Abstract
The crystal structure of the iodide salt of a quaternary ammonium derivative of cholic acid having a naphthalene group attached to the 3rd position of the steroid nucleus through an amide bond ([3β,5β,7α,12α]-3-(2-naphthyloylamino)-7,12-dihydroxycholan-24-triethylammonium iodide) has been resolved. The compound crystallizes in the P212121 orthorhombic space group (a/Å = 10.9458(3); b/Å = 12.1625(3); c/Å = 28.4706(7)). The lateral chain adopts a fully extended tttt conformation because the quaternary ammonium group cannot participate in the formation of hydrogen bonds. The iodide ion is involved in the formation of hydrogen bonds as well as the amide group and the two steroid hydroxy groups. Hirshfeld surface analysis confirms that these contacts, as well as the electrostatic interactions, stabilize the structure. The helixes around the 21 screw axis are right-handed ones.
1. Introduction
Bile salts are amphiphilic steroids which are of high interest for biology [1]. Their behaviors as surfactants in aqueous solutions have extensively been studied for some decades [2]. Many derivatives [3] which often modify the hydrophobic-hydrophilic balance through the insertion of substituents in the positions of the hydroxy groups of the original steroids have also been synthesized. In these derivatives, the carboxylic acid function of the side chain is preserved, although this chain has been sometimes lengthened or shortened [4].
The functionality of bile acids allows the synthesis of several cationic derivatives [5,6,7]. Many of them form gels [8,9,10] and also show a similar ability for the solubilization of cholesterol as that of natural bile salts [10]. Several studies about biophysical properties of cationic bile salts, such as membrane perturbation and protein solubilizing abilities, have been reported. For example, the hydrochloride salts of ethylenediamine deoxycholate are more effective than the natural bile salts in the release of the dye calcein from pre-encapsulated vesicles and in hemolytic activity; and possess different antimicrobial activities [11]. On the other hand, the fluorescence depolarization studies with both compounds suggested that the cationic bile salts would be useful as crystallizing agents for membrane proteins [11]. Other cationic derivatives are antibiotics [12] that mimic the properties of squalamine, a natural polyamine steroid isolated from tissues of sharks that exhibit potent bactericidal activity against both Gram-negative and Gram-positive bacteria [13]. In addition, squalamine is fungicidal and induces the osmotic lysis of protozoa. Recent studies show that squalamine can be considered a new frontier of medication since it is an antiangiogenic steroid that may be used to treat tumors and other diseases in the near future [14].
The study of the interactions between cationic surfactants and DNA has been the subject to research in the last two decades using different experimental techniques [15,16,17], with the interaction being dependent on the surfactant tails and heads [18]. Therefore, it is foreseeable that in the near future, the number of studies of cationic bile salt derivatives/DNA interactions will increase significantly.
The analysis of the steroid structures in the solid state provides information about the spatial arrangement, topology and geometry of cavities, and intermolecular interactions, therefore it, plays a key role in the understanding of recognition and binding to receptors. Crystal structures of bile acids and derivatives have been studied in detail [3,19,20]. In a much smaller number, crystal structures of anionic bile salts have been reported on, while the lack of these studies for cationic bile salts is notorious. In fact, 3-aminocholic, 24-aminocholic and 24-aminochenodeoxycholic acids, which recrystallize as ammonium salts, constitute as being a few exceptions. For the first compound, the counterion is the carboxylate in the 24th position (i e., the structure is zwitterionic) while in the other two, it is a carbamate formed by the spontaneous carbonation of the half of the steroid molecules [21].
In this paper, we analyze the crystal structure of the iodide salt of a quaternary ammonium derivative of cholic acid having a naphthalene group attached to the 3rd position of the steroid nucleus through an amide bond ([3β,5β,7α,12α]-3-(2-naphthyloylamino)-7,12-dihydroxycholan-24-triethylammonium iodide, 1, Figure 1a). The back (hydrophobic)-belly (hydrophilic), head (naphthyloyl moiety)-tail (quaternary ammonium group) and right-left (where, respectively, lie the O12 and O7 hydroxy groups) structure confers a three-axial chirality to the compound (Figure 1b) [3,19,20]. In addition, to covering the lack of information about these cationic derivatives, here presented results could contribute to the understanding of the supramolecular organization of the compound in an aqueous solution [6], and may be a preliminary step in the study of recognition and binding to receptors of these type of bile acid derivatives. This knowledge gained through the crystal structure can also be useful for the proposition of the structure of the micelles formed in aqueous solutions, a strategy largely followed by Giglio and co-workers [22].
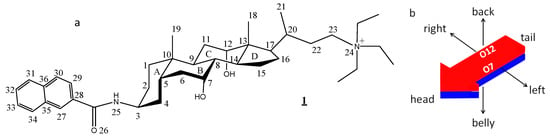
Figure 1.
(a) The chemical structure of the compound studied in this paper ([3β,5β,7α,12α]-3-(2-naphthyloylamino)-7,12-dihydroxycholan-24-triethylammonium iodide), and the numbering of selected atoms. (b) Resulting scheme of the three-axial characteristics of the molecule. The red and blue colors represent, respectively, the hydrophobic and the hydrophilic faces of the molecule.
The results and their discussion are organized in three successive subsections (molecular geometry, crystal packing and 21 helical assemblies) that can be considered as the primary, secondary and tertiary structure of the crystal structure.
2. Materials and Methods
2.1. Synthesis
The synthesis and characterization of 1 have been reported previously [6].
2.2. X-Ray Diffraction
Clear pale yellow prismatic crystals were grown from a solution of 1 in ethyl acetate:2-propanol (1:1 v/v). Data on these crystals were collected at 100 K on a Bruker AXS X8 APEX-II diffractometer (Bruker, Berlin, Germany ), with a radiation source consisting in a sealed X-ray tube SIEMENS KFN MO 2K-90, a graphite monochromator and a detector with a resolution of 16.6666 pixel mm−1. The absolute structure: flack was determined using 4279 quotients [(I+)–(I–)]/[(I+)+(I–)]. Hydrogen atoms were treated by a mixture of independent and constrained refinement. Molecular graphics were generated from Mercury (http://www.ccdc.cam.ac.uk/prods/mercury). A summary of the crystal data and relevant experimental details are listed in Table 1. CCDC 1891134 contains the supplementary crystallographic data for the crystal, which can be obtained free of charge from The Cambridge Crystallographic Data Centre via www.ccdc.cam.ac.uk/data_request/cif.

Table 1.
The crystal data, data collection and refinement for 1.
2.3. Hirshfeld Surface Analysis.
The Hirshfeld surface analysis was performed with CrystalExplorer [23].
3. Results and Discussion
3.1. Molecular Geometry
The compound crystallizes free of solvents in the orthorhombic space group P212121 with four molecules in the unit cell. The geometry of the steroid nucleus agrees well with that of the standard steroid recently proposed [3]. This standard steroid has been obtained from the data of bond lengths and the angles corresponding to the steroid nucleus in the crystal structures of 30 bile acids and esters (and substituted derivatives). The differences in the C-C bond distances for 1 are within the standard deviations of the reference steroid, with the exception of the C2-C3 bond (1.535 against 1.519 ± 0.011 Å). Although the bond angles are more likely to experience variations, only the values corresponding to C1-C2-C3, C13-C17-C20 and C22-C23-C24 (112.1, 117.7 and 108.2°) are outside of the standard deviations of the reference steroid (110.8 ± 1.1, 119.4 ± 1.1 and 113.7 ± 1.7°). The value of the phase angle (Δ = 12.1°) is indicative of a conformation close to the half-chair for the D ring. The angle between the horizontal (C1, C5-C17, and C20 carbon atoms) and vertical (C3, C10, C13, C17, C18, and C19 carbon atoms) planes of the molecule [24] is 88.2° (88.2 ± 0.5° for the standard steroid [3]). Finally, a fully extended (tttt) lateral chain is deduced since the torsion angles from C17 to C23 are 175.5, −172.1, 177.6 and 175.1°. In general, the resulting conformation depends on the hydrogen bonds formed (with other steroid molecules or with guests molecules) by the functional groups at the end of the chain, but the tttt conformation is common in bile acids derivatives [3]. The nitrogen atom, being tetra-alkyl substituted, cannot be involved in the formation of hydrogen bonds, a fact that favors this conformation.
Compound 1 has a naphthyloyl group linked to the C3 position of the cholic residue through an amide bond. The angle between the planes formed by the amide linking group and the naphthyl residue is 16.8°, a value which is close to the one observed for other aromatic amide derivatives of cholic acid. The values of 17.7° and 18.2° have been measured for the t-butylphenyl (recrystallized from chlorobenzene) and p-benzoic acid derivatives [3,25], respectively. On the other hand, the angles between the naphthyl plane and the horizontal and vertical planes [24] of 1 are 61.3 and 30.5°, respectively. Again, a comparison with these previous derivatives indicates that the naphthyl moiety is more twisted than the phenyl ones with respect to the steroid skeleton.
3.2. Crystal Packing
Views of the crystal packing of 1 along the crystallographic axes are shown in Figure 2a–c. It is noteworthy that no typical back-to-back hydrophobic interactions (nor belly-to-belly interactions) are present, and therefore interdigitation between the methyl groups C18 and C19 is not observed.
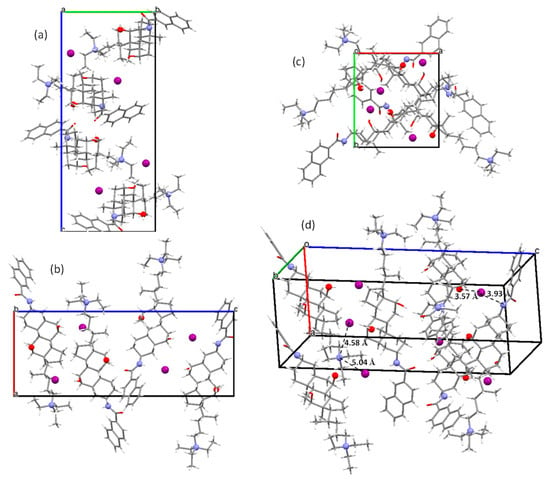
Figure 2.
The views of the crystal packing in 1 along the a (a), b (b) and c (c) crystallographic axes. (d) An arbitrary view that allows for the reflectance of relevant distances and interactions involving the iodide ion (see text).
Figure 2d has been constructed to highlight the following relevant distances in the crystal that involve the iodide ion and the quaternary nitrogen atom:
(i) The shortest distances found between an iodide ion and the charged nitrogen atom of steroid molecules are 4.58 and 5.04 Å, which correspond to electrostatic interactions (see references in Table 2). These values are similar to those measured from available cifs deposited at the Cambridge Crystallographic Data Centre for quaternary ammonium iodide salts (Table 2).

Table 2.
The distances found between an iodide ion and the charged nitrogen atom corresponding to the electrostatic interactions in ammonium iodide crystals.
(ii) On the other hand, the iodide ion in 1 is located at the distances of 3.57 and 3.93 Å from the O7 and N25 atoms, of the two other steroid molecules. These interactions correspond to hydrogen bonds. Their geometrical parameters being summarized in the first two rows of Table 3. Hydrogen bonds involving the iodide ion and hydroxyl groups (OH…I–) have been noticed for β-D-glucopyranosyl)-trimethyl- and (R,S)-N,N,N-trimethyl-1-hydroxy-3-methyl-2-butyl- ammonium iodides. Again, from the available cifs, the values of 3.45/3.48 Å, and 3.43/3.44 Å are obtained, which are in the range of the compound involving the hydroxyl group at O7 (Table 3). Likewise, the N25–H…I– hydrogen bond distance satisfactorily agrees with the values observed for the triethyl- (3.58 Å) [41], and 1-naphthyl- (3.54 and 3.70 Å) [42] ammonium iodides.

Table 3.
The hydrogen bond geometries found in the crystal structure of 1, D being the donor and A being the acceptor. Symmetry codes: (i) x – ½, –y + ½, –z + 1; (ii) –x + 2, y + ½, –z + ½.
Finally, the remaining hydrogen bond present in the crystal structure of 1 (last row in Table 3) is typical of other bile acids and esters bearing the amide group in the 3rd position of the steroid skeleton. In fact, 6 out of 8 such derivatives exhibit this hydrogen bond with a mean distance of 2.78 ± 0.10 Å [3], a value practically identical to the one shown in Table 3.
Molecular Hirshfeld surfaces visualize the intermolecular interactions in a crystal in a visual manner [43]. Two distances, de and di, mapped on the Hirshfeld surface, provide the three-dimensional picture of intermolecular close contacts in the crystal [44]. Here, di is the distance from the surface to the nearest atom interior of the surface, and de is the distance from the surface to the nearest atom exterior of the surface. Both are used to generate a two-dimensional plot (fingerprint plot) which is a summary of the intermolecular interactions in the crystal. Thus, while the Hirshfeld surface and properties defined by it emphasize a whole-of-molecule approach to understanding intermolecular interactions, they can also be used in conjunction with a more direct atom–atom based approach to gain a complete appreciation of the important interactions in a molecular crystal.
Furthermore, Spackman et al. [45] have defined a normalized contact distance, dnorm (Equation [1]), where rvdW is the van der Waals (vdW) radius of the appropriate atom internal or external to the surface. This distance is negative where the contacts shorter than van der Waals separations occur and positive for contacts greater than van der Waals separations, and is displayed using a red–white–blue color scheme (red highlights for shorter contacts, blue is for longer ones and white for contacts around the vdW separation). By using this information, it is possible to graphically highlight those regions of the surface involved in a specific type of intermolecular contact.
Figure 3 and Figure 4 resume the relative contributions to the Hirshfeld surface area for the various close intermolecular contacts in 1. The total surface is 682.41 Å2 and the volume is 876.38 Å3.
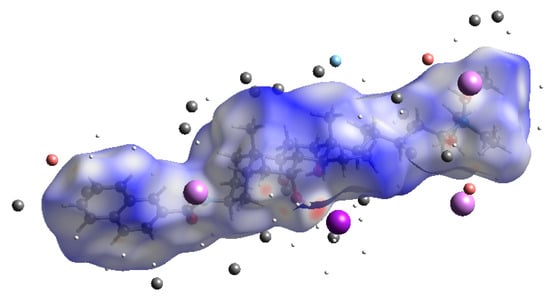
Figure 3.
The Hirshfeld surface area with atoms of external fragments.
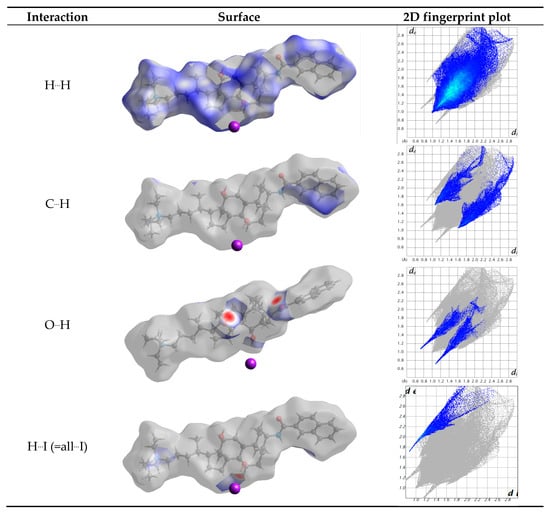
Figure 4.
The contributions to the Hirshfeld surface area for the various close intermolecular contacts in 1. dnorm surface of 1, highlighting the indicated intermolecular contacts, and corresponding 2D fingerprint plots. de and di in Å
Table 4 resumes the relative contributions to the Hirshfeld surface area for the various close intermolecular contacts in 1, H…H being the most common one since it represents 75% of the total Hirshfeld surface area. This value is logically lower than the observed in a cholest-5-ene crystal (=98.6%), where only the C-C and C-H bonds exist [46]. The percentage of O…H interactions (hydrogen bonds) is 6.5%. This value is equal to the sum of 3.4% and 3.1% since contributions of the atoms being inside or outside the surface have to be taken into consideration. Iodide (outside) only interacts with hydrogen, with the percentage being 4.8%. Therefore, the molecules are linked by a combination of strong (electrostatic and hydrogen bonds) interactions and weak (H...H and C...H) interactions.

Table 4.
The filtering fingerprint by element type. The surface area is included (as a percentage of the total surface area) for close contacts between atoms inside and outside the surface.
Strong interactions, as hydrogen bonds, are easily visualized in the fingerprint plots [45] as they appear as thin and long wings, parallel to the de-di diagonal. Figure 5 shows the presence of six “wings”. The interactions a (H(O7)…I), c (H(O12)…O26) and e (O26…H(O12)) correspond to conventional hydrogen bonds. However, fingerprint analysis is a helpful tool for revealing specific weak interactions. Thus the wing d shows H…H interactions with the hydrogen atoms bonded to C6 and to carbon atoms of the alkyl ammonium residue. Finally, the b and f wings reflect a H…H close contact, with hydrogen atoms corresponding to C19 (methyl group) and C36 (aromatic ring). The electrostatic interaction between the nitrogen atoms of the ammonium ion with the iodide anion is not visible through the Hirshfeld surface.
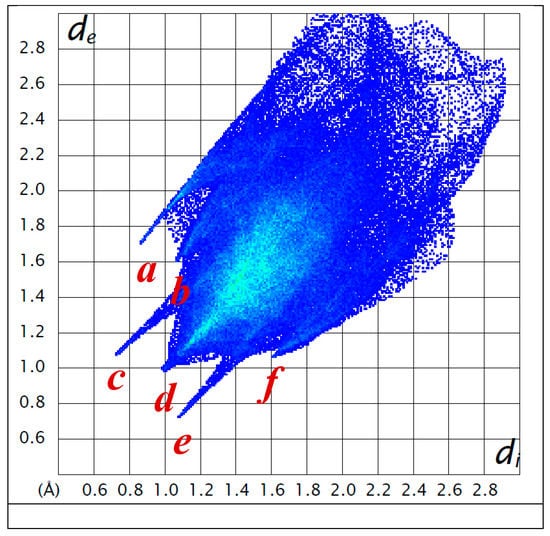
Figure 5.
Two-dimensional Hirshfeld fingerprint plots. The wings correspond to the following interactions present in 1: a: H(O7)…I; b and f: H…H with hydrogen atoms corresponding to C19 (methyl group) and C36 (aromatic ring); c: H(O12)…O26; d: H…H interactions with the hydrogen atoms bonded to C6 and to carbon atoms of the alkyl ammonium residue; e: O26…H-O12. de and di are given in Å.
The fingerprint analysis evidences a great spread of points, covering de and di values as large as 2.5–2.8 Å. The largest values on the diagonal of the de-di plot correspond to the naphthyl residue. This agrees with the 2D fingerprint plots for polycyclic aromatic hydrocarbons which exhibit C-H...π interactions [44]. In cholamide inclusion crystals with aromatic guests, Aburaya et al. [47] have shown that weak hydrogen bonds (N-H...π, C-H...π, and C-H...O) play a key role in linking the host and guest molecules. Although there is no π stacking in 1 (as observed for several bile acid derivatives [3,48]), there are H-C distances (both atoms of the naphthyl residue belonging to neighbouring molecules) as short as 2.87 Å, in agreement with the sum of the van der Waals radii for hydrogen (1.14(6) Å) and carbon (1.70(8) Å) [49] within the error associated to the average values.
The shortest iodine-hydrogen atoms distances (Figure 6) are 2.79 (O7-H…I–); 3.09 (N25-H…I–); 3.02 and 3.13 (–NCH2CH3); 3.11 (–NC23H2 steroid side chain); 3.18 (H-C27); 3.30 (H-C5, A ring) and 3.39 (H15 D ring); and 3.49 (–NCH2CH3)Å. Most of them are shorter than the sum of the van der Waals radii (= 3.26(9) Å; rIvdW= 2.12(7) Å; [49]). See the wing I in Figure 4 and Figure 5.
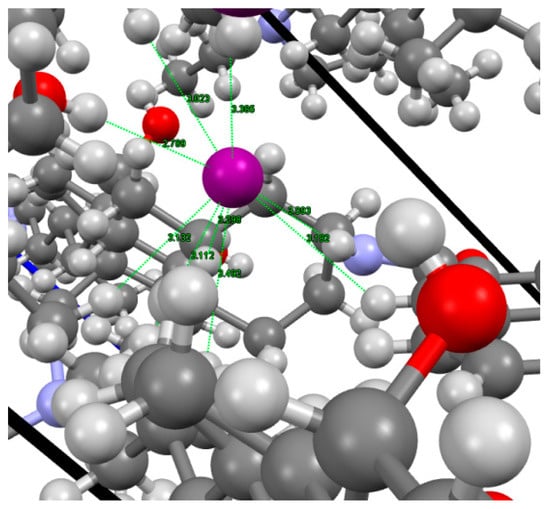
Figure 6.
The shortest H-I– distances observed in the crystal structure of 1.
3.3. 21 Helical Assemblies
According to Miyata et al. [50], crystals possessing 21 helical axes may be considered as formed by bundles of helixes with a given handedness. To determine this spatial tendency around the 21 symmetry element, one must focus one’s attention on the long molecular axis (tail-head) of the molecules with their bellies pointing to the axis. This arrangement appears in cholic and deoxycholic acids [50], both exhibiting right-handedness. However, this is not the case for 1 because the hydrogen bonds that take place cause the connected steroid molecules to adopt another spatial disposition. To resolve the problem of situations like this, an additional criterion was stablished [25] and, according to it, the steroid skeleton must be exposing its right side towards the 21 axis. Figure 7 is constructed by taking into account this criterion and from it, right-handedness is deduced. The measured value for the angle between the horizontal planes of two consecutive molecules used to define the handedness of a helix allows for the determination of the pitch and the tilt in the helical arrangement around the screw axis [25]. The application of this strategy in 1 affords a pitch angle of 57.1° and a tilt of 32.9°.
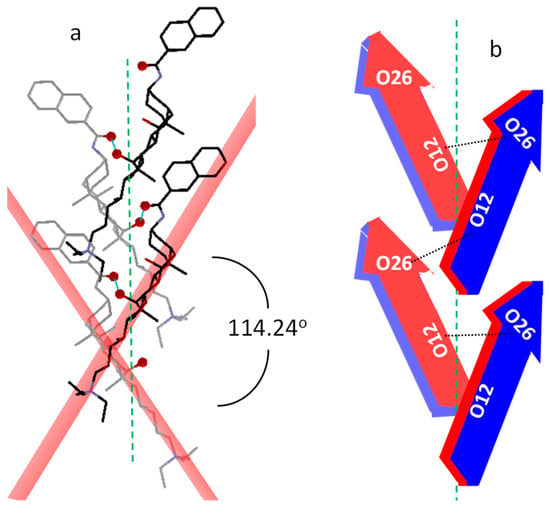
Figure 7.
(a) The packing of the molecules around the 21 axis to define the handedness of the molecular helical disposition. For clarity, hydrogen atoms and iodide ions have been hidden and O12 and O26 atoms highlighted. (b) The scheme of this arrangement based on the arrow model of Figure 1b.
Deoxycholic and cholic acids have very different tilts with respect to the 21 axis, being small for the first compound and laying the molecules of the second more perpendicular to that symmetry element (higher tilt). The inclination of the molecules with respect to the 21 axis that at first glance is observed in Figure 7a suggests that the degree of molecular tilt in 1 is intermediate to those ones. To evaluate the molecular tilt in the case of cholic acid, we have used the only available cif corresponding to its crystalline structure free of guests (solvents) [51], and a value of 41.5° was obtained. Although deoxycholic acid has been recrystallized in a large number of solvents, unfortunately, any crystal structure free of solvent molecules has been reported. Among those which recrystallize in the orthorhombic space group P212121 system, we have used the cif of the deoxycholic acid d-camphor clathrate crystal [52]. From it, we have estimated a value of 13.3° for the tilt angle, verifying that the value exhibited by 1 is indeed intermediate to both. The difference between the organizations of cholic and deoxycholic acids around the 21 axis (tilts) was established by Miyata et al. [50] as a consequence of the lack of the O7H. This situation also arises in the present case since the O7-H forms a hydrogen bond with the iodide ion and not with another bile salt molecule in the helix. Therefore, this fact could explain the intermediate value for the tilt angle in 1.
4. Conclusions
[3β,5β,7α,12α]-3-(2-naphthyloylamino)-7,12-dihydroxycholan-24-triethylammonium iodide crystallizes in the orthorhombic space group P212121. The primary crystal structure is characterized by the geometry (bond distances and angles) typical of similar steroids. The lateral chain adopts a fully extended tttt conformation because the quaternary ammonium group cannot participate in the formation of hydrogen bonds. However, the iodide ion is involved in the formation of hydrogen bonds as well as the amide group and the two steroid hydroxy groups. Hirshfeld surface analysis confirms these conventional hydrogen bonds as well as other weak H...H interactions which stabilize the secondary structure. Like other bile acids and derivatives with 21 axes of symmetry, it presents a structural motif based on 21 right-handed helixes, which can be considered as a tertiary structure. It is suggested that the hydrogen bonding network explains the tilt degree of the steroid molecules around that symmetry axis.
Author Contributions
Conceptualization, F.M. and J.V.T.; methodology, J.A.S.; software, M.P.V.-T. and J.A.S.; investigation, all authors; writing—original draft preparation, M.P.V.-T., J.A.S. and S.d.F.; writing—review and editing, F.M. and J.V.T.; supervision, J.V.T.; project administration, J.V.T.
Funding
This research was funded by the Ministerio de Ciencia y Tecnología, Spain (Project MAT2017-86109-P).
Acknowledgments
The authors thank the Ministerio de Ciencia y Tecnología (Project MAT2017-86109-P) for financial support. They also thank to Unidade de Raios X, RIAIDT, Universidade de Santiago de Compostela, for the resolution of the crystal.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Monte, M.J.; Garcia Marin, J.J.; Antelo, A.; Vázquez Tato, J. Bile acids, chemistry, physiology and pathophysiology. World J. Gastroenterol. 2009, 15, 804–816. [Google Scholar] [CrossRef]
- Carey, M.C.; Small, D.M. Micelle Formation by Bile Salts. Physical-Chemical and Thermodynamic Considerations. Arch. Intern. Med. 1972, 130, 506–527. [Google Scholar] [CrossRef] [PubMed]
- Meijide, F.; de Frutos, S.; Soto, V.H.; Jover, A.; Seijas, J.A.; Vázquez-Tato, M.P.; Fraga, F.; Vázquez Tato, J. A standard structure for bile acids and derivatives. Crystals 2018, 8, 86. [Google Scholar] [CrossRef]
- Kato, K.; Sugahara, M.; Tohnai, N.; Sada, K.; Miyata, M. Systematic structural study of asymmetric supramolecular assembly by a series of bile acid derivatives with different side-chain lengths. Cryst. Growth Des. 2004, 4, 263–272. [Google Scholar] [CrossRef]
- Terech, P.; Dourdain, S.; Bhat, S.; Maitra, U. Self-Assembly of Bile Steroid Analogues: Molecules, Fibers, and Networks. J. Phys. Chem. B 2009, 113, 8252–8267. [Google Scholar] [CrossRef] [PubMed]
- Trillo, J.V.; Meijide, F.; Jover, A.; Soto, V.H.; de Frutos, S.; di Gregorio, M.C.; Luciano, G.; Vázquez Tato, J. Self-aggregation mechanism of a naphthylamide cationic derivative of cholic acid. From fibers to tubules. RSC Adv. 2014, 4, 5598–5606. [Google Scholar] [CrossRef]
- Manghisi, N.; Galantini, L.; Leggio, C.; Jover, A.; Meijide, F.; Pavel, N.V.; Soto, V.H.; Vázquez Tato, J.; Agostino, R. Catanionic tubules with tunable charge. Angew. Chem. Int. Ed. 2010, 49, 6604–6607. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharya, S.; Maitra, U.; Mukhopadhyay, S.; Srivastava, A. Advances in molecular hydrogels. In Molecular Gels: Materials with Self-Assembled Fibrillar Networks; Weiss, G., Terech, P., Eds.; Springer: Dordrecht, The Netherlands, 2006; Chapter 17; pp. 613–647. [Google Scholar]
- Maitra, U.; Chakrabarty, A. Protonation and deprotonation induced organo/hydrogelation: Bile acid derived gelators containing a basic side chain. Beilstein J. Org. Chem. 2011, 7, 304–309. [Google Scholar] [CrossRef] [PubMed]
- Bhat, S.; Leikin-Gobbi, D.; Konikoff, F.M.; Maitra, U. Use of novel cationic bile salts in cholesterol crystallization and solubilization in vitro. Biochim. Biophys. Acta 2006, 1760, 1489–1496. [Google Scholar] [CrossRef] [PubMed]
- Araki, Y.-I.; Lee, A.; Sugihara, G.; Furuichi, M.; Yamashita, S.; Ohseto, F. New cationic surfactants derived from bile acids: Synthesis and thermodynamic and biophysicochemical properties such as membrane perturbation and protein solubilizing abilities. Colloids Surf. B 1996, 8, 81–92. [Google Scholar] [CrossRef]
- Savage, P.B. Cationic steroid antibiotics. Curr. Med. Chem.-Anti-Infect. Agents 2002, 1, 293–304. [Google Scholar] [CrossRef]
- Moore, K.S.; Wehrli, S.; Roder, H.; Rogers, M.; Forrest, J.N., Jr.; McCrimmon, D.; Zasloff, M. Squalamine: An aminosterol antibiotic from the shark. Proc. Natl. Acad. Sci. USA 1993, 90, 1354–1358. [Google Scholar] [CrossRef] [PubMed]
- Karmakar, S.; Pradhan, N.K. Squalamine is an antiangiogenic steroid: A future perspective of cancer drug. World J. Pharm. Pharm. Sci. 2016, 5, 576–592. [Google Scholar]
- Hianik, T.; Wang, X.; Tashlitsky, V.; Oretskaya, T.; Ponikova, S.; Antalik, M.; Ellis, J.S. Interaction of cationic surfactants with DNA detected by spectroscopic and acoustic wave techniques. Analyst 2010, 135, 980–986. [Google Scholar] [CrossRef] [PubMed]
- Husale, S.; Grange, W.; Karle, M.; Bürgi, S.; Hegner, M. Interaction of cationic surfactants with DNA: A single-molecule study. Nucl. Acids Res. 2008, 36, 1443–1449. [Google Scholar] [CrossRef]
- Yaseen, Z.; Rehman, S.U.; Tabish, M.; Din, K.U. Interaction between DNA and cationic diester-bonded Gemini surfactants. J. Mol. Liq. 2014, 197, 322–327. [Google Scholar] [CrossRef]
- Guo, Q.; Zhang, Z.; Song, Y.; Liu, S.; Gao, W.; Qiao, H.; Guo, L.; Wang, J. Investigation on interaction of DNA and several cationic surfactants with different head groups by spectroscopy, gel electrophoresis and viscosity technologies. Chemosphere 2017, 168, 599–605. [Google Scholar] [CrossRef] [PubMed]
- Miyata, M.; Tohnai, N.; Hisaki, I. Crystalline Host-Guest Assemblies of Steroidal and Related Molecules: Diversity, Hierarchy, and Supramolecular Chirality. Acc. Chem. Res. 2007, 40, 694–702. [Google Scholar] [CrossRef]
- Miyata, M.; Tohnai, N.; Hisaki, I. Supramolecular chirality in crystalline assemblies of bile acids and their derivatives; three-axial, tilt, helical, and bundle chirality. Molecules 2007, 12, 1973–2000. [Google Scholar] [CrossRef]
- Meijide, F.; Antelo, A.; Alvarez, M.; Soto, V.H.; Trillo, J.V.; Jover, A.; Vázquez Tato, J. Spontaneous formation in solid state of carbamate derivatives of bile acids. Cryst. Growth Des. 2011, 11, 356–361. [Google Scholar] [CrossRef]
- Campanelli, A.R.; Candeloro de Sanctis, S.; Giglio, E.; Viorel Pavel, N.; Quagliata, C. From crystal to micelle: A new approach to the micellar structure. J. Incl. Phenom. Macrocycl. Chem. 1989, 7, 391–400. [Google Scholar] [CrossRef]
- Turner, M.J.; McKinnon, J.J.; Wolff, S.K.; Grimwood, D.J.; Spackman, P.R.; Jayatilaka, D.; Spackman, M.A. CrystalExplorer, CrystalExplorer17 University of Western Australia. 2017. Available online: http://hirshfeldsurface.net (accessed on 12 January 2019).
- Alvarez, M.; Jover, A.; Carrazana, J.; Meijide, F.; Soto, V.H.; Vázquez Tato, J. Crystal structure of chenodeoxycholic acid, ursodeoxycholic acid and their two 3β,7α- and 3β,7β-dihydroxy epimers. Steroids 2007, 72, 535–544. [Google Scholar] [CrossRef] [PubMed]
- Meijide, F.; Trillo, J.V.; Soto, V.H.; Jover, A.; Vázquez Tato, J. Additional criterion for the determination of the handedness of 21 helices in crystals of bile acids: Crystal structure of a tert-butylphenyl derivative of cholic acid. Chirality 2011, 23, 940–947. [Google Scholar] [CrossRef] [PubMed]
- Iddon, L.; Bragg, R.A.; Harding, J.R.; Pidathala, C.; Bacsa, J.; Kirby, A.J.; Stachulski, A.V. Syntheses and structures of anomeric quaternary ammonium β-glucosides and comments on the anomeric C-N bond lengths. Tetrahedron 2009, 65, 6396–6402. [Google Scholar] [CrossRef]
- Flörke, U.; Ayaz, M.; Henkel, G. CCDC: 1446614. Experimental Crystal Structure Determination; CCDC: Cambridge, UK, 2016. [Google Scholar] [CrossRef]
- Brauer, D.J.; Buerger, H.; Grunwald, M.; Pawelke, G.; Wilke, J. Synthesis, vibrational spectra, and crystal structure analysis of di- and trifluoro-tetramethylammonium salts. Z. Anorg. Allgem. Chem. 1986, 537, 63–78. [Google Scholar] [CrossRef]
- Giese, M.; Albrecht, M.; Valkonen, A.; Rissanen, K. The pentafluorophenyl group as π-acceptor for anions: A case study. Chem. Sci. 2015, 6, 354–359. [Google Scholar] [CrossRef]
- Vincent, B.R.; Knop, O.; Linden, A.; Cameron, T.S.; Robertson, K.N. Crystal chemistry of tetraradial species. Part 2. Crystal structures of Et4NI, Ph4PBr·H2O, and Ph4PBr·2H2O. Can. J. Chem. 1988, 66, 3060–3069. [Google Scholar] [CrossRef]
- Herrschaft, G.; Hartl, H. The isomer pair tert-butylammonium iodide and tetramethylammonium iodide. Acta Cryst. 1989, C45, 1021–1024. [Google Scholar]
- Yoshida, T.; Nagata, K.; Yasuniwa, M.; Yoshimatsu, M. Low-temperature phase of tetrapropylammonium iodide. Acta Cryst. 1994, C50, 1758–1760. [Google Scholar] [CrossRef]
- Bardi, R.; Piazzesi, A.M.; Del Pra, A.; Villa, L. Structure of trimethyl[(cis-2-methyl-1,3-oxathiolan-5-yl)methyl]ammonium iodide, C8H18NOS+·I−. Acta Cryst. 1983, C39, 214–216. [Google Scholar] [CrossRef]
- Subramanian, P.K.; Ramalingam, K.; Pantaleo, N.S.; Van der Helm, D.; Satyamurthy, N.; Berlin, K.D. Studies on the conformations on 4-(N,N-dimethylamino)thianes and the corresponding methiodides. Evidence from a single crystal X-ray diffraction analysis that a twist-boat conformer exists in the solid state for trimethyl(2,2-dimethyl-trans-6-phenylthian-r-4-yl)ammonium iodide. Phosphorus Sulfur Relat. Elem. 1983, 17, 343–365. [Google Scholar]
- Kumar, V.; Pei, C.; Olsen, C.E.; Schaeffer, S.J.C.; Parmar, V.S.; Malhotra, S.V. Novel carbohydrate-based chiral ammonium ionic liquids derived from isomannide. Tetrahedron Asymmetry 2008, 19, 664–671. [Google Scholar] [CrossRef]
- Zhao, J.; Zheng, M.-X.; Lin, Y.-J.; Chen, Y.-C.; Ruan, Y.-P.; Zhang, H. CCDC: 757800. Experimental Crystal Structure Determination; CCDC: Cambridge, UK, 2014. [Google Scholar] [CrossRef]
- Hamor, T.A. Stereochemistry of anticholinergic agents. XIII. Structure of (dicyclohexylacetoxyethyl)triethylammonium iodide. Acta Cryst. 1980, B36, 99–102. [Google Scholar] [CrossRef]
- Munera-Orozco, C.; Ocampo-Cardona, R.; Cedeno, D.L.; Toscano, R.A.; Rios-Vasquez, L.A. Crystal structures of three new N-halomethylated quaternary ammonium salts. Acta Cryst. 2015, E71, 1230–1235. [Google Scholar]
- Mills, J.F.D.; Nyburg, S.C. Crystal structure of (−)-aspidospermine N(b)-methiodide. J. Chem. Soc. 1960, 1458–1463. [Google Scholar] [CrossRef]
- Kennard, O.; Kerr, K.A.; Watson, D.G.; Fawcett, J.K.; Riva di Sanseverino, L. X-ray analysis of the structure of (+-)-1-acetyl-3-methylaspidospermidine 9-methiodide. J. Chem. Soc. A 1970, 10, 1779–1783. [Google Scholar] [CrossRef]
- Liu, Y.-P.; Tan, Z.-C.; Di, Y.-Y.; Xing, Y.-T.; Zhang, P. Lattice potential energies and thermochemical properties of triethylammonium halides (Et3NHX) (X = Cl, Br, and I). J. Chem. Thermodyn. 2012, 45, 100–108. [Google Scholar] [CrossRef]
- Lemmerer, A.; Billing, D.G.; Robinson, J.M. Hydrogen-bonding one-dimensional chains containing the R42(8) motif in the ammonium salts 1-naphthylammonium iodide and naphthalene-1,8-diyldiammonium diiodide. Acta Cryst. 2008, C64, o481–o484. [Google Scholar]
- McKinnon, J.J.; Mitchell, A.S.; Spackman, M.A. Hirshfeld surfaces: A new tool for visualizing and exploring molecular crystals. Chem. Eur. J. 1998, 4, 2136–2141. [Google Scholar] [CrossRef]
- Spackman, M.A.; McKinnon, J.J. Fingerprinting intermolecular interactions in molecular crystals. CrystEngComm 2002, 4, 378–392. [Google Scholar] [CrossRef]
- McKinnon, J.J.; Jayatilaka, D.; Spackman, M.A. Towards quantitative analysis of intermolecular interactions with Hirshfeld surfaces. Chem. Commun. 2007, 37, 3814–3816. [Google Scholar] [CrossRef]
- Shamsuzzaman; Khanam, H.; Mashrai, A.; Asif, M.; Ali, A.; Barakat, A.; Mabkhot, Y.N. Synthesis, crystal structure, Hirshfeld surfaces, and thermal, mechanical and dielectrical properties of cholest-5-ene. J. Taibah Univ. Sci. 2017, 11, 141–150. [Google Scholar] [CrossRef]
- Aburaya, K.; Murai, T.; Hisaki, I.; Tohnai, N.; Miyata, M. Importance of Weak Hydrogen Bonds in the Formation of Cholamide Inclusion Crystals with Aromatic Guests. Cryst. Growth Des. 2008, 8, 1013–1022. [Google Scholar] [CrossRef]
- Mayorquin-Torres, M.C.; Arcos-Ramos, R.; Flores-Alamo, M.; Iglesias-Arteaga, M.A. Crystalline arrays of side chain modified bile acids derivatives. Two novel self-assemblies based on π-π and belly-to-belly interactions. Steroids 2016, 115, 169–176. [Google Scholar] [CrossRef] [PubMed]
- Hu, S.-Z.; Zhou, Z.-H.; Xie, Z.-X.; Robertson, B.E. A comparative study of crystallographic van der Waals radii. Z. Kristallogr. 2014, 229, 517–523. [Google Scholar] [CrossRef]
- Hisaki, I.; Sasaki, T.; Tohnai, N.; Miyata, M. Supramolecular-tilt-chirality on twofold helical assemblies. Chem. Eur. J. 2012, 18, 10066–10073. [Google Scholar] [CrossRef]
- Miki, K.; Kasai, N.; Shibakami, M.; Chirachanchai, S.; Takemoto, K.; Miyata, M. Crystal structure of cholic acid with no guest molecules. Acta Cryst. 1990, C46, 2442–2445. [Google Scholar] [CrossRef]
- Jones, J.G.; Schwarzbaum, S.; Lessinger, L.; Low, B.W. The structure of the 2:1 complex between the bile acid deoxycholic acid and (+)-camphor. Acta Cryst. 1982, B38, 1207–1215. [Google Scholar] [CrossRef]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).