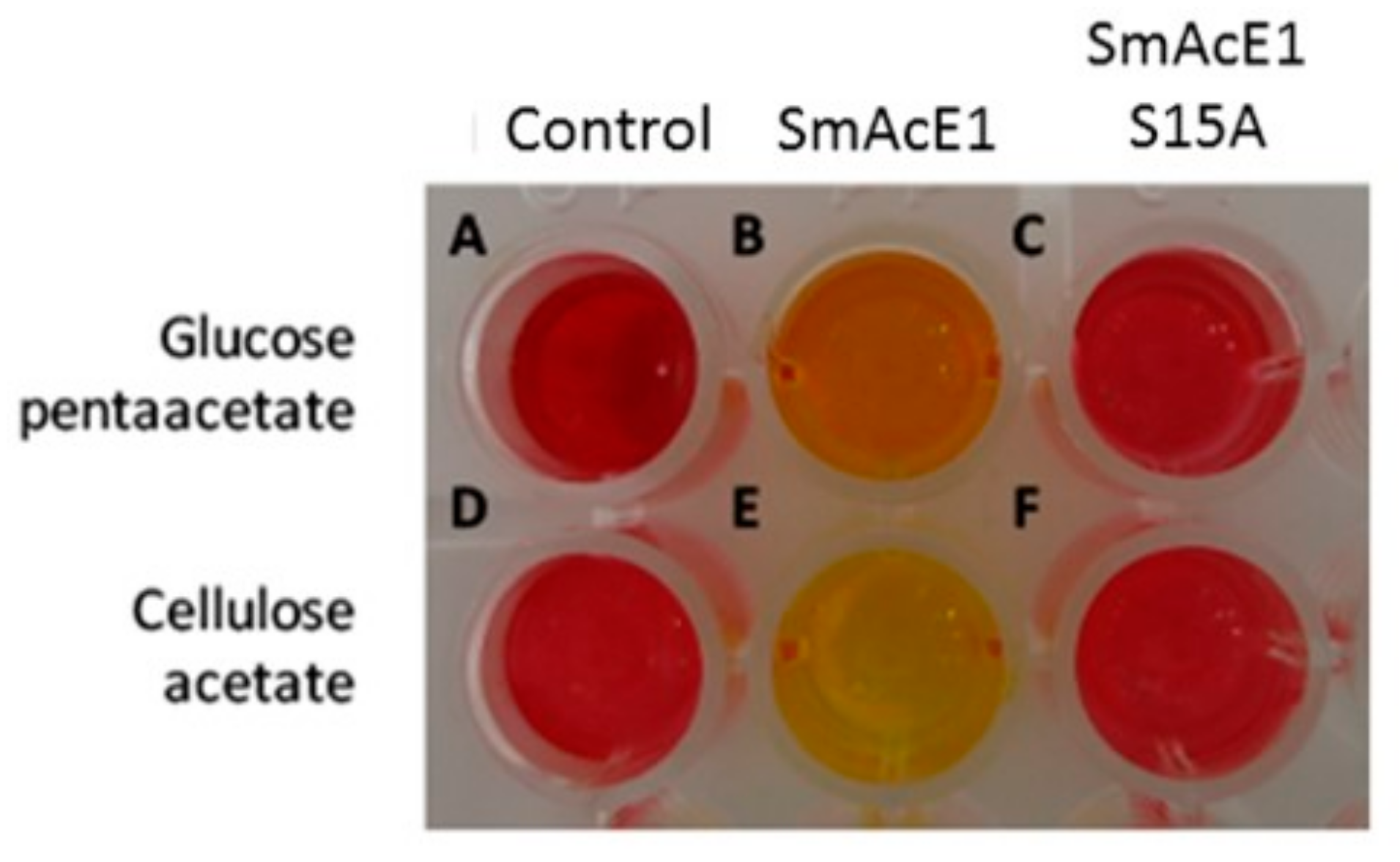
2.7. Enzyme Activity Assay Using Colorimetric Method
3. Results and Discussion
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Andre, I.; Potocki-Véronèse, G.; Barbe, S.; Moulis, C.; Remaud-Siméon, M. CAZyme discovery and design for sweet dreams. Curr. Opin. Chem. Biol. 2014, 19, 17–24. [Google Scholar] [CrossRef] [PubMed]
- Cantarel, B.L.; Coutinho, P.M.; Rancurel, C.; Bernard, T.; Lombard, V.; Henrissat, B. The Carbohydrate-Active EnZymes database (CAZy): An expert resource for Glycogenomics. Nucleic Acids Res. 2009, 37, D233–D238. [Google Scholar] [CrossRef] [PubMed]
- Coutinho, P.M.; Stam, M.; Blanc, E.; Henrissat, B. Why are there so many carbohydrate-active enzyme-related genes in plants? Trends Plant Sci. 2003, 8, 563–565. [Google Scholar] [CrossRef] [PubMed]
- Ekstrom, A.; Taujale, R.; McGinn, N.; Yin, Y. PlantCAZyme: A database for plant carbohydrate-active enzymes. Database 2014. [Google Scholar] [CrossRef] [PubMed]
- Adesioye, F.A.; Makhalanyane, T.P.; Biely, P.; Cowan, D.A. Phylogeny, classification and metagenomic bioprospecting of microbial acetyl xylan esterases. Enzyme Microb. Technol. 2016, 93–94, 79–91. [Google Scholar] [CrossRef] [PubMed]
- Wong, D.W. Feruloyl esterase: A key enzyme in biomass degradation. Appl. Biochem. Biotechnol. 2006, 133, 87–112. [Google Scholar] [CrossRef]
- Barad, S.; Sela, N.; Kumar, D.; Kumar-Dubey, A.; Glam-Matana, N.; Sherman, A.; Prusky, D. Fungal and host transcriptome analysis of pH-regulated genes during colonization of apple fruits by Penicillium expansum. BMC Genom. 2016, 17, 330. [Google Scholar] [CrossRef] [PubMed]
- Busse-Wicher, M.; Gomes, T.C.F.; Tryfona, T.; Nikolovski, N.; Stott, K.; Grantham, N.J.; Bolam, D.N.; Skaf, M.S.; Dupree, P. The pattern of xylan acetylation suggests xylan may interact with cellulose microfibrils as a twofold helical screw in the secondary plant cell wall of Arabidopsis thaliana. Plant J. 2014, 79, 492–506. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chebli, Y.; Geitmann, A. Cellular growth in plants requires regulation of cell wall biochemistry. Curr. Opin. Cell Biol. 2017, 44, 28–35. [Google Scholar] [CrossRef] [PubMed]
- Coque, L.; Neogi, P.; Pislariu, C.; Wilson, K.A.; Catalano, C.; Avadhani, M.; Sherrier, D.J.; Dickstein, R. Transcription of ENOD8 in medicago truncatula nodules directs ENOD8 esterase to developing and mature symbiosomes. Mol. Plant-Microbe Interact. 2008, 21, 404–410. [Google Scholar] [CrossRef] [PubMed]
- Dickstein, R. ENOD8, a novel early nodule-specific gene, is expressed in empty alfalfa nodules. Mol. Plant-Microbe Interact. 1993, 6, 715–721. [Google Scholar] [CrossRef] [PubMed]
- Grantham, N.J.; Wurman-Rodrich, J.; Terrett, O.M.; Lyczakowski, J.; Stott, K.; Iuga, D.; Simmons, T.J.; Durand-Tardif, M.; Brown, S.P.; Dupree, R.; et al. An even pattern of xylan substitution is critical for interaction with cellulose in plant cell walls. Nat. Plants 2017. [Google Scholar] [CrossRef] [PubMed]
- Jones, K.M.; Kobayashi, H.; Davies, B.W.; Taga, M.E.; Walker, G.C. How rhizobial symbionts invade plants: The Sinorhizobium–Medicago model. Nat. Rev. Microbiol. 2007, 5, 619–633. [Google Scholar] [CrossRef] [PubMed]
- Kabel, M.A.; Yeoman, C.J.; Han, Y.; Dodd, D.; Abbas, C.A.; de Bont, J.M.; Morrison, M.; Cann, I.K.O.; Mackie, R.I. Biochemical characterization and relative expression levels of multiple carbohydrate esterases of the xylanolytic rumen bacterium Prevotella ruminicola 23 grown on an ester-enriched substrate. Appl. Environ. Microbiol. 2011, 77, 5671–5681. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, B.; Zhang, L.; Li, F.; Zhang, D.; Liu, X.; Wang, H.; Xu, Z.; Chu, C.; Zhou, Y. Control of secondary cell wall patterning involves xylan deacetylation by a GDSL esterase. Nat. Plants 2017, 3, 17017. [Google Scholar] [CrossRef] [PubMed]
- Antonopoulou, I.; Varriale, S.; Topakas, E.; Rova, U.; Christakopoulos, P.; Faraco, V. Enzymatic synthesis of bioactive compounds with high potential for cosmeceutical application. Appl. Microbiol. Biotechnol. 2016, 100, 6519–6543. [Google Scholar] [CrossRef] [PubMed]
- Mathew, M.P.; Tan, E.; Shah, S.; Bhattacharya, R.; Adam Meledeo, M.; Huang, J.; Espinoza, F.A.; Yarema, K.J. Extracellular and intracellular esterase processing of SCFA–hexosamine analogs: Implications for metabolic glycoengineering and drug delivery. Bioorg. Med. Chem. Lett. 2012, 22, 6929–6933. [Google Scholar] [CrossRef] [PubMed]
- Yeoman, C.J.; Han, Y.; Dodd, D.; Schroeder, C.M.; Mackie, R.I.; Cann, I.K. Thermostable enzymes as biocatalysts in the biofuel industry. Adv. Appl. Microbiol. 2010, 70, 1–55. [Google Scholar] [PubMed]
- Kahya, F.H.; Andrew, P.W.; Yesilkaya, H. Deacetylation of sialic acid by esterases potentiates pneumococcal neuraminidase activity for mucin utilization, colonization and virulence. PLoS Pathog. 2017, 13, e1006263. [Google Scholar] [CrossRef] [PubMed]
- Steenbergen, M.S.; Jirik, J.L.; Vimr, E.R. YjhS (NanS) is required for Escherichia coli to grow on 9-O-acetylated N-acetylneuraminic acid. J. Bacteriol. 2009, 191, 7134–7139. [Google Scholar] [CrossRef] [PubMed]
- Pfeffer, J.M.; Weadge, J.T.; Clarke, A.J. Mechanism of action of Neisseria gonorrhoeae O-acetylpeptidoglycan esterase, an SGNH serine esterase. J. Biol. Chem. 2013, 288, 2605–2613. [Google Scholar] [CrossRef] [PubMed]
- Rangarajan, E.S.; Ruane, K.M.; Proteau, A.; Schrag, J.D.; Valladares, R.; Gonzalez, C.F.; Gilbert, M.; Yakunin, A.F.; Cygler, M. Structural and enzymatic characterization of NanS (YjhS), a 9-O-Acetyl N-acetylneuraminic acid esterase from Escherichia coli O157:H7. Protein Sci. 2011, 20, 1208–1219. [Google Scholar] [CrossRef] [PubMed]
- Till, M.; Goldstone, D.C.; Attwood, G.T.; Moon, C.D.; Kelly, W.J.; Arcus, V.L. Structure and function of an acetyl xylan esterase (Est2A) from the rumen bacterium Butyrivibrio proteoclasticus. Proteins Struct. Funct. BioInform. 2013, 81, 911–917. [Google Scholar] [CrossRef] [PubMed]
- Lansky, S.; Alalouf, O.; Solomon, H.V.; Alhassid, A.; Govada, L.; Chayen, N.E.; Belrhali, H.; Shoham, Y.; Shoham, G. A unique octameric structure of Axe2, an intracellular acetyl-xylooligosaccharide esterase from Geobacillus stearothermophilus. Acta Crystallogr. Sect. D Biol. Crystallogr. 2014, 70, 261–278. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.; Ryu, B.H.; Kim, S.S.; An, D.R.; Ngo, T.D.; Pandian, R.; Kim, K.K.; Kim, T.D. Structural and biochemical characterization of a carbohydrate acetylesterase from Sinorhizobium meliloti 1021. FEBS Lett. 2014, 589, 117–122. [Google Scholar] [CrossRef] [PubMed]
- Oh, C.; Ryu, B.H.; An, D.R.; Nguyen, D.D.; Yoo, W.; Kim, T.; Ngo, T.D.; Kim, H.S.; Kim, K.K.; Kim, T.D. Structural and biochemical characterization of an octameric carbohydrate acetylesterase from Sinorhizobium meliloti. FEBS Lett. 2016, 590, 1242–1252. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, M.; Fukada, H.; Inoue, H.; Ishikawa, K. Crystal structure of an acetylesterase from Talaromyces cellulolyticus and the importance of a disulfide bond near the active site. FEBS Lett. 2015, 589, 1200–1206. [Google Scholar] [CrossRef] [PubMed]
- Correia, M.A.S.; Prates, J.A.M.; Brás, J.; Fontes, C.M.G.A.; Newman, J.A.; Lewis, R.J.; Gilbert, H.J.; Flint, J.E. Crystal structure of a cellulosomal family 3 carbohydrate esterase from Clostridium thermocellum provides insights into the mechanism of substrate recognition. J. Mol. Biol. 2008, 379, 64–72. [Google Scholar] [CrossRef] [PubMed]
- Montanier, C.; Money, V.A.; Pires, V.M.R.; Flint, J.E.; Pinheiro, B.A.; Goyal, A.; Prates, J.A.M.; Izumi, A.; Stålbrand, H.; Morland, C.; et al. The active site of a carbohydrate esterase displays divergent catalytic and noncatalytic binding functions. PLoS Biol. 2009, 7, e1000071. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bae, S.Y.; Ryu, B.H.; Jang, E.; Kim, S.; Kim, T.D. Characterization and immobilization of a novel SGNH hydrolase (Est24) from Sinorhizobium meliloti. Appl. Microbiol. Biotechnol. 2013, 97, 1637–1647. [Google Scholar] [CrossRef] [PubMed]
- Hwang, H.; Kim, S.; Yoon, S.; Ryu, Y.; Lee, S.Y.; Kim, T.D. Characterization of a novel oligomeric SGNH-arylesterase from Sinorhizobium meliloti 1021. Int. J. Biol. Macromol. 2010, 46, 145–152. [Google Scholar] [CrossRef] [PubMed]
- Pei, J.; Kim, B.H.; Grishin, N.V. PROMALS3D: A tool for multiple protein sequence and structure alignments. Nucleic Acids Res. 2008, 36, 2295–2300. [Google Scholar] [CrossRef] [PubMed]
- Gouet, P.; Robert, X.; Courcelle, E. ESPript/ENDscript: Extracting and rendering sequence and 3D information from atomic structures of proteins. Nucleic Acids Res. 2003, 31, 3320–3323. [Google Scholar] [CrossRef] [PubMed]
- Otwinowski, Z.; Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997, 276, 307–326. [Google Scholar] [PubMed]
- Ngo, T.D.; Ryu, B.H.; Ju, H.; Jang, E.J.; Kim, K.K.; Kim, T.D. Crystallographic analysis and biochemical applications of a novel penicillin-binding protein/β-lactamase homologue from a metagenomic library. Acta Crystallogr. Sect. D Biol. Crystallogr. 2014, 70, 2455–2466. [Google Scholar] [CrossRef] [PubMed]
- Kantardjieff, A.K.; Rupp, B. Matthews coefficient probabilities: Improved estimates for unit cell contents of proteins, DNA, and protein-nucleic acid complex crystals. Protein Sci. 2003, 12, 1865–1871. [Google Scholar] [CrossRef] [PubMed]
- Matthews, B.W. Solvent content of protein crystals. J. Mol. Biol. 1968, 33, 491–497. [Google Scholar] [CrossRef]
- Weichenberger, X.C.; Rupp, B. Ten years of probabilistic estimates of biocrystal solvent content: New insights via nonparametric kernel density estimate. Acta Crystallogr. Sect. D Biol. Crystallogr. 2014, 70, 1579–1588. [Google Scholar] [CrossRef] [PubMed]
- Adxv. Adxv—A Program to Display X-ray Diffraction Images; The Scripps Research Institute: La Jolla, CA, USA, 2013. [Google Scholar]
- McCoy, A.J.; Grosse-Kunstleve, W.; Adams, P.D.; Winn, M.D.; Storoni, L.C.; Read, R.J. Phaser crystallographic software. J. Appl. Crystallogr. 2007, 40, 658–674. [Google Scholar] [CrossRef] [PubMed]
- Afonine, P.V.; Grosse-Kunstleve, R.W.; Echols, N.; Headd, J.J.; Moriarty, N.W.; Mustyakimov, M.; Terwilliger, T.C.; Urzhumtsev, A.; Zwart, P.H.; Adams, P.D. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. D Biol. Crystallogr. 2012, 68, 352–367. [Google Scholar] [CrossRef] [PubMed]
- Emsley, P.; Cowtan, K. Coot: Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004, 60, 2126–2132. [Google Scholar] [CrossRef] [PubMed]


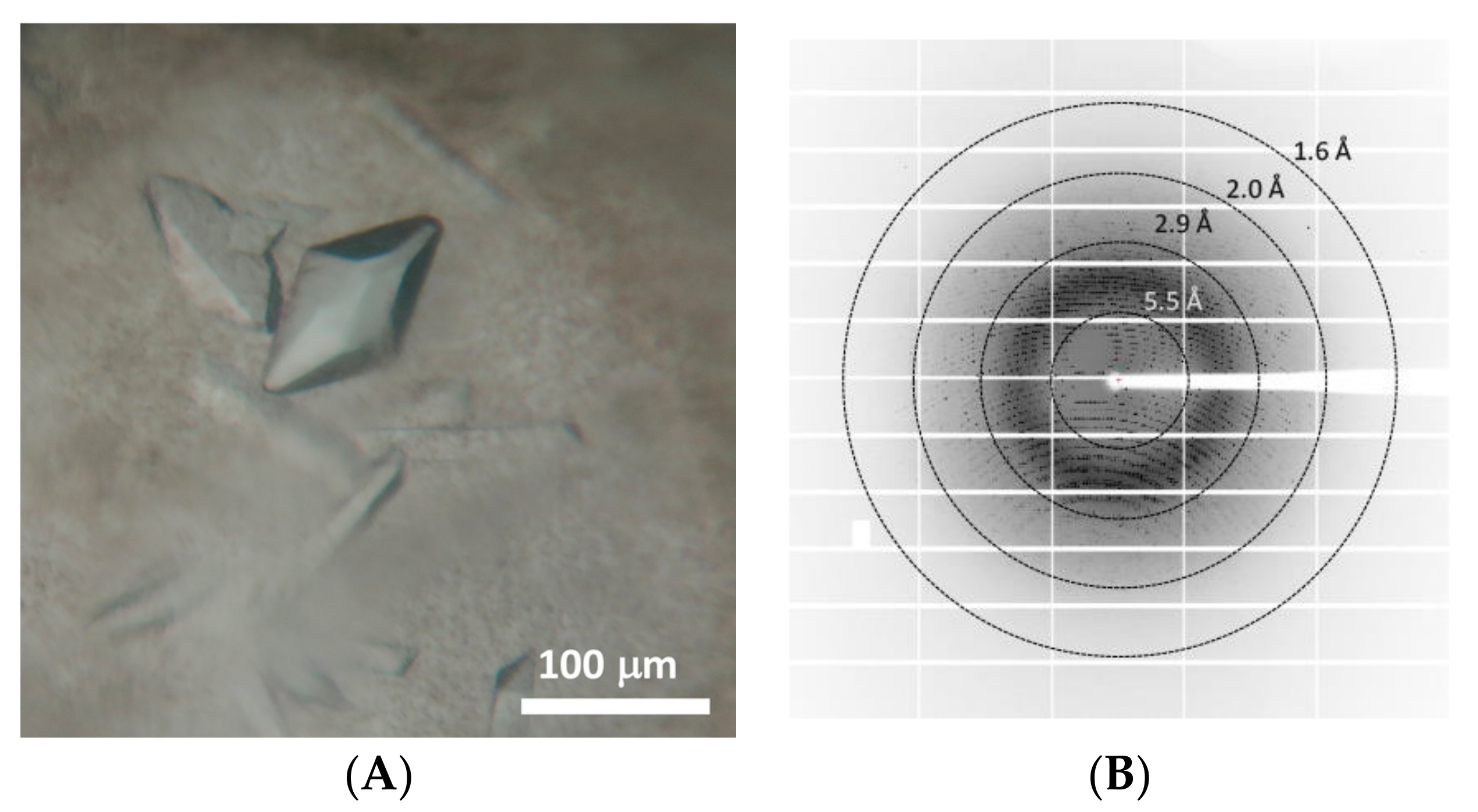

| Data Collection | |
|---|---|
| Space group | P212121 |
| Wavelength (Å) | 1.0000 |
| Resolution range (Å) | 47.35–2.05 (2.12–2.05) |
| Unit cell | |
| a, b, c (Å) | 99.12, 148.88, 149.84 |
| α, β, γ (°) | 90.00, 90.00, 90.00 |
| Rmerge (%) | 12.8 (51.05) |
| No. of unique reflections | 137748 (13410) |
| Completeness (%) | 98.91 (97.41) |
| Mean I/σ | 11.47 (3.28) |
| 1 CC1/2 | 0.996 (0.925) |
| 2 CC* | 0.999 (0.980) |
| Redundancy | 8.9 (7.8) |
| Wilson B-factor | 16.16 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Oh, C.; Ryu, B.H.; Yoo, W.; Nguyen, D.D.; Kim, T.; Ha, S.-C.; Kim, T.D.; Kim, K.K. Identification and Crystallographic Analysis of a New Carbohydrate Acetylesterase (SmAcE1) from Sinorhizobium meliloti. Crystals 2018, 8, 12. https://doi.org/10.3390/cryst8010012
Oh C, Ryu BH, Yoo W, Nguyen DD, Kim T, Ha S-C, Kim TD, Kim KK. Identification and Crystallographic Analysis of a New Carbohydrate Acetylesterase (SmAcE1) from Sinorhizobium meliloti. Crystals. 2018; 8(1):12. https://doi.org/10.3390/cryst8010012
Chicago/Turabian StyleOh, Changsuk, Bum Han Ryu, Wanki Yoo, Duy Duc Nguyen, Truc Kim, Sung-Chul Ha, T. Doohun Kim, and Kyeong Kyu Kim. 2018. "Identification and Crystallographic Analysis of a New Carbohydrate Acetylesterase (SmAcE1) from Sinorhizobium meliloti" Crystals 8, no. 1: 12. https://doi.org/10.3390/cryst8010012
APA StyleOh, C., Ryu, B. H., Yoo, W., Nguyen, D. D., Kim, T., Ha, S.-C., Kim, T. D., & Kim, K. K. (2018). Identification and Crystallographic Analysis of a New Carbohydrate Acetylesterase (SmAcE1) from Sinorhizobium meliloti. Crystals, 8(1), 12. https://doi.org/10.3390/cryst8010012







