Glycosylation of Cancer Extracellular Vesicles: Capture Strategies, Functional Roles and Potential Clinical Applications
Abstract
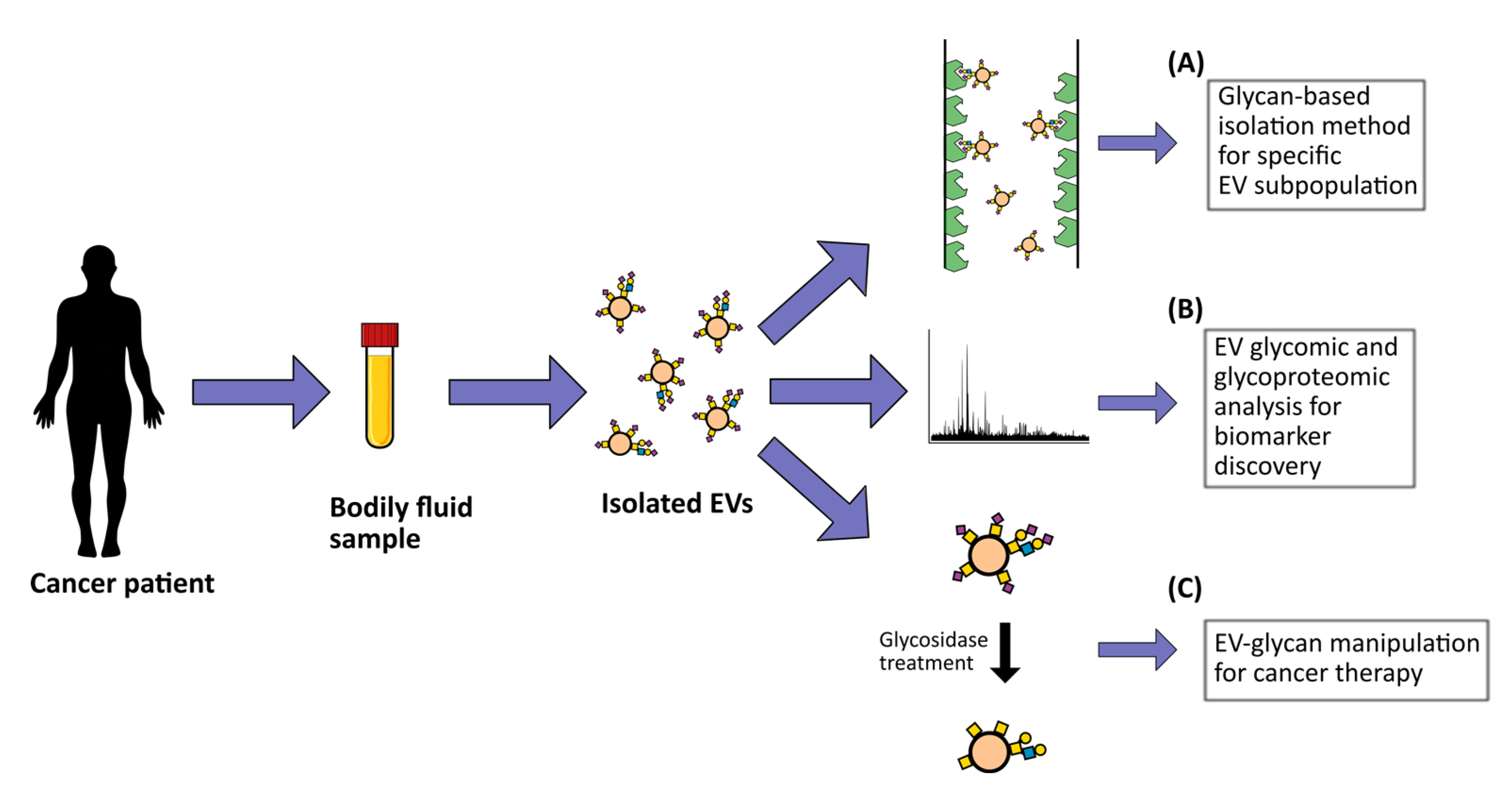
1. Introduction
2. The Impact of Glycosylation in Cancer Progression
2.1. Receptor Tyrosine Kinase Activation
2.2. Cell Adhesion Molecules
2.3. Immune Response Regulation
3. Shortcomings of Glycomic Approaches and Their Application in Extracellular Vesicle Glycome Analysis
4. Cancer Extracellular Vesicles Glycosylation
4.1. Extracellular Vesicle Surface Glycans as Relevant Markers for Its Detection and Isolation
4.2. The Functional Roles of Extracellular Vesicle Glycosylation in Cancer
4.2.1. EV Biodistribution and Uptake
4.2.2. Protein Sorting
4.2.3. Cell Behavior Modulation
4.3. The Potential Clinical Application of Cancer Extracellular Vesicles Glycosylation
4.3.1. EV Glycosylation as Therapeutic Delivery Tools
4.3.2. Cancer Biomarker Discovery
5. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AF4 | asymmetric-flow field-flow fractionation |
| AFP | alpha-fetoprotein |
| BGN | biglycan |
| CEA | carcinoembryonic antigen |
| CSPG4 | tumor antigen chondroitin sulfate proteoglycan 4 |
| DC-SIGN | dendritic cell-specific intercellular adhesion molecule-3-grabbing non-integrin |
| ECM | extracellular matrix |
| EFF | evanescent-field fluorescence-assisted |
| EGFR | epidermal growth factor receptor |
| EMMPRIN | extracellular matrix metalloproteinase inducer |
| ER | endoplasmic reticulum |
| ESCRT | endosomal sorting complex required for transport |
| EVs | extracellular vesicles |
| FasL | fas ligand |
| GA | golgi apparatus |
| GAGs | glycosaminoglycans |
| GPC1 | glypican-1 |
| GPI | glycosylphosphatidylinositol |
| HCC | hepatocellular carcinoma |
| HER2 | human epidermal growth factor receptor 2 |
| HGF | hepatocyte growth factor |
| HS | heparan Sulfate |
| HSPGs | heparan sulfate proteoglycans |
| IL-12 | interleukin 12 |
| iMAGE | integrated magnetic analysis of glycans in extracellular vesicles |
| LC/MS-MS | liquid chromatography/tandem mass spectrometry |
| LCN2 | lipocalin 2 |
| LGALS3BP | galactoside-binding soluble 3 binding protein |
| MALDI TOF | matrix-assisted laser desorption ionization time-of-flight |
| MAL-I | Maackia amurensis lectin I |
| MAPK | mitogen-activated protein kinase |
| MVBs | multivesicular bodies |
| NK | Natural Killer |
| NKG2D | Natural Killer group 2D |
| NP-TRFIA | nanoparticle-based time-resolved fluorescence immunoassay |
| PD-L1 | programmed death-ligand 1 |
| PNA | peanut agglutinin |
| PSA | prostate-specific antigen |
| RTK | receptor tyrosine kinase |
| SC | simple-cell |
| Siglecs | sialic acid binding immunoglobulin-type lectins |
| STn | Sialyl-Tn |
| TER ATPase | transitional endoplasmic reticulum ATPase |
| TGF-β | transforming growth factor-β |
| THP | Tamm–Horsfall protein |
| TMZ | alkylating agent temozolomide |
| TRAIL | TNF-related apoptosis-inducing ligand |
| VEGF | vascular endothelial growth factor |
| VEGFR | vascular endothelial growth factor receptor |
References
- Colombo, M.; Raposo, G.; Théry, C. Biogenesis, Secretion, and Intercellular Interactions of Exosomes and Other Extracellular Vesicles. Annu. Rev. Cell Dev. Biol. 2014, 30, 255–289. [Google Scholar] [CrossRef] [PubMed]
- Christiansen, M.N.; Chik, J.; Lee, L.; Anugraham, M.; Abrahams, J.L.; Packer, N.H. Cell Surface Protein Glycosylation in Cancer. Proteomics 2014, 14, 525–546. [Google Scholar] [CrossRef] [PubMed]
- Pinho, S.S.; Reis, C.A. Glycosylation in Cancer: Mechanisms and Clinical Implications. Nat. Rev. Cancer 2015, 15, 540–555. [Google Scholar] [CrossRef] [PubMed]
- Mereiter, S.; Balmaña, M.; Campos, D.; Gomes, J.; Reis, C.A. Glycosylation in the Era of Cancer-Targeted Therapy: Where Are We Heading? Cancer Cell 2019, 36, 6–16. [Google Scholar] [CrossRef] [PubMed]
- Surman, M.; Hoja-Łukowicz, D.; Szwed, S.; Drożdż, A.; Stępień, E.; Przybyło, M. Human Melanoma-Derived Ectosomes Are Enriched with Specific Glycan Epitopes. Life Sci. 2018, 207, 395–411. [Google Scholar] [CrossRef] [PubMed]
- Batista, B.S.; Eng, W.S.; Pilobello, K.T.; Hendricks-Muñoz, K.D.; Mahal, L.K. Identification of a Conserved Glycan Signature for Microvesicles. J. Proteome Res. 2011, 10, 4624–4633. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Wang, Y. Glycosylation Quality Control by the Golgi Structure. J. Mol. Biol. 2016, 428, 3183–3193. [Google Scholar] [CrossRef] [PubMed]
- Williams, C.; Pazos, R.; Royo, F.; González, E.; Roura-Ferrer, M.; Martinez, A.; Gamiz, J.; Reichardt, N.-C.; Falcón-Pérez, J.M. Assessing the Role of Surface Glycans of Extracellular Vesicles on Cellular Uptake. Sci. Rep. 2019, 9, 11920. [Google Scholar] [CrossRef]
- Christianson, H.C.; Svensson, K.J.; van Kuppevelt, T.H.; Li, J.-P.; Belting, M. Cancer Cell Exosomes Depend on Cell-Surface Heparan Sulfate Proteoglycans for Their Internalization and Functional Activity. Proc. Natl. Acad. Sci. USA 2013, 110, 17380–17385. [Google Scholar] [CrossRef]
- Chen, L.; Brigstock, D.R. Integrins and Heparan Sulfate Proteoglycans on Hepatic Stellate Cells (HSC) Are Novel Receptors for HSC-Derived Exosomes. FEBS Lett. 2016, 590, 4263–4274. [Google Scholar] [CrossRef]
- Franzen, C.A.; Simms, P.E.; Van Huis, A.F.; Foreman, K.E.; Kuo, P.C.; Gupta, G.N. Characterization of Uptake and Internalization of Exosomes by Bladder Cancer Cells. Biomed. Res. Int. 2014, 2014. [Google Scholar] [CrossRef] [PubMed]
- Cummings, R.D.; Pierce, J.M. The Challenge and Promise of Glycomics. Chem. Biol. 2014, 21, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Mereiter, S.; Balmaña, M.; Gomes, J.; Magalhães, A.; Reis, C.A. Glycomic Approaches for the Discovery of Targets in Gastrointestinal Cancer. Front. Oncol. 2016, 6, 55. [Google Scholar] [CrossRef] [PubMed]
- Varki, A. Biological Roles of Glycans. Glycobiology 2017, 27, 3–49. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, J.G.; Balmaña, M.; Macedo, J.A.; Poças, J.; Fernandes, Â.; de-Freitas-Junior, J.C.M.; Pinho, S.S.; Gomes, J.; Magalhães, A.; Gomes, C.; et al. Glycosylation in Cancer: Selected Roles in Tumour Progression, Immune Modulation and Metastasis. Cell Immunol. 2018, 333, 46–57. [Google Scholar] [CrossRef]
- Nagae, M.; Yamaguchi, Y. Function and 3D Structure of the N-Glycans on Glycoproteins. Int. J. Mol. Sci. 2012, 13, 8398–8429. [Google Scholar] [CrossRef]
- Gill, D.J.; Clausen, H.; Bard, F. Location, Location, Location: New Insights into O-GalNAc Protein Glycosylation. Trends Cell Biol. 2011, 21, 149–158. [Google Scholar] [CrossRef]
- Prabhakar, V.; Capila, I.; Sasisekharan, R. The Structural Elucidation of Glycosaminoglycans. Methods Mol. Biol. 2009, 534, 147–156. [Google Scholar] [CrossRef]
- Kumamoto, K.; Goto, Y.; Sekikawa, K.; Takenoshita, S.; Ishida, N.; Kawakita, M.; Kannagi, R. Increased Expression of UDP-Galactose Transporter Messenger RNA in Human Colon Cancer Tissues and Its Implication in Synthesis of Thomsen-Friedenreich Antigen and Sialyl Lewis A/X Determinants. Cancer Res. 2001, 61, 4620–4627. [Google Scholar]
- Marcos, N.T.; Bennett, E.P.; Gomes, J.; Magalhaes, A.; Gomes, C.; David, L.; Dar, I.; Jeanneau, C.; DeFrees, S.; Krustrup, D.; et al. ST6GalNAc-I Controls Expression of Sialyl-Tn Antigen in Gastrointestinal Tissues. Front. Biosci. (Elite Ed.) 2011, 3, 1443–1455. [Google Scholar] [CrossRef]
- Gill, D.J.; Chia, J.; Senewiratne, J.; Bard, F. Regulation of O-Glycosylation through Golgi-to-ER Relocation of Initiation Enzymes. J. Cell. Biol. 2010, 189, 843–858. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Ju, T.; Ding, X.; Xia, B.; Wang, W.; Xia, L.; He, M.; Cummings, R.D. Cosmc Is an Essential Chaperone for Correct Protein O-Glycosylation. Proc. Natl. Acad. Sci. USA 2010, 107, 9228–9233. [Google Scholar] [CrossRef] [PubMed]
- Lau, K.S.; Partridge, E.A.; Grigorian, A.; Silvescu, C.I.; Reinhold, V.N.; Demetriou, M.; Dennis, J.W. Complex N-Glycan Number and Degree of Branching Cooperate to Regulate Cell Proliferation and Differentiation. Cell 2007, 129, 123–134. [Google Scholar] [CrossRef] [PubMed]
- Croci, D.O.; Cerliani, J.P.; Pinto, N.A.; Morosi, L.G.; Rabinovich, G.A. Regulatory Role of Glycans in the Control of Hypoxia-Driven Angiogenesis and Sensitivity to Anti-Angiogenic Treatment. Glycobiology 2014, 24, 1283–1290. [Google Scholar] [CrossRef] [PubMed]
- Freitas, D.; Campos, D.; Gomes, J.; Pinto, F.; Macedo, J.A.; Matos, R.; Mereiter, S.; Pinto, M.T.; Polónia, A.; Gartner, F.; et al. O-Glycans Truncation Modulates Gastric Cancer Cell Signaling and Transcription Leading to a More Aggressive Phenotype. EBioMedicine 2019, 40, 349–362. [Google Scholar] [CrossRef] [PubMed]
- Häuselmann, I.; Borsig, L. Altered Tumor-Cell Glycosylation Promotes Metastasis. Front. Oncol. 2014, 4, 28. [Google Scholar] [CrossRef] [PubMed]
- Contessa, J.N.; Bhojani, M.S.; Freeze, H.H.; Rehemtulla, A.; Lawrence, T.S. Inhibition of N-Linked Glycosylation Disrupts Receptor Tyrosine Kinase Signaling in Tumor Cells. Cancer Res. 2008, 68, 3803–3809. [Google Scholar] [CrossRef]
- Gomes, C.; Osório, H.; Pinto, M.T.; Campos, D.; Oliveira, M.J.; Reis, C.A. Expression of ST3GAL4 Leads to SLe(x) Expression and Induces c-Met Activation and an Invasive Phenotype in Gastric Carcinoma Cells. PLoS One 2013, 8, e66737. [Google Scholar] [CrossRef]
- Mereiter, S.; Magalhães, A.; Adamczyk, B.; Jin, C.; Almeida, A.; Drici, L.; Ibáñez-Vea, M.; Gomes, C.; Ferreira, J.A.; Afonso, L.P.; et al. Glycomic Analysis of Gastric Carcinoma Cells Discloses Glycans as Modulators of RON Receptor Tyrosine Kinase Activation in Cancer. Biochim. Biophys. Acta 2016, 1860, 1795–1808. [Google Scholar] [CrossRef]
- Langer, M.D.; Guo, H.; Shashikanth, N.; Pierce, J.M.; Leckband, D.E. N-Glycosylation Alters Cadherin-Mediated Intercellular Binding Kinetics. J. Cell Sci. 2012, 125, 2478–2485. [Google Scholar] [CrossRef]
- Marsico, G.; Russo, L.; Quondamatteo, F.; Pandit, A. Glycosylation and Integrin Regulation in Cancer. Trends Cancer 2018, 4, 537–552. [Google Scholar] [CrossRef] [PubMed]
- Hang, Q.; Isaji, T.; Hou, S.; Wang, Y.; Fukuda, T.; Gu, J. A Key Regulator of Cell Adhesion: Identification and Characterization of Important N-Glycosylation Sites on Integrin A5 for Cell Migration. Mol. Cell Biol. 2017, 37. [Google Scholar] [CrossRef] [PubMed]
- Läubli, H.; Borsig, L. Altered Cell Adhesion and Glycosylation Promote Cancer Immune Suppression and Metastasis. Front. Immunol. 2019, 10, 2120. [Google Scholar] [CrossRef] [PubMed]
- Stowell, S.R.; Ju, T.; Cummings, R.D. Protein Glycosylation in Cancer. Annu. Rev. Pathol. 2015, 10, 473–510. [Google Scholar] [CrossRef] [PubMed]
- Lemmon, M.A.; Schlessinger, J. Cell Signaling by Receptor Tyrosine Kinases. Cell 2010, 141, 1117–1134. [Google Scholar] [CrossRef] [PubMed]
- Du, Z.; Lovly, C.M. Mechanisms of Receptor Tyrosine Kinase Activation in Cancer. Mol. Cancer 2018, 17, 58. [Google Scholar] [CrossRef] [PubMed]
- Butti, R.; Das, S.; Gunasekaran, V.P.; Yadav, A.S.; Kumar, D.; Kundu, G.C. Receptor Tyrosine Kinases (RTKs) in Breast Cancer: Signaling, Therapeutic Implications and Challenges. Mol. Cancer 2018, 17, 34. [Google Scholar] [CrossRef]
- Wang, C.; Yang, Y.; Yang, Z.; Liu, M.; Li, Z.; Sun, L.; Mei, C.; Chen, H.; Chen, L.; Wang, L.; et al. EGF-Mediated Migration Signaling Activated by N-Acetylglucosaminyltransferase-V via Receptor Protein Tyrosine Phosphatase Kappa. Arch. Biochem. Biophys. 2009, 486, 64–72. [Google Scholar] [CrossRef]
- Hang, Q.; Isaji, T.; Hou, S.; Zhou, Y.; Fukuda, T.; Gu, J. N-Glycosylation of Integrin A5 Acts as a Switch for EGFR-Mediated Complex Formation of Integrin A5β1 to A6β4. Sci. Rep. 2016, 6, 33507. [Google Scholar] [CrossRef]
- Britain, C.M.; Holdbrooks, A.T.; Anderson, J.C.; Willey, C.D.; Bellis, S.L. Sialylation of EGFR by the ST6Gal-I Sialyltransferase Promotes EGFR Activation and Resistance to Gefitinib-Mediated Cell Death. J. Ovarian Res. 2018, 11, 12. [Google Scholar] [CrossRef]
- Guo, H.-B.; Johnson, H.; Randolph, M.; Nagy, T.; Blalock, R.; Pierce, M. Specific Posttranslational Modification Regulates Early Events in Mammary Carcinoma Formation. Proc. Natl. Acad. Sci. USA 2010, 107, 21116–21121. [Google Scholar] [CrossRef] [PubMed]
- Scott, D.A.; Casadonte, R.; Cardinali, B.; Spruill, L.; Mehta, A.S.; Carli, F.; Simone, N.; Kriegsmann, M.; Del Mastro, L.; Kriegsmann, J.; et al. Increases in Tumor N-Glycan Polylactosamines Associated with Advanced HER2-Positive and Triple-Negative Breast Cancer Tissues. Proteom. Clin. Appl. 2019, 13, e1800014. [Google Scholar] [CrossRef] [PubMed]
- Duarte, H.O.; Balmaña, M.; Mereiter, S.; Osório, H.; Gomes, J.; Reis, C.A. Gastric Cancer Cell Glycosylation as a Modulator of the ErbB2 Oncogenic Receptor. Int. J. Mol. Sci. 2017, 18, 2262. [Google Scholar] [CrossRef] [PubMed]
- Markowska, A.I.; Jefferies, K.C.; Panjwani, N. Galectin-3 Protein Modulates Cell Surface Expression and Activation of Vascular Endothelial Growth Factor Receptor 2 in Human Endothelial Cells. J. Biol. Chem. 2011, 286, 29913–29921. [Google Scholar] [CrossRef] [PubMed]
- Croci, D.O.; Cerliani, J.P.; Dalotto-Moreno, T.; Méndez-Huergo, S.P.; Mascanfroni, I.D.; Dergan-Dylon, S.; Toscano, M.A.; Caramelo, J.J.; García-Vallejo, J.J.; Ouyang, J.; et al. Glycosylation-Dependent Lectin-Receptor Interactions Preserve Angiogenesis in Anti-VEGF Refractory Tumors. Cell 2014, 156, 744–758. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, I.G.; Pucci, M.; Venturi, G.; Malagolini, N.; Chiricolo, M.; Dall’Olio, F. Glycosylation as a Main Regulator of Growth and Death Factor Receptors Signaling. Int. J. Mol. Sci. 2018, 19, 580. [Google Scholar] [CrossRef]
- Cecchi, F.; Pajalunga, D.; Fowler, C.A.; Uren, A.; Rabe, D.C.; Peruzzi, B.; Macdonald, N.J.; Blackman, D.K.; Stahl, S.J.; Byrd, R.A.; et al. Targeted Disruption of Heparan Sulfate Interaction with Hepatocyte and Vascular Endothelial Growth Factors Blocks Normal and Oncogenic Signaling. Cancer Cell 2012, 22, 250–262. [Google Scholar] [CrossRef]
- Shintani, Y.; Takashima, S.; Asano, Y.; Kato, H.; Liao, Y.; Yamazaki, S.; Tsukamoto, O.; Seguchi, O.; Yamamoto, H.; Fukushima, T.; et al. Glycosaminoglycan Modification of Neuropilin-1 Modulates VEGFR2 Signaling. EMBO J. 2006, 25, 3045–3055. [Google Scholar] [CrossRef]
- Weber, G.F.; Bjerke, M.A.; DeSimone, D.W. Integrins and Cadherins Join Forces to Form Adhesive Networks. J. Cell Sci. 2011, 124, 1183–1193. [Google Scholar] [CrossRef]
- Paszek, M.J.; DuFort, C.C.; Rossier, O.; Bainer, R.; Mouw, J.K.; Godula, K.; Hudak, J.E.; Lakins, J.N.; Wijekoon, A.C.; Cassereau, L.; et al. The Cancer Glycocalyx Mechanically Primes Integrin-Mediated Growth and Survival. Nature 2014, 511, 319–325. [Google Scholar] [CrossRef]
- Yuan, Y.; Wu, L.; Shen, S.; Wu, S.; Burdick, M.M. Effect of Alpha 2,6 Sialylation on Integrin-Mediated Adhesion of Breast Cancer Cells to Fibronectin and Collagen IV. Life Sci. 2016, 149, 138–145. [Google Scholar] [CrossRef] [PubMed]
- Clément, M.; Rocher, J.; Loirand, G.; Le Pendu, J. Expression of Sialyl-Tn Epitopes on Beta1 Integrin Alters Epithelial Cell Phenotype, Proliferation and Haptotaxis. J. Cell Sci. 2004, 117, 5059–5069. [Google Scholar] [CrossRef] [PubMed]
- Pinho, S.S.; Figueiredo, J.; Cabral, J.; Carvalho, S.; Dourado, J.; Magalhães, A.; Gärtner, F.; Mendonfa, A.M.; Isaji, T.; Gu, J.; et al. E-Cadherin and Adherens-Junctions Stability in Gastric Carcinoma: Functional Implications of Glycosyltransferases Involving N-Glycan Branching Biosynthesis, N-Acetylglucosaminyltransferases III and V. Biochim. Biophys. Acta 2013, 1830, 2690–2700. [Google Scholar] [CrossRef] [PubMed]
- Borsig, L. Selectins in Cancer Immunity. Glycobiology 2018, 28, 648–655. [Google Scholar] [CrossRef] [PubMed]
- McEver, R.P. Selectins: Initiators of Leucocyte Adhesion and Signalling at the Vascular Wall. Cardiovasc. Res. 2015, 107, 331–339. [Google Scholar] [CrossRef]
- Ding, D.; Yao, Y.; Zhang, S.; Su, C.; Zhang, Y. C-Type Lectins Facilitate Tumor Metastasis. Oncol. Lett. 2017, 13, 13–21. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Esposito, M.; Mondal, N.; Greco, T.M.; Wei, Y.; Spadazzi, C.; Lin, S.-C.; Zheng, H.; Cheung, C.; Magnani, J.L.; Lin, S.-H.; et al. Bone Vascular Niche E-Selectin Induces Mesenchymal-Epithelial Transition and Wnt Activation in Cancer Cells to Promote Bone Metastasis. Nat. Cell Biol. 2019, 21, 627–639. [Google Scholar] [CrossRef]
- Jandus, C.; Boligan, K.F.; Chijioke, O.; Liu, H.; Dahlhaus, M.; Démoulins, T.; Schneider, C.; Wehrli, M.; Hunger, R.E.; Baerlocher, G.M.; et al. Interactions between Siglec-7/9 Receptors and Ligands Influence NK Cell-Dependent Tumor Immunosurveillance. J. Clin. Investig. 2014, 124, 1810–1820. [Google Scholar] [CrossRef]
- Hudak, J.E.; Canham, S.M.; Bertozzi, C.R. Glycocalyx Engineering Reveals a Siglec-Based Mechanism for NK Cell Immunoevasion. Nat. Chem. Biol. 2014, 10, 69–75. [Google Scholar] [CrossRef]
- Macauley, M.S.; Crocker, P.R.; Paulson, J.C. Siglec-Mediated Regulation of Immune Cell Function in Disease. Nat. Rev. Immunol. 2014, 14, 653–666. [Google Scholar] [CrossRef]
- Daley, D.; Mani, V.R.; Mohan, N.; Akkad, N.; Ochi, A.; Heindel, D.W.; Lee, K.B.; Zambirinis, C.P.; Pandian, G.S.B.; Savadkar, S.; et al. Dectin 1 Activation on Macrophages by Galectin 9 Promotes Pancreatic Carcinoma and Peritumoral Immune Tolerance. Nat. Med. 2017, 23, 556–567. [Google Scholar] [CrossRef] [PubMed]
- Kouo, T.; Huang, L.; Pucsek, A.B.; Cao, M.; Solt, S.; Armstrong, T.; Jaffee, E. Galectin-3 Shapes Antitumor Immune Responses by Suppressing CD8+ T Cells via LAG-3 and Inhibiting Expansion of Plasmacytoid Dendritic Cells. Cancer Immunol. Res. 2015, 3, 412–423. [Google Scholar] [CrossRef] [PubMed]
- Demotte, N.; Wieërs, G.; Van Der Smissen, P.; Moser, M.; Schmidt, C.; Thielemans, K.; Squifflet, J.-L.; Weynand, B.; Carrasco, J.; Lurquin, C.; et al. A Galectin-3 Ligand Corrects the Impaired Function of Human CD4 and CD8 Tumor-Infiltrating Lymphocytes and Favors Tumor Rejection in Mice. Cancer Res. 2010, 70, 7476–7488. [Google Scholar] [CrossRef] [PubMed]
- Merlotti, A.; Malizia, A.L.; Michea, P.; Bonte, P.-E.; Goudot, C.; Carregal, M.S.; Nuñez, N.; Sedlik, C.; Ceballos, A.; Soumelis, V.; et al. Aberrant Fucosylation Enables Breast Cancer Clusterin to Interact with Dendritic Cell-Specific ICAM-Grabbing Non-Integrin (DC-SIGN). Oncoimmunology 2019, 8, e1629257. [Google Scholar] [CrossRef] [PubMed]
- Kunej, T. Rise of Systems Glycobiology and Personalized Glycomedicine: Why and How to Integrate Glycomics with Multiomics Science? OMICS 2019, 23, 615–622. [Google Scholar] [CrossRef] [PubMed]
- Rudd, P.; Karlsson, N.G.; Khoo, K.-H.; Packer, N.H. Glycomics and Glycoproteomics, 3rd ed.; Cold Spring Harbor Laboratory Press: New York, NY, USA, 2015–2017; Chapter 51.
- Hirabayashi, J.; Kuno, A.; Tateno, H. Development and Applications of the Lectin Microarray. Top. Curr. Chem. 2015, 367, 105–124. [Google Scholar] [CrossRef]
- Wuhrer, M. Glycomics Using Mass Spectrometry. Glycoconj. J. 2013, 30, 11–22. [Google Scholar] [CrossRef]
- Gerlach, J.Q.; Griffin, M.D. Getting to Know the Extracellular Vesicle Glycome. Mol. Biosyst. 2016, 12, 1071–1081. [Google Scholar] [CrossRef]
- Gerlach, J.Q.; Krüger, A.; Gallogly, S.; Hanley, S.A.; Hogan, M.C.; Ward, C.J.; Joshi, L.; Griffin, M.D. Surface Glycosylation Profiles of Urine Extracellular Vesicles. PLoS ONE 2013, 8, e74801. [Google Scholar] [CrossRef]
- Costa, J.; Gatermann, M.; Nimtz, M.; Kandzia, S.; Glatzel, M.; Conradt, H.S. N-Glycosylation of Extracellular Vesicles from HEK-293 and Glioma Cell Lines. Anal. Chem. 2018, 90, 7871–7879. [Google Scholar] [CrossRef]
- Gomes, J.; Gomes-Alves, P.; Carvalho, S.B.; Peixoto, C.; Alves, P.M.; Altevogt, P.; Costa, J. Extracellular Vesicles from Ovarian Carcinoma Cells Display Specific Glycosignatures. Biomolecules 2015, 5, 1741–1761. [Google Scholar] [CrossRef] [PubMed]
- Williams, C.; Royo, F.; Aizpurua-Olaizola, O.; Pazos, R.; Boons, G.-J.; Reichardt, N.-C.; Falcon-Perez, J.M. Glycosylation of Extracellular Vesicles: Current Knowledge, Tools and Clinical Perspectives. J. Extracell. Vesicles 2018, 7, 1442985. [Google Scholar] [CrossRef] [PubMed]
- North, S.J.; Hitchen, P.G.; Haslam, S.M.; Dell, A. Mass Spectrometry in the Analysis of N-Linked and O-Linked Glycans. Curr. Opin. Struct. Biol. 2009, 19, 498–506. [Google Scholar] [CrossRef] [PubMed]
- Schulz, B.L.; Packer, N.H.; Karlsson, N.G. Small-Scale Analysis of O-Linked Oligosaccharides from Glycoproteins and Mucins Separated by Gel Electrophoresis. Anal. Chem. 2002, 74, 6088–6097. [Google Scholar] [CrossRef] [PubMed]
- Geyer, H.; Geyer, R. Strategies for Analysis of Glycoprotein Glycosylation. Biochim. Biophys. Acta 2006, 1764, 1853–1869. [Google Scholar] [CrossRef]
- Merry, A.H.; Neville, D.C.A.; Royle, L.; Matthews, B.; Harvey, D.J.; Dwek, R.A.; Rudd, P.M. Recovery of Intact 2-Aminobenzamide-Labeled O-Glycans Released from Glycoproteins by Hydrazinolysis. Anal. Biochem. 2002, 304, 91–99. [Google Scholar] [CrossRef]
- Kozak, R.P.; Royle, L.; Gardner, R.A.; Fernandes, D.L.; Wuhrer, M. Suppression of Peeling during the Release of O-Glycans by Hydrazinolysis. Anal. Biochem. 2012, 423, 119–128. [Google Scholar] [CrossRef]
- King, S.L.; Joshi, H.J.; Schjoldager, K.T.; Halim, A.; Madsen, T.D.; Dziegiel, M.H.; Woetmann, A.; Vakhrushev, S.Y.; Wandall, H.H. Characterizing the O-Glycosylation Landscape of Human Plasma, Platelets, and Endothelial Cells. Blood Adv. 2017, 1, 429–442. [Google Scholar] [CrossRef]
- Steentoft, C.; Vakhrushev, S.Y.; Joshi, H.J.; Kong, Y.; Vester-Christensen, M.B.; Schjoldager, K.T.-B.G.; Lavrsen, K.; Dabelsteen, S.; Pedersen, N.B.; Marcos-Silva, L.; et al. Precision Mapping of the Human O-GalNAc Glycoproteome through SimpleCell Technology. EMBO J. 2013, 32, 1478–1488. [Google Scholar] [CrossRef]
- Ye, Z.; Mao, Y.; Clausen, H.; Vakhrushev, S.Y. Glyco-DIA: A Method for Quantitative O-Glycoproteomics with in Silico-Boosted Glycopeptide Libraries. Nat. Methods 2019, 16, 902–910. [Google Scholar] [CrossRef]
- Campos, D.; Freitas, D.; Gomes, J.; Magalhães, A.; Steentoft, C.; Gomes, C.; Vester-Christensen, M.B.; Ferreira, J.A.; Afonso, L.P.; Santos, L.L.; et al. Probing the O-Glycoproteome of Gastric Cancer Cell Lines for Biomarker Discovery. Mol. Cell. Proteom. 2015, 14, 1616–1629. [Google Scholar] [CrossRef] [PubMed]
- Campos, D.; Freitas, D.; Gomes, J.; Reis, C.A. Glycoengineered Cell Models for the Characterization of Cancer O-Glycoproteome: An Innovative Strategy for Biomarker Discovery. Expert. Rev. Proteom. 2015, 12, 337–342. [Google Scholar] [CrossRef] [PubMed]
- Harada, Y.; Kizuka, Y.; Tokoro, Y.; Kondo, K.; Yagi, H.; Kato, K.; Inoue, H.; Taniguchi, N.; Maruyama, I. N-Glycome Inheritance from Cells to Extracellular Vesicles in B16 Melanomas. FEBS Lett. 2019, 593, 942–951. [Google Scholar] [CrossRef] [PubMed]
- Vermassen, T.; D’Herde, K.; Jacobus, D.; Van Praet, C.; Poelaert, F.; Lumen, N.; Callewaert, N.; Decaestecker, K.; Villeirs, G.; Hoebeke, P.; et al. Release of Urinary Extracellular Vesicles in Prostate Cancer Is Associated with Altered Urinary N-Glycosylation Profile. J. Clin. Pathol. 2017, 70, 838–846. [Google Scholar] [CrossRef] [PubMed]
- Freitas, D.; Balmaña, M.; Poças, J.; Campos, D.; Osório, H.; Konstantinidi, A.; Vakhrushev, S.Y.; Magalhães, A.; Reis, C.A. Different Isolation Approaches Lead to Diverse Glycosylated Extracellular Vesicle Populations. J. Extracell. Vesicles 2019, 8, 1621131. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.; Guo, Y.; Li, Y.; Tao, J.; Ding, L.; Wu, J.; Ju, H. Lectin-Mediated in Situ Rolling Circle Amplification on Exosomes for Probing Cancer-Related Glycan Pattern. Anal. Chim. Acta 2018, 1039, 108–115. [Google Scholar] [CrossRef]
- Chaiyawat, P.; Weeraphan, C.; Netsirisawan, P.; Chokchaichamnankit, D.; Srisomsap, C.; Svasti, J.; Champattanachai, V. Elevated O-GlcNAcylation of Extracellular Vesicle Proteins Derived from Metastatic Colorectal Cancer Cells. Cancer Genom. Proteom. 2016, 13, 387–398. [Google Scholar]
- Zou, G.; Benktander, J.D.; Gizaw, S.T.; Gaunitz, S.; Novotny, M.V. Comprehensive Analytical Approach toward Glycomic Characterization and Profiling in Urinary Exosomes. Anal. Chem. 2017, 89, 5364–5372. [Google Scholar] [CrossRef]
- Peixoto, A.; Relvas-Santos, M.; Azevedo, R.; Santos, L.L.; Ferreira, J.A. Protein Glycosylation and Tumor Microenvironment Alterations Driving Cancer Hallmarks. Front. Oncol. 2019, 9. [Google Scholar] [CrossRef]
- Zhang, H.; Freitas, D.; Kim, H.S.; Fabijanic, K.; Li, Z.; Chen, H.; Mark, M.T.; Molina, H.; Martin, A.B.; Bojmar, L.; et al. Identification of Distinct Nanoparticles and Subsets of Extracellular Vesicles by Asymmetric Flow Field-Flow Fractionation. Nat. Cell. Biol. 2018, 20, 332–343. [Google Scholar] [CrossRef]
- Doyle, L.M.; Wang, M.Z. Overview of Extracellular Vesicles, Their Origin, Composition, Purpose, and Methods for Exosome Isolation and Analysis. Cells 2019, 8, 727. [Google Scholar] [CrossRef] [PubMed]
- Van Niel, G.; D’Angelo, G.; Raposo, G. Shedding Light on the Cell Biology of Extracellular Vesicles. Nat. Rev. Mol. Cell Biol. 2018, 19, 213–228. [Google Scholar] [CrossRef] [PubMed]
- Bebelman, M.P.; Smit, M.J.; Pegtel, D.M.; Baglio, S.R. Biogenesis and Function of Extracellular Vesicles in Cancer. Pharmacol. Ther. 2018, 188, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Willms, E.; Johansson, H.J.; Mäger, I.; Lee, Y.; Blomberg, K.E.M.; Sadik, M.; Alaarg, A.; Smith, C.I.E.; Lehtiö, J.; EL Andaloussi, S.; et al. Cells Release Subpopulations of Exosomes with Distinct Molecular and Biological Properties. Sci. Rep. 2016, 6, 22519. [Google Scholar] [CrossRef] [PubMed]
- Géminard, C.; De Gassart, A.; Blanc, L.; Vidal, M. Degradation of AP2 during Reticulocyte Maturation Enhances Binding of Hsc70 and Alix to a Common Site on TFR for Sorting into Exosomes. Traffic 2004, 5, 181–193. [Google Scholar] [CrossRef] [PubMed]
- Stuffers, S.; Sem Wegner, C.; Stenmark, H.; Brech, A. Multivesicular Endosome Biogenesis in the Absence of ESCRTs. Traffic 2009, 10, 925–937. [Google Scholar] [CrossRef]
- Trajkovic, K.; Hsu, C.; Chiantia, S.; Rajendran, L.; Wenzel, D.; Wieland, F.; Schwille, P.; Brügger, B.; Simons, M. Ceramide Triggers Budding of Exosome Vesicles into Multivesicular Endosomes. Science 2008, 319, 1244–1247. [Google Scholar] [CrossRef] [PubMed]
- Andreu, Z.; Yáñez-Mó, M. Tetraspanins in Extracellular Vesicle Formation and Function. Front. Immunol. 2014, 5. [Google Scholar] [CrossRef]
- Escola, J.M.; Kleijmeer, M.J.; Stoorvogel, W.; Griffith, J.M.; Yoshie, O.; Geuze, H.J. Selective Enrichment of Tetraspan Proteins on the Internal Vesicles of Multivesicular Endosomes and on Exosomes Secreted by Human B-Lymphocytes. J. Biol. Chem. 1998, 273, 20121–20127. [Google Scholar] [CrossRef]
- Østergaard, O.; Nielsen, C.T.; Iversen, L.V.; Jacobsen, S.; Tanassi, J.T.; Heegaard, N.H.H. Quantitative Proteome Profiling of Normal Human Circulating Microparticles. J. Proteome. Res. 2012, 11, 2154–2163. [Google Scholar] [CrossRef]
- Heijnen, H.F.; Schiel, A.E.; Fijnheer, R.; Geuze, H.J.; Sixma, J.J. Activated Platelets Release Two Types of Membrane Vesicles: Microvesicles by Surface Shedding and Exosomes Derived from Exocytosis of Multivesicular Bodies and Alpha-Granules. Blood 1999, 94, 3791–3799. [Google Scholar] [CrossRef] [PubMed]
- Théry, C.; Boussac, M.; Véron, P.; Ricciardi-Castagnoli, P.; Raposo, G.; Garin, J.; Amigorena, S. Proteomic Analysis of Dendritic Cell-Derived Exosomes: A Secreted Subcellular Compartment Distinct from Apoptotic Vesicles. J. Immunol. 2001, 166, 7309–7318. [Google Scholar] [CrossRef] [PubMed]
- Vig, S.; Buitinga, M.; Rondas, D.; Crèvecoeur, I.; van Zandvoort, M.; Waelkens, E.; Eizirik, D.L.; Gysemans, C.; Baatsen, P.; Mathieu, C.; et al. Cytokine-Induced Translocation of GRP78 to the Plasma Membrane Triggers a pro-Apoptotic Feedback Loop in Pancreatic Beta Cells. Cell Death Dis. 2019, 10, 309. [Google Scholar] [CrossRef] [PubMed]
- Campanella, C.; D’Anneo, A.; Gammazza, A.M.; Bavisotto, C.C.; Barone, R.; Emanuele, S.; Lo Cascio, F.; Mocciaro, E.; Fais, S.; De Macario, E.C.; et al. The Histone Deacetylase Inhibitor SAHA Induces HSP60 Nitration and Its Extracellular Release by Exosomal Vesicles in Human Lung-Derived Carcinoma Cells. Oncotarget 2015, 7, 28849–28867. [Google Scholar] [CrossRef] [PubMed]
- Kusuma, G.D.; Barabadi, M.; Tan, J.L.; Morton, D.A.V.; Frith, J.E.; Lim, R. To Protect and to Preserve: Novel Preservation Strategies for Extracellular Vesicles. Front. Pharmacol. 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- Kalra, H.; Drummen, G.P.C.; Mathivanan, S. Focus on Extracellular Vesicles: Introducing the Next Small Big Thing. Int. J. Mol. Sci. 2016, 17, 170. [Google Scholar] [CrossRef]
- O’Brien, K.; Breyne, K.; Ughetto, S.; Laurent, L.C.; Breakefield, X.O. RNA Delivery by Extracellular Vesicles in Mammalian Cells and Its Applications. Nat. Rev. Mol. Cell Biol. 2020, 21, 585–606. [Google Scholar] [CrossRef]
- Momen-Heravi, F.; Getting, S.J.; Moschos, S.A. Extracellular Vesicles and Their Nucleic Acids for Biomarker Discovery. Pharmacol. Ther. 2018, 192, 170–187. [Google Scholar] [CrossRef]
- Massaro, C.; Sgueglia, G.; Frattolillo, V.; Baglio, S.R.; Altucci, L.; Dell’Aversana, C. Extracellular Vesicle-Based Nucleic Acid Delivery: Current Advances and Future Perspectives in Cancer Therapeutic Strategies. Pharmaceutics 2020, 12, 980. [Google Scholar] [CrossRef]
- Escrevente, C.; Grammel, N.; Kandzia, S.; Zeiser, J.; Tranfield, E.M.; Conradt, H.S.; Costa, J. Sialoglycoproteins and N-Glycans from Secreted Exosomes of Ovarian Carcinoma Cells. PLoS ONE 2013, 8, e78631. [Google Scholar] [CrossRef]
- Becker, A.; Thakur, B.K.; Weiss, J.M.; Kim, H.S.; Peinado, H.; Lyden, D. Extracellular Vesicles in Cancer: Cell-to-Cell Mediators of Metastasis. Cancer Cell 2016, 30, 836–848. [Google Scholar] [CrossRef] [PubMed]
- King, H.W.; Michael, M.Z.; Gleadle, J.M. Hypoxic Enhancement of Exosome Release by Breast Cancer Cells. BMC Cancer 2012, 12, 421. [Google Scholar] [CrossRef] [PubMed]
- Wysoczynski, M.; Ratajczak, M.Z. Lung Cancer Secreted Microvesicles: Underappreciated Modulators of Microenvironment in Expanding Tumors. Int. J. Cancer 2009, 125, 1595–1603. [Google Scholar] [CrossRef] [PubMed]
- Parolini, I.; Federici, C.; Raggi, C.; Lugini, L.; Palleschi, S.; De Milito, A.; Coscia, C.; Iessi, E.; Logozzi, M.; Molinari, A.; et al. Microenvironmental PH Is a Key Factor for Exosome Traffic in Tumor Cells. J. Biol. Chem. 2009, 284, 34211–34222. [Google Scholar] [CrossRef] [PubMed]
- Haraszti, R.A.; Miller, R.; Dubuke, M.L.; Rockwell, H.E.; Coles, A.H.; Sapp, E.; Didiot, M.-C.; Echeverria, D.; Stoppato, M.; Sere, Y.Y.; et al. Serum Deprivation of Mesenchymal Stem Cells Improves Exosome Activity and Alters Lipid and Protein Composition. iScience 2019, 16, 230–241. [Google Scholar] [CrossRef] [PubMed]
- Garcia, N.A.; Ontoria-Oviedo, I.; González-King, H.; Diez-Juan, A.; Sepúlveda, P. Glucose Starvation in Cardiomyocytes Enhances Exosome Secretion and Promotes Angiogenesis in Endothelial Cells. PLoS ONE 2015, 10, e0138849. [Google Scholar] [CrossRef] [PubMed]
- Hedlund, M.; Nagaeva, O.; Kargl, D.; Baranov, V.; Mincheva-Nilsson, L. Thermal- and Oxidative Stress Causes Enhanced Release of NKG2D Ligand-Bearing Immunosuppressive Exosomes in Leukemia/Lymphoma T and B Cells. PLoS ONE 2011, 6, e16899. [Google Scholar] [CrossRef]
- Eldh, M.; Ekström, K.; Valadi, H.; Sjöstrand, M.; Olsson, B.; Jernås, M.; Lötvall, J. Exosomes Communicate Protective Messages during Oxidative Stress; Possible Role of Exosomal Shuttle RNA. PLoS ONE 2010, 5, e15353. [Google Scholar] [CrossRef]
- Lv, L.-H.; Wan, Y.-L.; Lin, Y.; Zhang, W.; Yang, M.; Li, G.-L.; Lin, H.-M.; Shang, C.-Z.; Chen, Y.-J.; Min, J. Anticancer Drugs Cause Release of Exosomes with Heat Shock Proteins from Human Hepatocellular Carcinoma Cells That Elicit Effective Natural Killer Cell Antitumor Responses in Vitro. J. Biol. Chem. 2012, 287, 15874–15885. [Google Scholar] [CrossRef]
- Kucharzewska, P.; Belting, M. Emerging Roles of Extracellular Vesicles in the Adaptive Response of Tumour Cells to Microenvironmental Stress. J. Extracell. Vesicles 2013, 2. [Google Scholar] [CrossRef]
- Guo, Y.; Tao, J.; Li, Y.; Feng, Y.; Ju, H.; Wang, Z.; Ding, L. Quantitative Localized Analysis Reveals Distinct Exosomal Protein-Specific Glycosignatures: Implications in Cancer Cell Subtyping, Exosome Biogenesis, and Function. J. Am. Chem. Soc. 2020, 142, 7404–7412. [Google Scholar] [CrossRef] [PubMed]
- Surman, M.; Hoja-Łukowicz, D.; Szwed, S.; Kędracka-Krok, S.; Jankowska, U.; Kurtyka, M.; Drożdż, A.; Lityńska, A.; Stępień, E.; Przybyło, M. An Insight into the Proteome of Uveal Melanoma-Derived Ectosomes Reveals the Presence of Potentially Useful Biomarkers. Int. J. Mol. Sci. 2019, 20, 3789. [Google Scholar] [CrossRef] [PubMed]
- Shimoda, A.; Tahara, Y.; Sawada, S.-I.; Sasaki, Y.; Akiyoshi, K. Glycan Profiling Analysis Using Evanescent-Field Fluorescence-Assisted Lectin Array: Importance of Sugar Recognition for Cellular Uptake of Exosomes from Mesenchymal Stem Cells. Biochem. Biophys. Res. Commun. 2017, 491, 701–707. [Google Scholar] [CrossRef] [PubMed]
- Théry, C.; Witwer, K.W.; Aikawa, E.; Alcaraz, M.J.; Anderson, J.D.; Andriantsitohaina, R.; Antoniou, A.; Arab, T.; Archer, F.; Atkin-Smith, G.K.; et al. Minimal Information for Studies of Extracellular Vesicles 2018 (MISEV2018): A Position Statement of the International Society for Extracellular Vesicles and Update of the MISEV2014 Guidelines. J. Extracell. Vesicles 2018, 7. [Google Scholar] [CrossRef] [PubMed]
- Ramirez, M.I.; Amorim, M.G.; Gadelha, C.; Milic, I.; Welsh, J.A.; Freitas, V.M.; Nawaz, M.; Akbar, N.; Couch, Y.; Makin, L.; et al. Technical Challenges of Working with Extracellular Vesicles. Nanoscale 2018, 10, 881–906. [Google Scholar] [CrossRef] [PubMed]
- Williams, C.; Palviainen, M.; Reichardt, N.-C.; Siljander, P.R.-M.; Falcón-Pérez, J.M. Metabolomics Applied to the Study of Extracellular Vesicles. Metabolites 2019, 9, 276. [Google Scholar] [CrossRef]
- Tang, Y.-T.; Huang, Y.-Y.; Zheng, L.; Qin, S.-H.; Xu, X.-P.; An, T.-X.; Xu, Y.; Wu, Y.-S.; Hu, X.-M.; Ping, B.-H.; et al. Comparison of Isolation Methods of Exosomes and Exosomal RNA from Cell Culture Medium and Serum. Int. J. Mol. Med. 2017, 40, 834–844. [Google Scholar] [CrossRef]
- Royo, F.; Zuñiga-Garcia, P.; Sanchez-Mosquera, P.; Egia, A.; Perez, A.; Loizaga, A.; Arceo, R.; Lacasa, I.; Rabade, A.; Arrieta, E.; et al. Different EV Enrichment Methods Suitable for Clinical Settings Yield Different Subpopulations of Urinary Extracellular Vesicles from Human Samples. J. Extracell. Vesicles 2016, 5, 29497. [Google Scholar] [CrossRef]
- Van Deun, J.; Mestdagh, P.; Sormunen, R.; Cocquyt, V.; Vermaelen, K.; Vandesompele, J.; Bracke, M.; De Wever, O.; Hendrix, A. The Impact of Disparate Isolation Methods for Extracellular Vesicles on Downstream RNA Profiling. J. Extracell. Vesicles 2014, 3. [Google Scholar] [CrossRef]
- Matsuda, A.; Kuno, A.; Yoshida, M.; Wagatsuma, T.; Sato, T.; Miyagishi, M.; Zhao, J.; Suematsu, M.; Kabe, Y.; Narimatsu, H. Comparative Glycomic Analysis of Exosome Subpopulations Derived from Pancreatic Cancer Cell Lines. J. Proteome Res. 2020, 19, 2516–2524. [Google Scholar] [CrossRef]
- Tauro, B.J.; Greening, D.W.; Mathias, R.A.; Ji, H.; Mathivanan, S.; Scott, A.M.; Simpson, R.J. Comparison of Ultracentrifugation, Density Gradient Separation, and Immunoaffinity Capture Methods for Isolating Human Colon Cancer Cell Line LIM1863-Derived Exosomes. Methods 2012, 56, 293–304. [Google Scholar] [CrossRef] [PubMed]
- Théry, C.; Amigorena, S.; Raposo, G.; Clayton, A. Isolation and Characterization of Exosomes from Cell Culture Supernatants and Biological Fluids. Curr. Protoc. Cell Biol. 2006, 30, 3–22. [Google Scholar] [CrossRef] [PubMed]
- Salih, M.; Zietse, R.; Hoorn, E.J. Urinary Extracellular Vesicles and the Kidney: Biomarkers and Beyond. Am. J. Physiol. Renal. Physiol. 2014, 306, F1251–F1259. [Google Scholar] [CrossRef] [PubMed]
- Taylor, D.D.; Shah, S. Methods of Isolating Extracellular Vesicles Impact Down-Stream Analyses of Their Cargoes. Methods 2015, 87, 3–10. [Google Scholar] [CrossRef] [PubMed]
- Taylor, D.D.; Zacharias, W.; Gercel-Taylor, C. Exosome Isolation for Proteomic Analyses and RNA Profiling. Methods Mol. Biol. 2011, 728, 235–246. [Google Scholar] [CrossRef]
- Gámez-Valero, A.; Monguió-Tortajada, M.; Carreras-Planella, L.; la Franquesa, M.; Beyer, K.; Borràs, F.E. Size-Exclusion Chromatography-Based Isolation Minimally Alters Extracellular Vesicles’ Characteristics Compared to Precipitating Agents. Sci. Rep. 2016, 6, 33641. [Google Scholar] [CrossRef]
- Karttunen, J.; Heiskanen, M.; Navarro-Ferrandis, V.; Das Gupta, S.; Lipponen, A.; Puhakka, N.; Rilla, K.; Koistinen, A.; Pitkänen, A. Precipitation-Based Extracellular Vesicle Isolation from Rat Plasma Co-Precipitate Vesicle-Free MicroRNAs. J. Extracell. Vesicles 2018, 8. [Google Scholar] [CrossRef]
- Bickmore, D.C.; Miklavcic, J.J. Characterization of Extracellular Vesicles Isolated From Human Milk Using a Precipitation-Based Method. Front. Nutr. 2020, 7. [Google Scholar] [CrossRef]
- He, M.; Crow, J.; Roth, M.; Zeng, Y.; Godwin, A.K. Integrated Immunoisolation and Protein Analysis of Circulating Exosomes Using Microfluidic Technology. Lab. Chip. 2014, 14, 3773–3780. [Google Scholar] [CrossRef]
- Kanwar, S.S.; Dunlay, C.J.; Simeone, D.M.; Nagrath, S. Microfluidic Device (ExoChip) for on-Chip Isolation, Quantification and Characterization of Circulating Exosomes. Lab. Chip. 2014, 14, 1891–1900. [Google Scholar] [CrossRef]
- Greening, D.W.; Xu, R.; Ji, H.; Tauro, B.J.; Simpson, R.J. A Protocol for Exosome Isolation and Characterization: Evaluation of Ultracentrifugation, Density-Gradient Separation, and Immunoaffinity Capture Methods. Methods Mol. Biol. 2015, 1295, 179–209. [Google Scholar] [CrossRef] [PubMed]
- Oksvold, M.P.; Neurauter, A.; Pedersen, K.W. Magnetic Bead-Based Isolation of Exosomes. Methods Mol. Biol. 2015, 1218, 465–481. [Google Scholar] [CrossRef] [PubMed]
- Gerlach, J.Q.; Maguire, C.M.; Krüger, A.; Joshi, L.; Prina-Mello, A.; Griffin, M.D. Urinary Nanovesicles Captured by Lectins or Antibodies Demonstrate Variations in Size and Surface Glycosylation Profile. Nanomedicine (Lond.) 2017, 12, 1217–1229. [Google Scholar] [CrossRef] [PubMed]
- Echevarria, J.; Royo, F.; Pazos, R.; Salazar, L.; Falcon-Perez, J.M.; Reichardt, N.-C. Microarray-Based Identification of Lectins for the Purification of Human Urinary Extracellular Vesicles Directly from Urine Samples. Chembiochem 2014, 15, 1621–1626. [Google Scholar] [CrossRef] [PubMed]
- Wachalska, M.; Koppers-Lalic, D.; van Eijndhoven, M.; Pegtel, M.; Geldof, A.A.; Lipinska, A.D.; van Moorselaar, R.J.; Bijnsdorp, I.V. Protein Complexes in Urine Interfere with Extracellular Vesicle Biomarker Studies. J. Circ. Biomark. 2016, 5, 4. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, M.; Harada, Y.; Suzuki, T.; Fukushige, T.; Yamakuchi, M.; Kanekura, T.; Dohmae, N.; Hori, K.; Maruyama, I. Application of High-Mannose-Type Glycan-Specific Lectin from Oscillatoria Agardhii for Affinity Isolation of Tumor-Derived Extracellular Vesicles. Anal. Biochem. 2019, 580, 21–29. [Google Scholar] [CrossRef]
- Islam, M.K.; Syed, P.; Lehtinen, L.; Leivo, J.; Gidwani, K.; Wittfooth, S.; Pettersson, K.; Lamminmäki, U. A Nanoparticle-Based Approach for the Detection of Extracellular Vesicles. Sci. Rep. 2019, 9, 10038. [Google Scholar] [CrossRef]
- Herrero, C.; de la Fuente, A.; Casas-Arozamena, C.; Sebastian, V.; Prieto, M.; Arruebo, M.; Abalo, A.; Colás, E.; Moreno-Bueno, G.; Gil-Moreno, A.; et al. Extracellular Vesicles-Based Biomarkers Represent a Promising Liquid Biopsy in Endometrial Cancer. Cancers (Basel) 2019, 11, 2000. [Google Scholar] [CrossRef]
- Royo, F.; Cossío, U.; Ruiz de Angulo, A.; Llop, J.; Falcon-Perez, J.M. Modification of the Glycosylation of Extracellular Vesicles Alters Their Biodistribution in Mice. Nanoscale 2019, 11, 1531–1537. [Google Scholar] [CrossRef]
- Lv, J.; Wang, Z.; Li, F.; Zhang, Y.; Lu, H. Reverse Capture for Selectively and Sensitively Revealing the N-Glycome of Serum Exosomes. Chem. Commun. (Camb.) 2019, 55, 14339–14342. [Google Scholar] [CrossRef]
- Escrevente, C.; Keller, S.; Altevogt, P.; Costa, J. Interaction and Uptake of Exosomes by Ovarian Cancer Cells. BMC Cancer 2011, 11, 108. [Google Scholar] [CrossRef] [PubMed]
- Horrevorts, S.K.; Stolk, D.A.; van de Ven, R.; Hulst, M.; van Het Hof, B.; Duinkerken, S.; Heineke, M.H.; Ma, W.; Dusoswa, S.A.; Nieuwland, R.; et al. Glycan-Modified Apoptotic Melanoma-Derived Extracellular Vesicles as Antigen Source for Anti-Tumor Vaccination. Cancers (Basel) 2019, 11, 1266. [Google Scholar] [CrossRef]
- Netsirisawan, P.; Chokchaichamnankit, D.; Srisomsap, C.; Svasti, J.; Champattanachai, V. Proteomic Analysis Reveals Aberrant O-GlcNAcylation of Extracellular Proteins from Breast Cancer Cell Secretion. Cancer Genom. Proteom. 2015, 12, 201–209. [Google Scholar]
- Melo, S.A.; Luecke, L.B.; Kahlert, C.; Fernandez, A.F.; Gammon, S.T.; Kaye, J.; LeBleu, V.S.; Mittendorf, E.A.; Weitz, J.; Rahbari, N.; et al. Glypican-1 Identifies Cancer Exosomes and Detects Early Pancreatic Cancer. Nature 2015, 523, 177–182. [Google Scholar] [CrossRef] [PubMed]
- Sharma, P.; Ludwig, S.; Muller, L.; Hong, C.S.; Kirkwood, J.M.; Ferrone, S.; Whiteside, T.L. Immunoaffinity-Based Isolation of Melanoma Cell-Derived Exosomes from Plasma of Patients with Melanoma. J. Extracell. Vesicles 2018, 7, 1435138. [Google Scholar] [CrossRef] [PubMed]
- Mulcahy, L.A.; Pink, R.C.; Carter, D.R.F. Routes and Mechanisms of Extracellular Vesicle Uptake. J. Extracell. Vesicles 2014, 3. [Google Scholar] [CrossRef]
- Klaas, M.; Crocker, P.R. Sialoadhesin in Recognition of Self and Non-Self. Semin. Immunopathol. 2012, 34, 353–364. [Google Scholar] [CrossRef]
- Saunderson, S.C.; Dunn, A.C.; Crocker, P.R.; McLellan, A.D. CD169 Mediates the Capture of Exosomes in Spleen and Lymph Node. Blood 2014, 123, 208–216. [Google Scholar] [CrossRef]
- Ko, S.Y.; Naora, H. Extracellular Vesicle Membrane-Associated Proteins: Emerging Roles in Tumor Angiogenesis and Anti-Angiogenesis Therapy Resistance. Int. J. Mol. Sci. 2020, 21, 5418. [Google Scholar] [CrossRef]
- Liang, Y.; Eng, W.S.; Colquhoun, D.R.; Dinglasan, R.R.; Graham, D.R.; Mahal, L.K. Complex N-Linked Glycans Serve as a Determinant for Exosome/Microvesicle Cargo Recruitment. J. Biol. Chem. 2014, 289, 32526–32537. [Google Scholar] [CrossRef]
- Clark, D.J.; Schnaubelt, M.; Hoti, N.; Hu, Y.; Zhou, Y.; Gooya, M.; Zhang, H. Impact of Increased FUT8 Expression on the Extracellular Vesicle Proteome in Prostate Cancer Cells. J. Proteome Res. 2020, 19, 2195–2205. [Google Scholar] [CrossRef] [PubMed]
- Gao, T.; Wen, T.; Ge, Y.; Liu, J.; Yang, L.; Jiang, Y.; Dong, X.; Liu, H.; Yao, J.; An, G. Disruption of Core 1-Mediated O-Glycosylation Oppositely Regulates CD44 Expression in Human Colon Cancer Cells and Tumor-Derived Exosomes. Biochem. Biophys. Res. Commun. 2020, 521, 514–520. [Google Scholar] [CrossRef] [PubMed]
- Dall’Olio, F. The Sialyl-Alpha2,6-Lactosaminyl-Structure: Biosynthesis and Functional Role. Glycoconj. J. 2000, 17, 669–676. [Google Scholar] [CrossRef]
- Jung, Y.R.; Park, J.-J.; Jin, Y.B.; Cao, Y.J.; Park, M.-J.; Kim, E.J.; Lee, M. Silencing of ST6Gal I Enhances Colorectal Cancer Metastasis by Down-Regulating KAI1 via Exosome-Mediated Exportation and Thereby Rescues Integrin Signaling. Carcinogenesis 2016, 37, 1089–1097. [Google Scholar] [CrossRef] [PubMed]
- Zöller, M. Tetraspanins: Push and Pull in Suppressing and Promoting Metastasis. Nat. Rev. Cancer 2009, 9, 40–55. [Google Scholar] [CrossRef] [PubMed]
- Borkham-Kamphorst, E.; Van de Leur, E.; Meurer, S.K.; Buhl, E.M.; Weiskirchen, R. N-Glycosylation of Lipocalin 2 Is Not Required for Secretion or Exosome Targeting. Front. Pharmacol. 2018, 9, 426. [Google Scholar] [CrossRef]
- Yáñez-Mó, M.; Siljander, P.R.-M.; Andreu, Z.; Zavec, A.B.; Borràs, F.E.; Buzas, E.I.; Buzas, K.; Casal, E.; Cappello, F.; Carvalho, J.; et al. Biological Properties of Extracellular Vesicles and Their Physiological Functions. J. Extracell. Vesicles 2015, 4, 27066. [Google Scholar] [CrossRef] [PubMed]
- Xu, R.; Rai, A.; Chen, M.; Suwakulsiri, W.; Greening, D.W.; Simpson, R.J. Extracellular Vesicles in Cancer—Implications for Future Improvements in Cancer Care. Nat. Rev. Clin. Oncol. 2018, 15, 617–638. [Google Scholar] [CrossRef]
- Menck, K.; Scharf, C.; Bleckmann, A.; Dyck, L.; Rost, U.; Wenzel, D.; Dhople, V.M.; Siam, L.; Pukrop, T.; Binder, C.; et al. Tumor-Derived Microvesicles Mediate Human Breast Cancer Invasion through Differentially Glycosylated EMMPRIN. J. Mol. Cell. Biol. 2015, 7, 143–153. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Xu, J.; Li, L.; Ianni, A.; Kumari, P.; Liu, S.; Sun, P.; Braun, T.; Tan, X.; Xiang, R.; et al. MGAT3-Mediated Glycosylation of Tetraspanin CD82 at Asparagine 157 Suppresses Ovarian Cancer Metastasis by Inhibiting the Integrin Signaling Pathway. Theranostics 2020, 10, 6467–6482. [Google Scholar] [CrossRef]
- Zhang, Q.; Higginbotham, J.N.; Jeppesen, D.K.; Yang, Y.-P.; Li, W.; McKinley, E.T.; Graves-Deal, R.; Ping, J.; Britain, C.M.; Dorsett, K.A.; et al. Transfer of Functional Cargo in Exomeres. Cell Rep. 2019, 27, 940–954.e6. [Google Scholar] [CrossRef] [PubMed]
- Tang, K.; Zhang, Y.; Zhang, H.; Xu, P.; Liu, J.; Ma, J.; Lv, M.; Li, D.; Katirai, F.; Shen, G.-X.; et al. Delivery of Chemotherapeutic Drugs in Tumour Cell-Derived Microparticles. Nat. Commun. 2012, 3, 1282. [Google Scholar] [CrossRef] [PubMed]
- Sun, D.; Zhuang, X.; Xiang, X.; Liu, Y.; Zhang, S.; Liu, C.; Barnes, S.; Grizzle, W.; Miller, D.; Zhang, H.-G. A Novel Nanoparticle Drug Delivery System: The Anti-Inflammatory Activity of Curcumin Is Enhanced When Encapsulated in Exosomes. Mol. Ther. 2010, 18, 1606–1614. [Google Scholar] [CrossRef] [PubMed]
- Armstrong, J.P.K.; Holme, M.N.; Stevens, M.M. Re-Engineering Extracellular Vesicles as Smart Nanoscale Therapeutics. ACS Nano 2017, 11, 69–83. [Google Scholar] [CrossRef] [PubMed]
- Villata, S.; Canta, M.; Cauda, V. EVs and Bioengineering: From Cellular Products to Engineered Nanomachines. Int. J. Mol. Sci. 2020, 21, 6048. [Google Scholar] [CrossRef] [PubMed]
- Patel, J.M.; Vartabedian, V.F.; Bozeman, E.N.; Caoyonan, B.E.; Srivatsan, S.; Pack, C.D.; Dey, P.; D’Souza, M.J.; Yang, L.; Selvaraj, P. Plasma Membrane Vesicles Decorated with Glycolipid-Anchored Antigens and Adjuvants via Protein Transfer as an Antigen Delivery Platform for Inhibition of Tumor Growth. Biomaterials 2016, 74, 231–244. [Google Scholar] [CrossRef] [PubMed]
- Kooijmans, S.A.A.; Aleza, C.G.; Roffler, S.R.; van Solinge, W.W.; Vader, P.; Schiffelers, R.M. Display of GPI-Anchored Anti-EGFR Nanobodies on Extracellular Vesicles Promotes Tumour Cell Targeting. J. Extracell. Vesicles 2016, 5, 31053. [Google Scholar] [CrossRef] [PubMed]
- Nagarajan, S.; Selvaraj, P. Human Tumor Membrane Vesicles Modified to Express Glycolipid-Anchored IL-12 by Protein Transfer Induce T Cell Proliferation in Vitro: A Potential Approach for Local Delivery of Cytokines during Vaccination. Vaccine 2006, 24, 2264–2274. [Google Scholar] [CrossRef]
- Lee, T.S.; Kim, Y.; Zhang, W.; Song, I.H.; Tung, C.-H. Facile Metabolic Glycan Labeling Strategy for Exosome Tracking. Biochim. Biophys. Acta. Gen. Subj. 2018, 1862, 1091–1100. [Google Scholar] [CrossRef]
- Hammond, E.; Khurana, A.; Shridhar, V.; Dredge, K. The Role of Heparanase and Sulfatases in the Modification of Heparan Sulfate Proteoglycans within the Tumor Microenvironment and Opportunities for Novel Cancer Therapeutics. Front. Oncol. 2014, 4, 195. [Google Scholar] [CrossRef]
- Atai, N.A.; Balaj, L.; van Veen, H.; Breakefield, X.O.; Jarzyna, P.A.; Van Noorden, C.J.F.; Skog, J.; Maguire, C.A. Heparin Blocks Transfer of Extracellular Vesicles between Donor and Recipient Cells. J. Neurooncol. 2013, 115, 343–351. [Google Scholar] [CrossRef] [PubMed]
- Sento, S.; Sasabe, E.; Yamamoto, T. Application of a Persistent Heparin Treatment Inhibits the Malignant Potential of Oral Squamous Carcinoma Cells Induced by Tumor Cell-Derived Exosomes. PLoS ONE 2016, 11, e0148454. [Google Scholar] [CrossRef] [PubMed]
- Dredge, K.; Hammond, E.; Handley, P.; Gonda, T.J.; Smith, M.T.; Vincent, C.; Brandt, R.; Ferro, V.; Bytheway, I. PG545, a Dual Heparanase and Angiogenesis Inhibitor, Induces Potent Anti-Tumour and Anti-Metastatic Efficacy in Preclinical Models. Br. J. Cancer 2011, 104, 635–642. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Roy, S.; Cochran, E.; Zouaoui, R.; Chu, C.L.; Duffner, J.; Zhao, G.; Smith, S.; Galcheva-Gargova, Z.; Karlgren, J.; et al. M402, a Novel Heparan Sulfate Mimetic, Targets Multiple Pathways Implicated in Tumor Progression and Metastasis. PLoS ONE 2011, 6, e21106. [Google Scholar] [CrossRef] [PubMed]
- Ritchie, J.P.; Ramani, V.C.; Ren, Y.; Naggi, A.; Torri, G.; Casu, B.; Penco, S.; Pisano, C.; Carminati, P.; Tortoreto, M.; et al. SST0001, a Chemically Modified Heparin, Inhibits Myeloma Growth and Angiogenesis via Disruption of the Heparanase/Syndecan-1 Axis. Clin. Cancer Res. 2011, 17, 1382–1393. [Google Scholar] [CrossRef] [PubMed]
- Basche, M.; Gustafson, D.L.; Holden, S.N.; O’Bryant, C.L.; Gore, L.; Witta, S.; Schultz, M.K.; Morrow, M.; Levin, A.; Creese, B.R.; et al. A Phase I Biological and Pharmacologic Study of the Heparanase Inhibitor PI-88 in Patients with Advanced Solid Tumors. Clin. Cancer Res. 2006, 12, 5471–5480. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.-J.; Lee, P.-H.; Lin, D.-Y.; Wu, C.-C.; Jeng, L.-B.; Lin, P.-W.; Mok, K.-T.; Lee, W.-C.; Yeh, H.-Z.; Ho, M.-C.; et al. Heparanase Inhibitor PI-88 as Adjuvant Therapy for Hepatocellular Carcinoma after Curative Resection: A Randomized Phase II Trial for Safety and Optimal Dosage. J. Hepatol. 2009, 50, 958–968. [Google Scholar] [CrossRef]
- Naggi, A.; Casu, B.; Perez, M.; Torri, G.; Cassinelli, G.; Penco, S.; Pisano, C.; Giannini, G.; Ishai-Michaeli, R.; Vlodavsky, I. Modulation of the Heparanase-Inhibiting Activity of Heparin through Selective Desulfation, Graded N-Acetylation, and Glycol Splitting. J. Biol. Chem. 2005, 280, 12103–12113. [Google Scholar] [CrossRef]
- Pala, D.; Rivara, S.; Mor, M.; Milazzo, F.M.; Roscilli, G.; Pavoni, E.; Giannini, G. Kinetic Analysis and Molecular Modeling of the Inhibition Mechanism of Roneparstat (SST0001) on Human Heparanase. Glycobiology 2016, 26, 640–654. [Google Scholar] [CrossRef]
- Zhang, L.; Ngo, J.A.; Wetzel, M.D.; Marchetti, D. Heparanase Mediates a Novel Mechanism in Lapatinib-Resistant Brain Metastatic Breast Cancer. Neoplasia 2015, 17, 101–113. [Google Scholar] [CrossRef]
- Dusoswa, S.A.; Horrevorts, S.K.; Ambrosini, M.; Kalay, H.; Paauw, N.J.; Nieuwland, R.; Pegtel, M.D.; Würdinger, T.; Van Kooyk, Y.; Garcia-Vallejo, J.J. Glycan Modification of Glioblastoma-Derived Extracellular Vesicles Enhances Receptor-Mediated Targeting of Dendritic Cells. J. Extracell. Vesicles 2019, 8, 1648995. [Google Scholar] [CrossRef] [PubMed]
- Marleau, A.M.; Chen, C.-S.; Joyce, J.A.; Tullis, R.H. Exosome Removal as a Therapeutic Adjuvant in Cancer. J. Transl. Med. 2012, 10, 134. [Google Scholar] [CrossRef] [PubMed]
- Tullis, R.H.; Duffin, R.P.; Handley, H.H.; Sodhi, P.; Menon, J.; Joyce, J.A.; Kher, V. Reduction of Hepatitis C Virus Using Lectin Affinity Plasmapheresis in Dialysis Patients. Blood Purif. 2009, 27, 64–69. [Google Scholar] [CrossRef] [PubMed]
- Samuel, P.; Fabbri, M.; Carter, D.R.F. Mechanisms of Drug Resistance in Cancer: The Role of Extracellular Vesicles. Proteomics 2017, 17. [Google Scholar] [CrossRef] [PubMed]
- Ifergan, I.; Scheffer, G.L.; Assaraf, Y.G. Novel Extracellular Vesicles Mediate an ABCG2-Dependent Anticancer Drug Sequestration and Resistance. Cancer Res. 2005, 65, 10952–10958. [Google Scholar] [CrossRef] [PubMed]
- Shedden, K.; Xie, X.T.; Chandaroy, P.; Chang, Y.T.; Rosania, G.R. Expulsion of Small Molecules in Vesicles Shed by Cancer Cells: Association with Gene Expression and Chemosensitivity Profiles. Cancer Res. 2003, 63, 4331–4337. [Google Scholar] [PubMed]
- Muralidharan-Chari, V.; Kohan, H.G.; Asimakopoulos, A.G.; Sudha, T.; Sell, S.; Kannan, K.; Boroujerdi, M.; Davis, P.J.; Mousa, S.A. Microvesicle Removal of Anticancer Drugs Contributes to Drug Resistance in Human Pancreatic Cancer Cells. Oncotarget 2016, 7, 50365–50379. [Google Scholar] [CrossRef]
- Corcoran, C.; Rani, S.; O’Brien, K.; O’Neill, A.; Prencipe, M.; Sheikh, R.; Webb, G.; McDermott, R.; Watson, W.; Crown, J.; et al. Docetaxel-Resistance in Prostate Cancer: Evaluating Associated Phenotypic Changes and Potential for Resistance Transfer via Exosomes. PLoS ONE 2012, 7, e50999. [Google Scholar] [CrossRef]
- Zhang, F.; Zhu, Y.; Zhao, Q.; Yang, D.; Dong, Y.; Jiang, L.; Xing, W.; Li, X.; Xing, H.; Shi, M.; et al. Microvesicles Mediate Transfer of P-Glycoprotein to Paclitaxel-Sensitive A2780 Human Ovarian Cancer Cells, Conferring Paclitaxel-Resistance. Eur. J. Pharmacol. 2014, 738, 83–90. [Google Scholar] [CrossRef] [PubMed]
- Bebawy, M.; Combes, V.; Lee, E.; Jaiswal, R.; Gong, J.; Bonhoure, A.; Grau, G.E.R. Membrane Microparticles Mediate Transfer of P-Glycoprotein to Drug Sensitive Cancer Cells. Leukemia 2009, 23, 1643–1649. [Google Scholar] [CrossRef] [PubMed]
- Lv, M.; Zhu, X.; Chen, W.; Zhong, S.; Hu, Q.; Ma, T.; Zhang, J.; Chen, L.; Tang, J.; Zhao, J. Exosomes Mediate Drug Resistance Transfer in MCF-7 Breast Cancer Cells and a Probable Mechanism Is Delivery of P-Glycoprotein. Tumour Biol. 2014, 35, 10773–10779. [Google Scholar] [CrossRef] [PubMed]
- Yu, D.-D.; Wu, Y.; Zhang, X.-H.; Lv, M.-M.; Chen, W.-X.; Chen, X.; Yang, S.-J.; Shen, H.; Zhong, S.-L.; Tang, J.-H.; et al. Exosomes from Adriamycin-Resistant Breast Cancer Cells Transmit Drug Resistance Partly by Delivering MiR-222. Tumour Biol. 2016, 37, 3227–3235. [Google Scholar] [CrossRef] [PubMed]
- Mikamori, M.; Yamada, D.; Eguchi, H.; Hasegawa, S.; Kishimoto, T.; Tomimaru, Y.; Asaoka, T.; Noda, T.; Wada, H.; Kawamoto, K.; et al. MicroRNA-155 Controls Exosome Synthesis and Promotes Gemcitabine Resistance in Pancreatic Ductal Adenocarcinoma. Sci. Rep. 2017, 7, 42339. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Zhou, J.; Mei, S.; Wu, D.; Mu, Z.; Chen, B.; Xie, Y.; Ye, Y.; Liu, J. Circulating Exosomal MicroRNA-96 Promotes Cell Proliferation, Migration and Drug Resistance by Targeting LMO7. J. Cell. Mol. Med. 2017, 21, 1228–1236. [Google Scholar] [CrossRef] [PubMed]
- Mittal, S.; Gupta, P.; Chaluvally-Raghavan, P.; Pradeep, S. Emerging Role of Extracellular Vesicles in Immune Regulation and Cancer Progression. Cancers (Basel) 2020, 12, 3563. [Google Scholar] [CrossRef]
- Xie, F.; Zhou, X.; Fang, M.; Li, H.; Su, P.; Tu, Y.; Zhang, L.; Zhou, F. Extracellular Vesicles in Cancer Immune Microenvironment and Cancer Immunotherapy. Adv. Sci. (Weinh) 2019, 6. [Google Scholar] [CrossRef]
- Costa, J. Glycoconjugates from Extracellular Vesicles: Structures, Functions and Emerging Potential as Cancer Biomarkers. Biochim. Biophys. Acta Rev. Cancer 2017, 1868, 157–166. [Google Scholar] [CrossRef]
- Reis, C.A.; Osorio, H.; Silva, L.; Gomes, C.; David, L. Alterations in Glycosylation as Biomarkers for Cancer Detection. J. Clin. Pathol. 2010, 63, 322–329. [Google Scholar] [CrossRef]
- Chen, Z.; Liang, Q.; Zeng, H.; Zhao, Q.; Guo, Z.; Zhong, R.; Xie, M.; Cai, X.; Su, J.; He, Z.; et al. Exosomal CA125 as A Promising Biomarker for Ovarian Cancer Diagnosis. J. Cancer 2020, 11, 6445–6453. [Google Scholar] [CrossRef]
- Yokose, T.; Kabe, Y.; Matsuda, A.; Kitago, M.; Matsuda, S.; Hirai, M.; Nakagawa, T.; Masugi, Y.; Hishiki, T.; Nakamura, Y.; et al. O-Glycan-Altered Extracellular Vesicles: A Specific Serum Marker Elevated in Pancreatic Cancer. Cancers (Basel) 2020, 12, 2469. [Google Scholar] [CrossRef]
- Sancho-Albero, M.; Sebastián, V.; Sesé, J.; Pazo-Cid, R.; Mendoza, G.; Arruebo, M.; Martín-Duque, P.; Santamaría, J. Isolation of Exosomes from Whole Blood by a New Microfluidic Device: Proof of Concept Application in the Diagnosis and Monitoring of Pancreatic Cancer. J. Nanobiotechnol. 2020, 18, 150. [Google Scholar] [CrossRef] [PubMed]
- Sakaue, T.; Koga, H.; Iwamoto, H.; Nakamura, T.; Ikezono, Y.; Abe, M.; Wada, F.; Masuda, A.; Tanaka, T.; Fukahori, M.; et al. Glycosylation of Ascites-Derived Exosomal CD133: A Potential Prognostic Biomarker in Patients with Advanced Pancreatic Cancer. Med. Mol. Morphol. 2019, 52, 198–208. [Google Scholar] [CrossRef] [PubMed]
- Llorente, A.; Skotland, T.; Sylvänne, T.; Kauhanen, D.; Róg, T.; Orłowski, A.; Vattulainen, I.; Ekroos, K.; Sandvig, K. Molecular Lipidomics of Exosomes Released by PC-3 Prostate Cancer Cells. Biochim. Biophys. Acta 2013, 1831, 1302–1309. [Google Scholar] [CrossRef] [PubMed]
- Hoshino, A.; Kim, H.S.; Bojmar, L.; Gyan, K.E.; Cioffi, M.; Hernandez, J.; Zambirinis, C.P.; Rodrigues, G.; Molina, H.; Heissel, S.; et al. Extracellular Vesicle and Particle Biomarkers Define Multiple Human Cancers. Cell 2020, 182, 1044–1061.e18. [Google Scholar] [CrossRef] [PubMed]
- Menck, K.; Bleckmann, A.; Wachter, A.; Hennies, B.; Ries, L.; Schulz, M.; Balkenhol, M.; Pukrop, T.; Schatlo, B.; Rost, U.; et al. Characterisation of Tumour-Derived Microvesicles in Cancer Patients’ Blood and Correlation with Clinical Outcome. J. Extracell. Vesicles 2017, 6, 1340745. [Google Scholar] [CrossRef]
- Walker, S.A.; Aguilar Díaz De León, J.S.; Busatto, S.; Wurtz, G.A.; Zubair, A.C.; Borges, C.R.; Wolfram, J. Glycan Node Analysis of Plasma-Derived Extracellular Vesicles. Cells 2020, 9, 1946. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Sun, X.; Natalia, A.; Tang, C.S.L.; Ang, C.B.T.; Ong, C.-A.J.; Teo, M.C.C.; So, J.B.Y.; Shao, H. Dual-Selective Magnetic Analysis of Extracellular Vesicle Glycans. Matter 2020, 2, 150–166. [Google Scholar] [CrossRef]
- Etxebarria, J.; Reichardt, N.-C. Methods for the Absolute Quantification of N-Glycan Biomarkers. Biochim. Biophys. Acta 2016, 1860, 1676–1687. [Google Scholar] [CrossRef]
- Thobhani, S.; Yuen, C.-T.; Bailey, M.J.A.; Jones, C. Identification and Quantification of N-Linked Oligosaccharides Released from Glycoproteins: An Inter-Laboratory Study. Glycobiology 2009, 19, 201–211. [Google Scholar] [CrossRef]
- Schjoldager, K.T.; Narimatsu, Y.; Joshi, H.J.; Clausen, H. Global View of Human Protein Glycosylation Pathways and Functions. Nat. Rev. Mol. Cell. Biol. 2020. [Google Scholar] [CrossRef]
- Soekmadji, C.; Li, B.; Huang, Y.; Wang, H.; An, T.; Liu, C.; Pan, W.; Chen, J.; Cheung, L.; Falcon-Perez, J.M.; et al. The Future of Extracellular Vesicles as Theranostics—An ISEV Meeting Report. J. Extracell. Vesicles 2020, 9, 1809766. [Google Scholar] [CrossRef] [PubMed]

| Glycan Structure | Cancer Type | Sample | Relevance/Potential Impact | References |
|---|---|---|---|---|
| GalNAcα-Thr/Ser (Tn antigen) | Cervical | Cell lines | EV biomarker potential | [87] |
| Neu5Acα(2,6)-GalNAcα-Thr/Ser (STn antigen) | Gastric, cervical | Cell lines | EV biomarker potential | [86,87] |
| Gal-β(1,3)-GalNAcα-Thr/Ser (T antigen) | Ovarian, cervical | Cell lines | EV detection EV biomarker potential | [72,87] |
| Sialylation (α(2,6)- or α(2,3)-linked) | Hepatic, melanoma, cervical, pancreatic, ovarian, colorectal, glioma, breast, gastric | Cell lines, human serum, healthy individuals’ urine | EV detection and capture EV biomarker potential EV uptake EV biodistribution | [5,6,8,70,71,72,84,86,87,89,91,111,122,123,131,144,148,150,151,152] |
| Core fucosylated N-glycans | Hepatocellular, breast, glioma, gastric melanoma | Cell lines, human serum | EV detection EV biomarker potential | [71,84,86,87,91,151] |
| Terminal fucosylation | Melanoma, breast | Cell lines | EV detection | [5,91] |
| Complex N-glycans | Melanoma, colorectal, hepatocellular | Cell lines, human serum, healthy individuals’ urine | EV detection EV biomarker potential | [6,70,84,89,151] |
| Branched N-glycans | Pancreatic, melanoma, breast, ovarian, gastric | Cell lines | EV detection EV biomarker potential | [5,84,86,91,111,123,131] |
| Bisected N-glycans | Melanoma, pancreatic, ovarian, gastric | Cell lines, healthy individuals’ urine | EV detection EV biomarker potential | [5,70,72,86,91,123] |
| High mannose N-glycans | Melanoma, glioblastoma, lung, colorectal, ovarian, hepatocellular | Cell lines, human serum, healthy individuals’ urine | EV detection and capture EV therapy potential EV biomarker potential EV uptake | [6,70,72,89,111,123,144,151,153] |
| Polylactosamine | Colorectal, melanoma | Cell lines | EV detection | [6] |
| O-GlcNAc | Breast, colorectal | Cell lines | EV biomarker potential | [88,154] |
| GAGs | Endometrial, ovarian, breast | Cell lines | EV capture | [149] |
| Proteoglycans | Pancreatic, melanoma, breast | Cell lines, human and mice serum, human plasma | EV capture EV biomarker potential | [155,156] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Martins, Á.M.; Ramos, C.C.; Freitas, D.; Reis, C.A. Glycosylation of Cancer Extracellular Vesicles: Capture Strategies, Functional Roles and Potential Clinical Applications. Cells 2021, 10, 109. https://doi.org/10.3390/cells10010109
Martins ÁM, Ramos CC, Freitas D, Reis CA. Glycosylation of Cancer Extracellular Vesicles: Capture Strategies, Functional Roles and Potential Clinical Applications. Cells. 2021; 10(1):109. https://doi.org/10.3390/cells10010109
Chicago/Turabian StyleMartins, Álvaro M., Cátia C. Ramos, Daniela Freitas, and Celso A. Reis. 2021. "Glycosylation of Cancer Extracellular Vesicles: Capture Strategies, Functional Roles and Potential Clinical Applications" Cells 10, no. 1: 109. https://doi.org/10.3390/cells10010109
APA StyleMartins, Á. M., Ramos, C. C., Freitas, D., & Reis, C. A. (2021). Glycosylation of Cancer Extracellular Vesicles: Capture Strategies, Functional Roles and Potential Clinical Applications. Cells, 10(1), 109. https://doi.org/10.3390/cells10010109







