A Novel Substrate-Binding Site in the X-ray Structure of an Oxidized E. coli Glyceraldehyde 3-Phosphate Dehydrogenase Elucidated by Single-Wavelength Anomalous Dispersion
Abstract
:1. Introduction
2. Materials and Methods
2.1. Preparation of EcGAPDH
2.2. Protein Purification
2.3. Crystallization
2.4. Soaking Experiments
2.5. Data Collection and Processing
2.6. Model Refinement
2.7. PDB Search and Comparison
2.8. Matrix-Assisted Laser Desorption/Ionization-Time of Flight (MALDI-TOF)
3. Results
3.1. Crystals
3.2. Identification of GAPDH
3.3. GAPDH Structures
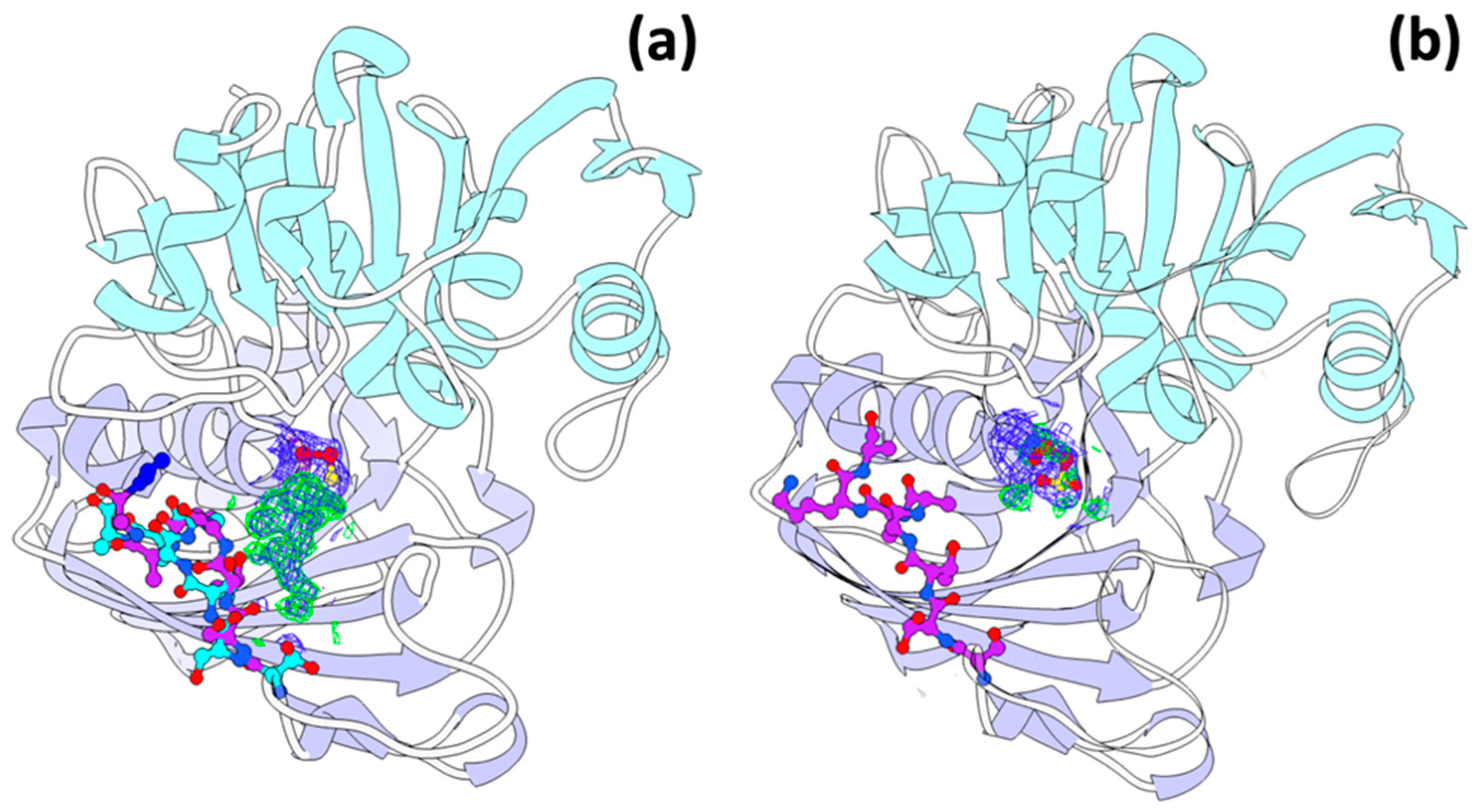
3.4. Oxidized Cys149 in the GAPDH Model
3.5. Double Conformation of the Helix (Residues 206–214)
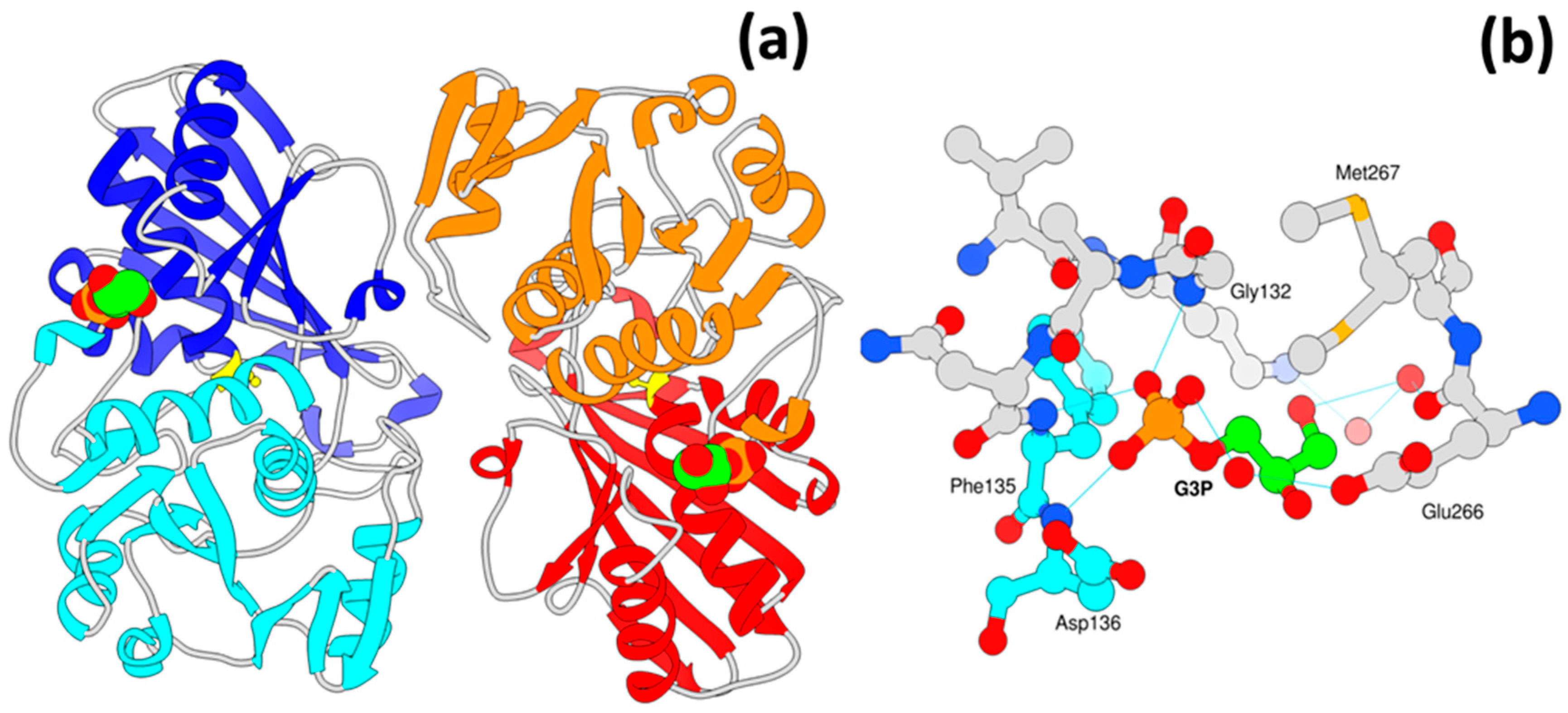
3.6. A New Binding Site for Glyceraldehyde 3-Phosphate
3.7. Trehalose Binding Site
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Boyer, P.D. (Ed.) Oxidation Reduction Part C; Academic Press, Inc. (London): Cambridge, MA, USA, 1976; Volume XIII. [Google Scholar]
- Gustafsson, A.C.; Kupershmidt, I.; Edlundh-Rose, E.; Greco, G.; Serafino, A.; Krasnowska, E.K.; Lundeberg, T.; Bracci-Laudiero, L.; Romano, M.C.; Parasassi, T.; et al. Global gene expression analysis in time series following N-acetyl L-cysteine induced epithelial differentiation of human normal and cancer cells in vitro. BMC Cancer 2005, 5. [Google Scholar] [CrossRef] [PubMed]
- Sirover, M.A. New insights into an old protein: The functional diversity of mammalian glyceraldehyde-3-phosphate dehydrogenase. BBA Protein Struct. Mol. Enzymol. 1999, 1432, 159–184. [Google Scholar] [CrossRef]
- Azam, S.; Jouvet, N.; Jilani, A.; Vongsamphanh, R.; Yang, X.M.; Yang, S.; Ramotar, D. Human Glyceraldehyde-3-phosphate Dehydrogenase Plays a Direct Role in Reactivating Oxidized Forms of the DNA Repair Enzyme APE1. J. Biol. Chem. 2008, 283, 30632–30641. [Google Scholar] [CrossRef] [PubMed]
- Butera, G.; Mullappilly, N.; Masetto, F.; Palmieri, M.; Scupoli, M.T.; Pacchiana, R.; Donadelli, M. Regulation of Autophagy by Nuclear GAPDH and Its Aggregates in Cancer and Neurodegenerative Disorders. Int. J. Mol. Sci. 2019, 20, 2062. [Google Scholar] [CrossRef]
- Butterfield, D.A.; Hardas, S.S.; Lange, M.L.B. Oxidatively Modified Glyceraldehyde-3-Phosphate Dehydrogenase (GAPDH) and Alzheimer’s Disease: Many Pathways to Neurodegeneration. J. Alzheimers Dis. 2010, 20, 369–393. [Google Scholar] [CrossRef]
- Dastoor, Z.; Dreyer, J.L. Potential role of nuclear translocation of glyceraldehyde-3-phosphate dehydrogenase in apoptosis and oxidative stress. J. Cell Sci. 2001, 114, 1643–1653. [Google Scholar]
- Heard, K.S.; Diguette, M.; Heard, A.C.; Carruthers, A. Membrane-bound glyceraldehyde-3-phosphate dehydrogenase and multiphasic erythrocyte sugar transport. Exp. Physiol. 1998, 83, 195–202. [Google Scholar] [CrossRef]
- Ferreira, E.; Gimenez, R.; Aguilera, L.; Guzman, K.; Aguilar, J.; Badia, J.; Baldoma, L. Protein interaction studies point to new functions for Escherichia coli glyceraldehyde-3-phosphate dehydrogenase. Res. Microbiol. 2013, 164, 145–154. [Google Scholar] [CrossRef]
- Aguilera, L.; Ferreira, E.; Gimenez, R.; Fernandez, F.J.; Taules, M.; Aguilar, J.; Vega, M.C.; Badia, J.; Baldoma, L. Secretion of the housekeeping protein glyceraldehyde-3-phosphate dehydrogenase by the LEE-encoded type III secretion system in enteropathogenic Escherichia coli. Int. J. Biochem. Cell B 2012, 44, 955–962. [Google Scholar] [CrossRef]
- Lama, A.; Kucknoor, A.; Mundodi, V.; Alderete, J.F. Glyceraldehyde-3-Phosphate Dehydrogenase Is a Surface-Associated, Fibronectin-Binding Protein of Trichomonas vaginalis. Infect. Immun. 2009, 77, 2703–2711. [Google Scholar] [CrossRef]
- Querol-Garcia, J.; Fernandez, F.J.; Marin, A.V.; Gomez, S.; Fulla, D.; Melchor-Tafur, C.; Franco-Hidalgo, V.; Albert, S.; Juanhuix, J.; de Cordoba, S.R.; et al. Crystal Structure of Glyceraldehyde-3-Phosphate Dehydrogenase from the Gram-Positive Bacterial Pathogen A. vaginae, an Immunoevasive Factor that Interacts with the Human C5a Anaphylatoxin. Front. Microbiol. 2017, 8. [Google Scholar] [CrossRef]
- Sanchez, B.; Schmitter, J.M.; Urdaci, M.C. Identification of novel proteins secreted by Lactobacillus rhamnosus GG grown in de Mann-Rogosa-Sharpe broth. Lett. Appl. Microbiol. 2009, 48, 618–622. [Google Scholar] [CrossRef]
- Ferreira, E.; Gimenez, R.; Canas, M.A.; Aguilera, L.; Aguilar, J.; Badia, J.; Baldoma, L. Glyceraldehyde-3-phosphate dehydrogenase is required for efficient repair of cytotoxic DNA lesions in Escherichia coli. Int. J. Biochem. Cell B 2015, 60, 202–212. [Google Scholar] [CrossRef]
- Schuppekoistinen, I.; Moldeus, P.; Bergman, T.; Cotgreave, I.A. S-Thiolation of Human Endothelial-Cell Glyceraldehyde-3-Phosphate Dehydrogenase after Hydrogen-Peroxide Treatment. Eur. J. Biochem. 1994, 221, 1033–1037. [Google Scholar] [CrossRef]
- Shenton, D.; Grant, C.M. Protein S-thiolation targets glycolysis and protein synthesis in response to oxidative stress in the yeast Saccharomyces cerevisiae. Biochem. J. 2003, 374, 513–519. [Google Scholar] [CrossRef]
- Tristan, C.; Shahani, N.; Sedlak, T.W.; Sawa, A. The diverse functions of GAPDH: Views from different subcellular compartments. Cell. Signal. 2011, 23, 317–323. [Google Scholar] [CrossRef]
- Park, J.; Han, D.; Kim, K.; Kang, Y.; Kim, Y. O-GlcNAcylation disrupts glyceraldehyde-3-phosphate dehydrogenase homo-tetramer formation and mediates its nuclear translocation. BBA Proteins Proteom. 2009, 1794, 254–262. [Google Scholar] [CrossRef] [PubMed]
- Lind, C.; Gerdes, R.; Schuppe-Koistinen, I.; Cotgreave, I.A. Studies on the mechanism of oxidative modification of human glyceraldehyde-3-phosphate dehydrogenase by glutathione: Catalysis by glutaredoxin. Biochem. Biophys. Res. Commun. 1998, 247, 481–486. [Google Scholar] [CrossRef] [PubMed]
- Ralser, M.; Wamelink, M.M.; Kowald, A.; Gerisch, B.; Heeren, G.; Struys, E.A.; Klipp, E.; Jakobs, C.; Breitenbach, M.; Lehrach, H.; et al. Dynamic rerouting of the carbohydrate flux is key to counteracting oxidative stress. J. Biol. 2007, 6, 10. [Google Scholar] [CrossRef]
- Aroca, A.; Schneider, M.; Scheibe, R.; Gotor, C.; Romero, L.C. Hydrogen Sulfide Regulates the Cytosolic/Nuclear Partitioning of Glyceraldehyde-3-Phosphate Dehydrogenase by Enhancing its Nuclear Localization. Plant Cell Physiol. 2017, 58, 983–992. [Google Scholar] [CrossRef] [PubMed]
- Piattoni, C.V.; Ferrero, D.M.L.; Dellaferrera, I.; Vegetti, A.; Iglesias, A.A. Cytosolic Glyceraldehyde-3-Phosphate Dehydrogenase Is Phosphorylated during Seed Development. Front. Plant Sci. 2017, 8. [Google Scholar] [CrossRef] [PubMed]
- Imber, M.; Huyen, N.T.T.; Pietrzyk-Brzezinska, A.J.; Loi, V.V.; Hillion, M.; Bernhardt, J.; Tharichen, L.; Kolsek, K.; Saleh, M.; Hamilton, C.J.; et al. Protein S-Bacillithiolation Functions in Thiol Protection and Redox Regulation of the Glyceraldehyde-3-Phosphate Dehydrogenase Gap in Staphylococcus aureus Under Hypochlorite Stress. Antioxid. Redox Signal. 2018, 28, 410–430. [Google Scholar] [CrossRef] [PubMed]
- Tsuji, K.; Yoon, K.S.; Ogo, S. Glyceraldehyde-3-phosphate dehydrogenase from Citrobacter sp. S-77 is post-translationally modified by CoA (protein CoAlation) under oxidative stress. FEBS Open Bio 2019, 9, 53–73. [Google Scholar] [CrossRef]
- Nakajima, H.; Amano, W.; Fujita, A.; Fukuhara, A.; Azuma, Y.T.; Hata, F.; Inui, T.; Takeuchi, T. The active site cysteine of the proapoptotic protein glyceraldehyde-3-phosphate dehydrogenase is essential in oxidative stress-induced aggregation and cell death. J. Biol. Chem. 2007, 282. [Google Scholar] [CrossRef] [PubMed]
- Minor, W.; Cymborowski, M.; Otwinowski, Z.; Chruszcz, M. HKL-3000: The integration of data reduction and structure solution-from diffraction images to an initial model in minutes. Acta Crystallogr. D 2006, 62, 859–866. [Google Scholar] [CrossRef] [PubMed]
- Evans, P. Scaling and assessment of data quality. Acta Crystallogr. Sect. D Struct. Biol. 2006, 62, 72–82. [Google Scholar] [CrossRef] [PubMed]
- Terwilliger, T.C.; Adams, P.D.; Read, R.J.; Mccoy, A.J.; Moriarty, N.W.; Grosse-Kunstleve, R.W.; Afonine, P.V.; Zwart, P.H.; Hung, L.W. Decision-making in structure solution using Bayesian estimates of map quality: The PHENIX AutoSol wizard. Acta Crystallogr. D 2009, 65, 582–601. [Google Scholar] [CrossRef]
- Emsley, P.; Cowtan, K. Coot: Model-building tools for molecular graphics. Acta Crystallogr. D 2004, 60, 2126–2132. [Google Scholar] [CrossRef]
- Bunkoczi, G.; Echols, N.; McCoy, A.J.; Oeffner, R.D.; Adams, P.D.; Read, R.J. Phaser.MRage: Automated molecular replacement. Acta Crystallogr. Sect. D Struct. Biol. 2013, 69, 2276–2286. [Google Scholar] [CrossRef]
- Afonine, P.V.; Grosse-Kunstleve, R.W.; Echols, N.; Headd, J.J.; Moriarty, N.W.; Mustyakimov, M.; Terwilliger, T.C.; Urzhumtsev, A.; Zwart, P.H.; Adams, P.D. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. Sect. D Struct. Biol. 2012, 68, 352–367. [Google Scholar] [CrossRef]
- Sierra-Gomez, Y.; Rodriguez-Hernandez, A.; Cano-Sanchez, P.; Gomez-Velasco, H.; Hernandez-Santoyo, A.; Siliqi, D.; Rodriguez-Romero, A. A biophysical and structural study of two chitinases from Agave tequilana and their potential role as defense proteins. FEBS J. 2019. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.J. A cryoprotectant induces conformational change in glyceraldehyde-3-phosphate dehydrogenase. Acta Crystallogr. Sect. F Struct. Biol. Commun. 2018, 74, 277–282. [Google Scholar] [CrossRef] [PubMed]
- Sirover, M.A. Structural analysis of glyceraldehyde-3-phosphate dehydrogenase functional diversity. Int. J. Biochem. Cell B 2014, 57, 20–26. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bedhomme, M.; Adamo, M.; Marchand, C.H.; Couturier, J.; Rouhier, N.; Lemaire, S.D.; Zaffagnini, M.; Trost, P. Glutathionylation of cytosolic glyceraldehyde-3-phosphate dehydrogenase from the model plant Arabidopsis thaliana is reversed by both glutaredoxins and thioredoxins in vitro. Biochem. J. 2012, 445, 337–347. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Muronetz, V.I.; Melnikova, A.K.; Saso, L.; Schmalhausen, E.V. Influence of Oxidative Stress on Catalytic and Non-glycolytic Functions of Glyceraldehyde-3-phosphate dehydrogenase. Curr. Med. Chem. 2018. [Google Scholar] [CrossRef]
- Hoffmann, F.; Rinas, U. Stress induced by recombinant protein production in Escherichia coli. Adv. Biochem. Eng. Biotechnol. 2004, 89, 73–92. [Google Scholar]
- Duee, E.; OlivierDeyris, L.; Fanchon, E.; Corbier, C.; Branlant, G.; Dideberg, O. Comparison of the structures of wild-type and a N313T mutant of Escherichia coli glyceraldehyde 3-phosphate dehydrogenases: Implication for NAD binding and cooperativity. J. Mol. Biol. 1996, 257, 814–838. [Google Scholar] [CrossRef]
- Wang, H.B.; Wang, M.J.; Yang, X.M.; Xu, X.H.; Hao, Q.; Yan, A.X.; Hu, M.L.; Lobinski, R.; Li, H.Y.; Sun, H.Z. Antimicrobial silver targets glyceraldehyde-3-phosphate dehydrogenase in glycolysis of E. coli. Chem. Sci. 2019, 10, 7193–7199. [Google Scholar] [CrossRef] [Green Version]
- Patel, D.K.; Shah, K.R.; Pappachan, A.; Gupta, S.; Singh, D.D. Cloning, expression and characterization of a mucin-binding GAPDH from Lactobacillus acidophilus. Int. J. Biol. Macromol. 2016, 91, 338–346. [Google Scholar] [CrossRef]
- Terrasse, R.; Amoroso, A.; Vernet, T.; Di Guilmi, A.M. Streptococcus pneumoniae GAPDH Is Released by Cell Lysis and Interacts with Peptidoglycan. PLoS ONE 2015, 10, e0125377. [Google Scholar] [CrossRef] [Green Version]
- Perez-Casal, J.; Potter, A.A. Glyceradehyde-3-phosphate dehydrogenase as a suitable vaccine candidate for protection against bacterial and parasitic diseases. Vaccine 2016, 34, 1012–1017. [Google Scholar] [CrossRef] [PubMed]
- Gerszon, J.; Rodacka, A. Oxidatively modified glyceraldehyde-3-phosphate dehydrogenase in neurodegenerative processes and the role of low molecular weight compounds in counteracting its aggregation and nuclear translocation. Ageing Res. Rev. 2018, 48, 21–31. [Google Scholar] [CrossRef] [PubMed]
- Muronetz, V.I.; Barinova, K.V.; Stroylova, Y.Y.; Semenyuk, P.I.; Schmalhausen, E.V. Glyceraldehyde-3-phosphate dehydrogenase: Aggregation mechanisms and impact on amyloid neurodegenerative diseases. Int. J. Biol. Macromol. 2017, 100, 55–66. [Google Scholar] [CrossRef] [PubMed]
- Eby, D.; Kirtley, M.E. Role of Lysine Residues in the Binding of Glyceraldehyde-3-Phosphate Dehydrogenase to Human-Erythrocyte Membranes. Biochem. Biophys. Res. Commun. 1983, 116, 423–427. [Google Scholar] [CrossRef]
- Hara, M.R.; Agrawal, N.; Kim, S.F.; Cascio, M.B.; Fujimuro, M.; Ozeki, Y.; Takahashi, M.; Cheah, J.H.; Tankou, S.K.; Hester, L.D.; et al. S-nitrosylated GAPDH initiates apoptotic cell death by nuclear translocation following Siah1 binding. Nat. Cell Biol. 2005, 7, 665–674. [Google Scholar] [CrossRef]
- Hillion, M.; Imber, M.; Pedre, B.; Bernhardt, J.; Saleh, M.; Van Loi, V.; Maass, S.; Becher, D.; Rosado, L.A.; Adrian, L.; et al. The glyceraldehyde-3-phosphate dehydrogenase GapDH of Corynebacterium diphtheriae is redox-controlled by protein S-mycothiolation under oxidative stress. Sci. Rep. UK 2017, 7. [Google Scholar] [CrossRef] [Green Version]
- Arutyunova, E.I.; Danshina, P.V.; Domnina, L.V.; Pleten, A.P.; Muronetz, V.I. Oxidation of glyceraidehyde-3-phosphate dehydrogenase enhances its binding to nucleic acids. Biochem. Biophys. Res. Commun. 2003, 307, 547–552. [Google Scholar] [CrossRef]
- Frayne, J.; Taylor, A.; Cameron, G.; Hadfield, A.T. Structure of Insoluble Rat Sperm Glyceraldehyde-3-phosphate Dehydrogenase (GAPDH) via Heterotetramer Formation with Escherichia coli GAPDH Reveals Target for Contraceptive Design. J. Biol. Chem. 2009, 284, 22703–22712. [Google Scholar] [CrossRef] [Green Version]



| Parameter | Se-Modified | Se-Modified NAD+ | Native |
|---|---|---|---|
| Data collection | |||
| Wavelength (Å) | 1.54 | 1.54 | 1.54 |
| Space group | C2221 | C2221 | C2221 |
| Cell dimensions a, b, c (Å) | 77.27 | 77.67 | 77.91 |
| 186.71 | 187.35 | 186.97 | |
| 122.13 | 121.67 | 121.75 | |
| Resolution range (Å) | 50–1.64 | 38.8–1.79 | 46.7–2.14 |
| Outer shell | 1.67–1.64 | 1.82–1.79 | 2.17–2.14 |
| Total reflections | 1,157,097 | 746,389 | 48,546 |
| Unique reflections | 107,066 | 83,268 | 14,278 |
| Completeness (%) | 99.6 (84.5) | 99.8 (99.1) | 97.9 (89.3) |
| Multiplicity | 11.6 (6.7) | 9.0 (6.3) | 3.4 (2.0) |
| Mean I/σ(I) | 44.7 (2.7) | 37.7 (3.0) | 31.3 (2.4) |
| Rmerge | 0.76 (0.05) | 0.73 (0.11) | 0.29 (.078) |
| Rmeas | 0.82 (0.06) | 0.80 (0.12) | 0.37 (0.09) |
| Rpim | 0.31 (0.02) | 0.31 (0.036) | |
| Wilson B-factor (Å2) | 15.0 | 16.3 | 28.4 |
| CC1/2 (high resolution shell) | 0.86 | 0.87 | 0.86 |
| Refinement | |||
| Rwork/Rfree | 0.17/0.19 | 0.18/0.20 | 0.16/0.20 |
| Average B, all atoms (Å2) | 16.7 | 21.0 | 31.0 |
| Total atoms | 5825 | 5598 | 5373 |
| Protein | 5126 | 4981 | 4989 |
| Ligands | 71 | 71 | 35 |
| Water | 628 | 546 | 349 |
| R.m.s. deviations | |||
| Bond lengths (Å) | 0.008 | 0.015 | 0.009 |
| Bond angles (°) | 1.040 | 1.324 | 1.034 |
| Ramachandran plot | |||
| Favored (%) | 96.04 | 96.31 | 95.43 |
| Allowed (%) | 3.66 | 3.38 | 4.12 |
| Outliers (%) | 0.30 | 0.31 | 0.46 |
| PDB entry | 6UTO | 6UTN | 6UTM |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Annia, R.-H.; Romo-Arévalo, E.; Rodríguez-Romero, A. A Novel Substrate-Binding Site in the X-ray Structure of an Oxidized E. coli Glyceraldehyde 3-Phosphate Dehydrogenase Elucidated by Single-Wavelength Anomalous Dispersion. Crystals 2019, 9, 622. https://doi.org/10.3390/cryst9120622
Annia R-H, Romo-Arévalo E, Rodríguez-Romero A. A Novel Substrate-Binding Site in the X-ray Structure of an Oxidized E. coli Glyceraldehyde 3-Phosphate Dehydrogenase Elucidated by Single-Wavelength Anomalous Dispersion. Crystals. 2019; 9(12):622. https://doi.org/10.3390/cryst9120622
Chicago/Turabian StyleAnnia, Rodríguez-Hernández, Enrique Romo-Arévalo, and Adela Rodríguez-Romero. 2019. "A Novel Substrate-Binding Site in the X-ray Structure of an Oxidized E. coli Glyceraldehyde 3-Phosphate Dehydrogenase Elucidated by Single-Wavelength Anomalous Dispersion" Crystals 9, no. 12: 622. https://doi.org/10.3390/cryst9120622





