Mimicking Tumor Hypoxia in Non-Small Cell Lung Cancer Employing Three-Dimensional In Vitro Models
Abstract
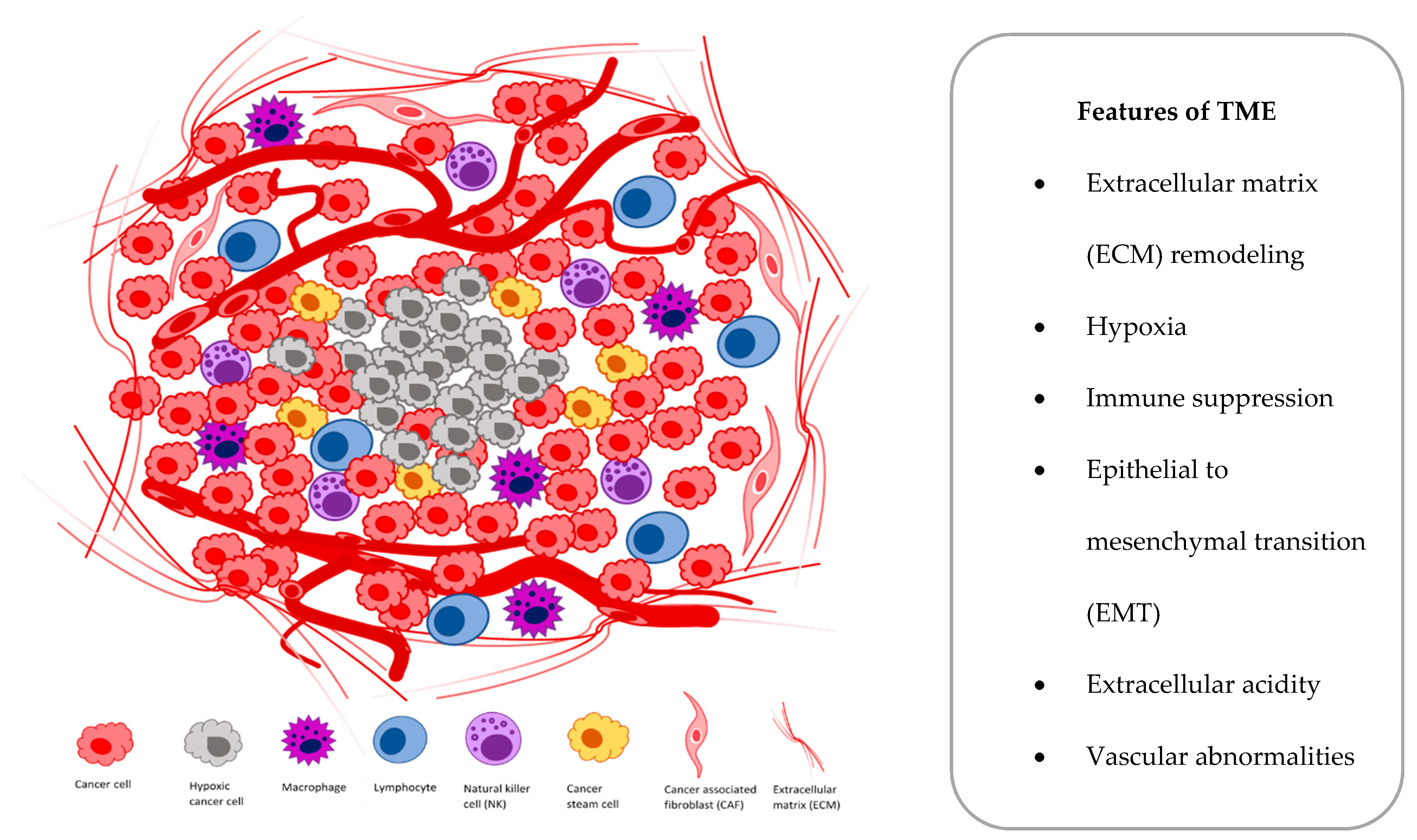
:1. Introduction
2. Lung Tumor Hypoxia
2.1. Biology
2.2. Consequences at Cellular and Molecular Level
3. Role of HIFs in Lung Cancer
3.1. In Vitro Studies
3.2. In Vivo Studies
4. Hypoxia Assessment in NSCLC
4.1. Oxygen Electrodes
4.2. Imaging Hypoxia
4.3. Detection of Hypoxia-Induced Markers
5. Clinical Implications
6. 2D Model Limitations in Studying Hypoxia Biology
7. Multicellular Lung Tumor Spheroids
8. Organoids
9. D Scaffolds and Hydrogels
10. Microfluidic Devices
11. 3D Bioprinting
12. Fluorescence Imaging of 3D Lung Cancer Models
13. Conclusions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, T.-Y.D.; Cramb, S.M.; Baade, P.D.; Youlden, D.R.; Nwogu, C.; Reid, M.E. The International Epidemiology of Lung Cancer: Latest Trends, Disparities, and Tumor Characteristics. J. Thorac. Oncol. 2016, 11, 1653–1671. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Molina, J.R.; Yang, P.; Cassivi, S.D.; Schild, S.E.; Adjei, A.A. Non-small cell lung cancer: Epidemiology, risk factors, treatment, and survivorship. Mayo Clin. Proc. 2008, 83, 584–594. [Google Scholar] [CrossRef]
- Travis, W.D.; Brambilla, E.; Noguchi, M.; Nicholson, A.G.; Geisinger, K.; Yatabe, Y.; Powell, C.A.; Beer, D.; Riely, G.; Garg, K.; et al. International Association for the Study of Lung Cancer/American Thoracic Society/European Respiratory Society: International Multidisciplinary Classification of Lung Adenocarcinoma: Executive Summary. Proc. Am. Thorac. Soc. 2011, 8, 381–385. [Google Scholar] [CrossRef]
- Jing, X.; Yang, F.; Shao, C.; Wei, K.; Xie, M.; Shen, H.; Shu, Y. Role of hypoxia in cancer therapy by regulating the tumor microenvironment. Mol. Cancer 2019, 18, 157. [Google Scholar] [CrossRef] [Green Version]
- Marhuenda, E.; Campillo, N.; Gabasa, M.; Martínez-García, M.A.; Campos-Rodríguez, F.; Gozal, D.; Navajas, D.; Al-caraz, J.; Farré, R.; Almendros, I. Effects of Sustained and Intermittent Hypoxia on Human Lung Cancer Cells. Am. J. Respir. Cell Mol. Biol. 2019, 61, 540–544. [Google Scholar] [CrossRef]
- Yu, B.; Shah, A.; Wang, B.; Rajaram, N.; Wang, Q.; Ramanujam, N.; Palmer, G.M.; Dewhirst, M.W. Measuring tumor cy-cling hypoxia and angiogenesis using a side-firing fiber optic probe. J. Biophotonics 2012, 7, 552–564. [Google Scholar] [CrossRef]
- McKeown, S.R. Defining normoxia, physoxia and hypoxia in tumours—implications for treatment response. Br. J. Radiol. 2014, 87, 20130676. [Google Scholar] [CrossRef] [Green Version]
- Le, Q.-T.; Chen, E.; Salim, A.; Cao, H.; Kong, C.S.; Whyte, R.; Donington, J.; Cannon, W.A.; Wakelee, H.; Tibshirani, R.; et al. An Evaluation of Tumor Oxygenation and Gene Expression in Patients with Early Stage Non-Small Cell Lung Cancers. Clin. Cancer Res. 2006, 12, 1507–1514. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Song, X.; Wang, X.; Wei, L.; Liu, X.; Yuan, S.; Lv, L. Effect of chronic intermittent hypoxia on biological behavior and hypoxia-associated gene expression in lung cancer cells. J. Cell. Biochem. 2010, 111, 554–563. [Google Scholar] [CrossRef]
- Salem, A.; Asselin, M.-C.; Reymen, B.; Jackson, A.; Lambin, P.; West, C.M.L.; O’Connor, J.P.; Faivre-Finn, C. Targeting Hypoxia to Improve Non-Small Cell Lung Cancer Outcome. J. Natl. Cancer Inst. 2017, 110, 14–30. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cooper, C.; Liu, G.-Y.; Niu, Y.-L.; Santos, S.; Murphy, L.C.; Watson, P.H. Intermittent Hypoxia Induces Pro-teasome-Dependent Down-Regulation of Estrogen Receptor α in Human Breast Carcinoma. Clin. Cancer Res. 2004, 10, 8720–8727. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vaupel, P.; Harrison, L. Tumor Hypoxia: Causative Factors, Compensatory Mechanisms, and Cellular Response. Oncologist 2004, 9, 4–9. [Google Scholar] [CrossRef] [Green Version]
- Williams, K.J.; Cowen, L.; Stratford, I.J. Hypoxia and oxidative stress. Tumour hypoxia--therapeutic considera-tions. Breast Cancer Res. 2001, 3, 328–331. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Al Tameemi, W.; Dale, T.P.; Al-Jumaily, R.M.K.; Forsyth, N. Hypoxia-Modified Cancer Cell Metabolism. Front. Cell Dev. Biol. 2019, 7, 4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Berchner-Pfannschmidt, U.; Frede, S.; Wotzlaw, C.; Fandrey, J. Imaging of the hypoxia-inducible factor pathway: Insights into oxygen sensing. Eur. Respir. J. 2008, 32, 210–217. [Google Scholar] [CrossRef] [PubMed]
- Mole, D.R.; Blancher, C.; Copley, R.R.; Pollard, P.J.; Gleadle, J.M.; Ragoussis, J.; Ratcliffe, P.J. Genome-wide Association of Hypoxia-inducible Factor (HIF)-1α and HIF-2α DNA Binding with Expression Profiling of Hypoxia-inducible Tran-scripts. J. Biol. Chem. 2009, 284, 16767–16775. [Google Scholar] [CrossRef] [Green Version]
- Wang, G.L.; Jiang, B.H.; Rue, E.A.; Semenza, G.L. Hypoxia-inducible factor 1 is a basic-helix-loop-helix-PAS heterodimer regulated by cellular O2 tension. Proc. Natl. Acad. Sci. USA 1995, 92, 5510–5514. [Google Scholar] [CrossRef] [Green Version]
- Semenza, G.L. Hypoxia-inducible factor 1: Master regulator of O2 homeostasis. Curr. Opin. Genet. Dev. 1998, 8, 588–594. [Google Scholar] [CrossRef]
- Pientka, F.K.; Hu, J.; Schindler, S.G.; Brix, B.; Thiel, A.; Jöhren, O.; Fandrey, J.; Berchner-Pfannschmidt, U.; Depping, R. Oxygen sensing by the prolyl-4-hydroxylase PHD2 within the nuclear compartment and the influence of compartmental-isation on HIF-1 signalling. J. Cell Sci. 2012, 125, 5168–5176. [Google Scholar] [CrossRef] [Green Version]
- Metzen, E.; Ratcliffe, P.J.; Ratcliffe, E.M.P.J. HIF hydroxylation and cellular oxygen sensing. Biol. Chem. 2004, 385, 223–230. [Google Scholar] [CrossRef] [PubMed]
- Kaelin, W.G., Jr.; Ratcliffe, P.J.; Semenza, G.L. Pathways for Oxygen Regulation and Homeostasis: The 2016 Albert Lasker Basic Medical Research Award. JAMA 2016, 316, 1252–1253. [Google Scholar] [CrossRef] [PubMed]
- Semenza, G.L. Oxygen homeostasis. Wiley Interdiscip. Rev. Syst. Biol. Med. 2010, 2, 336–361. [Google Scholar] [CrossRef] [PubMed]
- Wenger, R.H.; Gassmann, M. Oxygen(es) and the hypoxia-inducible factor-1. Biol. Chem. 1997, 378, 609–616. [Google Scholar]
- Liu, W.; Shen, S.-M.; Zhao, X.-Y.; Chen, G.-Q. Targeted genes and interacting proteins of hypoxia inducible factor-1. Int. J. Biochem. Mol. Boil. 2012, 3, 165–178. [Google Scholar]
- Wang, G.L.; Semenza, G.L. Purification and Characterization of Hypoxia-inducible Factor 1. J. Biol. Chem. 1995, 270, 1230–1237. [Google Scholar] [CrossRef] [Green Version]
- Ye, I.C.; Fertig, E.J.; Digiacomo, J.W.; Considine, M.; Godet, I.; Gilkes, D.M. Molecular Portrait of Hypoxia in Breast Cancer: A Prognostic Signature and Novel HIF-Regulated Genes. Mol. Cancer Res. 2018, 16, 1889–1901. [Google Scholar] [CrossRef] [Green Version]
- Semenza, G.L. Targeting HIF-1 for cancer therapy. Nat. Rev. Cancer 2003, 3, 721–732. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef] [Green Version]
- Shimoda, L.A.; Semenza, G.L. HIF and the lung: Role of hypoxia-inducible factors in pulmonary development and disease. Am. J. Respir Crit Care Med. 2011, 183, 152–156. [Google Scholar] [CrossRef]
- Giatromanolaki, A.I.; Koukourakis, M.; Sivridis, E.; Turley, H.; Talks, K.; Pezzella, F.; Gatter, K.C.; Harris, A.L. Relation of hypoxia inducible factor 1α and 2α in operable non-small cell lung cancer to angiogenic/molecular profile of tumours and survival. Br. J. Cancer 2001, 85, 881–890. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, X.-H.; Qian, C.; Yuan, K. Correlations of hypoxia-inducible factor-1?/hypoxia-inducible factor-2? expression with angiogenesis factors expression and prognosis in non-small cell lung cancer. Chin. Med. J. 2011, 124, 11–18. [Google Scholar] [PubMed]
- Thul, P.; Stenström, L.; Wiking, M.; Mahdessian, D.; Geladaki, A.; Blal, H.A.; Alm, T.; Asplund, A.; Björk, L.; Breckels, L.M.; et al. A subcellular map of the human proteome. Science 2017, 356, eaal3321. [Google Scholar] [CrossRef]
- Uhlén, M.; Zhang, C.; Lee, S.; Sjöstedt, E.; Fagerberg, L.; Bidkhori, G.; Benfeitas, R.; Arif, M.; Liu, Z.; Edfors, F.; et al. A pathology atlas of the human cancer transcriptome. Science 2017, 357, eaan2507. [Google Scholar] [CrossRef] [Green Version]
- Koshikawa, N.; Iyozumi, A.; Gassmann, M.; Takenaga, K. Constitutive upregulation of hypoxia-inducible factor-1α mRNA occurring in highly metastatic lung carcinoma cells leads to vascular endothelial growth factor overexpression upon hypoxic exposure. Oncogene 2003, 22, 6717–6724. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jackson, A.L.; Zhou, B.; Kim, W.Y. HIF, hypoxia and the role of angiogenesis in non-small cell lung cancer. Expert Opin. Ther. Targets 2010, 14, 1047–1057. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roig, E.M.; Yaromina, A.; Houben, R.; Groot, A.J.; Dubois, L.; Vooijs, M.A.G.G. Prognostic Role of Hypoxia-Inducible Factor-2α Tumor Cell Expression in Cancer Patients: A Meta-Analysis. Front. Oncol. 2018, 8, 224. [Google Scholar] [CrossRef]
- Gao, Z.-J.; Wang, Y.; Yuan, W.-D.; Yuan, J.-Q.; Yuan, K. HIF-2α not HIF-1α overexpression confers poor prognosis in non–small cell lung cancer. Tumor Biol. 2017, 39, 1010428317709637. [Google Scholar] [CrossRef] [Green Version]
- Sun, J.; He, F.; Yi, W.; Wan, M.; Li, R.; Wei, X.; Wu, R.; Niu, D. High expression of HIF-2α and its anti-radiotherapy effect in lung cancer stem cells. Genet. Mol. Res. 2015, 14, 18110–18120. [Google Scholar] [CrossRef]
- Bertout, J.A.; Majmundar, A.J.; Gordan, J.D.; Lam, J.C.; Ditsworth, D.; Keith, B.; Brown, E.J.; Nathanson, K.L.; Simon, M.C. HIF2 inhibition promotes p53 pathway activity, tumor cell death, and radiation responses. Proc. Natl. Acad. Sci. USA 2009, 106, 14391–14396. [Google Scholar] [CrossRef] [Green Version]
- Roig, E.M.; Groot, A.J.; Yaromina, A.; Hendrickx, T.C.; Barbeau, L.M.; Giuranno, L.; Dams, G.; Ient, J.; Pimentel, V.O.; Van Gisbergen, M.; et al. HIF-1α and HIF-2α Differently Regulate the Radiation Sensitivity of NSCLC Cells. Cells 2019, 8, 45. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, A.Y.; Shimoda, L.A.; Iyer, N.V.; Huso, D.L.; Sun, X.; McWilliams, R.; Beaty, T.; Sham, J.S.K.; Wiener, C.M.; Sylvester, J.T.; et al. Impaired physiological responses to chronic hypoxia in mice partially deficient for hypoxia-inducible factor 1α. J. Clin. Investig. 1999, 103, 691–696. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Bosch-Marce, M.; Shimoda, L.A.; Tan, Y.S.; Baek, J.H.; Wesley, J.B.; Gonzalez, F.J.; Semenza, G.L. Mitochon-drial Autophagy Is an HIF-1-dependent Adaptive Metabolic Response to Hypoxia. J. Biol. Chem. 2008, 283, 10892–10903. [Google Scholar] [CrossRef] [Green Version]
- Franovic, A.; Holterman, C.E.; Payette, J.; Lee, S. Human cancers converge at the HIF-2α oncogenic axis. Proc. Natl. Acad. Sci. USA 2009, 106, 21306–21311. [Google Scholar] [CrossRef] [Green Version]
- Liao, D.; Corle, C.; Seagroves, T.N.; Johnson, R.S. Hypoxia-Inducible Factor-1α Is a Key Regulator of Metastasis in a Transgenic Model of Cancer Initiation and Progression. Cancer Res. 2007, 67, 563–572. [Google Scholar] [CrossRef] [Green Version]
- Jacoby, J.J.; Erez, B.; Korshunova, M.V.; Williams, R.R.; Furutani, K.; Takahashi, O.; Kirkpatrick, L.; Lippman, S.M.; Powis, G.; Oʼreilly, M.S.; et al. Treatment with HIF-1α Antagonist PX-478 Inhibits Progression and Spread of Orthotopic Human Small Cell Lung Cancer and Lung Adenocarcinoma in Mice. J. Thorac. Oncol. 2010, 5, 940–949. [Google Scholar] [CrossRef] [Green Version]
- Han, J.-Y.; Oh, S.H.; Morgillo, F.; Myers, J.N.; Kim, E.; Hong, W.K.; Lee, H.-Y. Hypoxia-inducible Factor 1α and Antian-giogenic Activity of Farnesyltransferase Inhibitor SCH66336 in Human Aerodigestive Tract Cancer. J. Natl. Cancer Inst. 2005, 97, 1272–1286. [Google Scholar] [CrossRef]
- Kim, W.Y.; Perera, S.; Zhou, B.; Carretero, J.; Yeh, J.J.; Heathcote, S.A.; Jackson, A.L.; Nikolinakos, P.; Ospina, B.; Naumov, G.; et al. HIF2alpha cooperates with RAS to promote lung tumorigenesis in mice. J. Clin. Invest. 2009, 119, 2160–2170. [Google Scholar] [CrossRef] [Green Version]
- Mazumdar, J.; Hickey, M.M.; Pant, D.K.; Durham, A.C.; Sweet-Cordero, A.; Vachani, A.; Jacks, T.E.; Chodosh, L.A.; Kis-sil, J.L.; Simon, M.C.; et al. HIF-2 deletion promotes Kras-driven lung tumor development. Proc. Natl. Acad. Sci. USA 2010, 107, 14182–14187. [Google Scholar] [CrossRef] [Green Version]
- Chen, W.; Hill, H.; Christie, A.; Kim, M.S.; Holloman, E.; Pavia-Jimenez, A.; Homayoun, F.; Ma, Y.; Patel, N.; Yell, P.; et al. Targeting renal cell carcinoma with a HIF-2 antagonist. Nat. Cell Biol. 2016, 539, 112–117. [Google Scholar] [CrossRef] [Green Version]
- Ellinghaus, P.; Heisler, I.; Unterschemmann, K.; Haerter, M.; Beck, H.; Greschat, S.; Ehrmann, A.; Summer, H.; Flamme, I.; Oehme, F.; et al. BAY 87-2243, a highly potent and selective inhibitor of hypoxia-induced gene activation has anti-tumor activities by inhibition of mitochondrial complex I. Cancer Med. 2013, 2, 611–624. [Google Scholar] [CrossRef] [PubMed]
- Graves, E.E.; Maity, A.; Le, Q.-T. The Tumor Microenvironment in Non-Small-Cell Lung Cancer. Semin. Radiat. Oncol. 2010, 20, 156–163. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Walsh, J.C.; Lebedev, A.; Aten, E.; Madsen, K.; Marciano, L.; Kolb, H.C. The Clinical Importance of Assessing Tumor Hypoxia: Relationship of Tumor Hypoxia to Prognosis and Therapeutic Opportunities. Antioxid. Redox Signal. 2014, 21, 1516–1554. [Google Scholar] [CrossRef]
- Challapalli, A.; Carroll, L.; Aboagye, E.O. Molecular mechanisms of hypoxia in cancer. Clin. Transl. Imaging 2017, 5, 225–253. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Even, A.J.; Reymen, B.; La Fontaine, M.D.; Das, M.; Jochems, A.; Mottaghy, F.M.; Belderbos, J.S.A.; De Ruysscher, D.; Lambin, P.; Van Elmpt, W. Predicting tumor hypoxia in non-small cell lung cancer by combining CT, FDG PET and dy-namic contrast-enhanced CT. Acta Oncol. 2017, 56, 1591–1596. [Google Scholar] [CrossRef] [Green Version]
- Peeters, S.G.J.A.; Zegers, C.M.L.; Lieuwes, N.G.; Van Elmpt, W.; Eriksson, J.; Van Dongen, G.A.; Dubois, L.; Lambin, P. A Comparative Study of the Hypoxia PET Tracers [18F]HX4, [18F]FAZA, and [18F]FMISO in a Preclinical Tumor Model. Int. J. Radiat. Oncol. 2015, 91, 351–359. [Google Scholar] [CrossRef] [Green Version]
- Bollineni, V.R.; Kerner, G.S.M.A.; Pruim, J.; Steenbakkers, R.J.H.M.; Wiegman, E.M.; Koole, M.J.; De Groot, E.H.; Willem-sen, A.T.; Luurtsema, G.; Widder, J.; et al. PET Imaging of Tumor Hypoxia Using 18F-Fluoroazomycin Arabinoside in Stage III-IV Non-Small Cell Lung Cancer Patients. J. Nucl. Med. 2013, 54, 1175–1180. [Google Scholar] [CrossRef] [Green Version]
- Hu, M.; Xing, L.; Mu, D.; Yang, W.; Yang, G.; Kong, L.; Yu, J. Hypoxia Imaging With 18F-Fluoroerythronitroimidazole Integrated PET/CT and Immunohistochemical Studies in Non-Small Cell Lung Cancer. Clin. Nucl. Med. 2013, 38, 591–596. [Google Scholar] [CrossRef]
- Dehdashti, F.; Mintun, M.A.; Lewis, J.S.; Bradley, J.; Govindan, R.; Laforest, R.; Welch, M.J.; Siegel, B.A. In vivo assess-ment of tumor hypoxia in lung cancer with 60Cu-ATSM. Eur. J. Nucl. Med. Mol. Imaging 2003, 30, 844–850. [Google Scholar] [CrossRef]
- Raleigh, J.A.; Chou, S.-C.; Arteel, G.E.; Horsman, M.R. Comparisons among Pimonidazole Binding, Oxygen Electrode Measurements, and Radiation Response in C3H Mouse Tumors. Radiat. Res. 1999, 151, 580–589. [Google Scholar] [CrossRef]
- Mandeville, H.; Ng, Q.S.; Daley, F.M.; Barber, P.R.; Pierce, G.; Finch, J.; Burke, M.; Bell, A.; Townsend, E.R.; Kozarski, R.; et al. Operable Non-Small Cell Lung Cancer: Correlation of Volumetric Helical Dynamic Contrast-enhanced CT Param-eters with Immunohistochemical Markers of Tumor Hypoxia. Radiology 2012, 264, 581–589. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koukourakis, M.I.; Giatromanolaki, A.; Sivridis, E.; Bougioukas, G.; Didilis, V.; Gatter, K.C.; Harris, A.L.; Tumour and Angiogenesis Research Group. Lactate dehydrogenase-5 (LDH-5) overexpression in non-small-cell lung cancer tis-sues is linked to tumour hypoxia, angiogenic factor production and poor prognosis. Br. J. Cancer 2003, 89, 877–885. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.J.; Rabbani, Z.N.; Vollmer, R.T.; Schreiber, E.-G.; Oosterwijk, E.; Dewhirst, M.W.; Vujaskovic, Z.; Kelley, M.J. Carbonic Anhydrase IX in Early-Stage Non-Small Cell Lung Cancer. Clin. Cancer Res. 2004, 10, 7925–7933. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Giatromanolaki, A.; Koukourakis, M.I.; Sowter, H.M.; Sivridis, E.; Gibson, S.; Gatter, K.C.; Harris, A.L. BNIP3 expression is linked with hypoxia-regulated protein expression and with poor prognosis in non-small cell lung cancer. Clin. Cancer Res. 2004, 10, 5566–5571. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Andersen, S.; Donnem, T.; Al-Saad, S.; Al-Shibli, K.; Busund, L.-T.; Bremnes, R.M. Angiogenic Markers Show High Prognostic Impact on Survival in Marginally Operable Non-Small Cell Lung Cancer Patients Treated with Adjuvant Ra-diotherapy. J. Thorac. Oncol. 2009, 4, 463–471. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Giatromanolaki, A.; Koukourakis, M.I.; Sivridis, E.; Pastorek, J.; Wykoff, C.C.; Gatter, K.C.; Harris, A.L. Expression of hypoxia-inducible carbonic anhydrase-9 relates to angiogenic pathways and independently to poor outcome in non-small cell lung cancer. Cancer Res. 2001, 61, 7992–7998. [Google Scholar] [PubMed]
- Dagnon, K.; Pacary, E.; Commo, F.; Antoine, M.; Bernaudin, M.; Bernaudin, J.-F.; Callard, P. Expression of erythropoietin and erythropoietin receptor in non-small cell lung carcinomas. Clin. Cancer Res. 2005, 11, 993–999. [Google Scholar]
- Cui, W.; Wu, F.; Ma, L. Hypoxia associated biomarkers in lung cancer—An update. Eur. Rev. Med. Pharmacol. Sci. 2017, 21, 43–46. [Google Scholar]
- Ostheimer, C.; Bache, M.; Güttler, A.; Kotzsch, M.; Vordermark, D. A pilot study on potential plasma hypoxia markers in the radiotherapy of non-small cell lung cancer. Strahlenther. und Onkol. 2013, 190, 276–282. [Google Scholar] [CrossRef]
- Shi, Y.; Fan, S.; Wu, M.; Zuo, Z.; Li, X.; Jiang, L.; Shen, Q.; Xu, P.; Zeng, L.; Zhou, Y.; et al. YTHDF1 links hypoxia adap-tation and non-small cell lung cancer progression. Nat. Commun. 2019, 10, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Grosso, S.; Doyen, J.; Parks, S.K.; Bertero, T.; Paye, A.; Cardinaud, B.; Gounon, P.; Lacas-Gervais, S.; Noël, A.; Pouys-ségur, J.; et al. MiR-210 promotes a hypoxic phenotype and increases radioresistance in human lung cancer cell lines. Cell Death Dis. 2013, 4, e544. [Google Scholar] [CrossRef] [PubMed]
- Osugi, J.; Kimura, Y.; Owada, Y.; Inoue, T.; Watanabe, Y.; Yamaura, T.; Fukuhara, M.; Muto, S.; Okabe, N.; Matsumura, Y.; et al. Prognostic Impact of Hypoxia-Inducible miRNA-210 in Patients with Lung Adenocarcinoma. J. Oncol. 2015, 2015, 316745–316748. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.-H.; Zhang, H.; Yang, Z.-G.; Wen, G.-Q.; Cui, Y.-B.; Shao, G.-G. Prognostic significance of serum microRNA-210 levels in nonsmall-cell lung cancer. J. Int. Med. Res. 2013, 41, 1437–1444. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Onnis, B.; Rapisarda, A.; Melillo, G. Development of HIF-1 inhibitors for cancer therapy. J. Cell. Mol. Med. 2009, 13, 2780–2786. [Google Scholar] [CrossRef]
- Aggarwal, V.; Miranda, O.; Johnston, P.A.; Sant, S. Three dimensional engineered models to study hypoxia biology in breast cancer. Cancer Lett. 2020, 490, 124–142. [Google Scholar] [CrossRef]
- Takagi, A.; Watanabe, M.; Ishii, Y.; Morita, J.; Hirokawa, Y.; Matsuzaki, T.; Shiraishi, T. Three-dimensional cellular sphe-roid formation provides human prostate tumor cells with tissue-like features. Anticancer. Res. 2007, 27, 45–53. [Google Scholar]
- Nath, S.; Devi, G.R. Three-dimensional culture systems in cancer research: Focus on tumor spheroid model. Pharmacol. Ther. 2016, 163, 94–108. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arai, K.; Eguchi, T.; Rahman, M.M.; Sakamoto, R.; Masuda, N.; Nakatsura, T.; Calderwood, S.K.; Kozaki, K.-I.; Itoh, M. A Novel High-Throughput 3D Screening System for EMT Inhibitors: A Pilot Screening Discovered the EMT Inhibitory Activity of CDK2 Inhibitor SU9516. PLoS ONE 2016, 11, e0162394. [Google Scholar] [CrossRef]
- Amann, A.; Zwierzina, M.; Koeck, S.; Gamerith, G.; Pechriggl, E.; Huber, J.M.; Lorenz, E.; Kelm, J.M.; Hilbe, W.; Zwier-zina, H.; et al. Development of a 3D angiogenesis model to study tumour-endothelial cell interactions and the effects of anti-angiogenic drugs. Sci. Rep. 2017, 7, 2963. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.-Q.; Kiefl, R.; Roskopf, C.; Tian, F.; Huber, R.M. Interactions among Lung Cancer Cells, Fibroblasts, and Macro-phages in 3D Co-Cultures and the Impact on MMP-1 and VEGF Expression. PLoS ONE 2016, 11, e0156268. [Google Scholar] [CrossRef]
- Zhang, Z.; Wang, H.; Ding, Q.; Xing, Y.; Xu, Z.; Lu, C.; Luo, D.; Xu, L.; Xia, W.; Zhou, C.; et al. Establishment of patient-derived tumor spheroids for non-small cell lung cancer. PLoS ONE 2018, 13, e0194016. [Google Scholar] [CrossRef] [PubMed]
- Sachs, N.; Papaspyropoulos, A.; Ommen, D.D.Z.; Heo, I.; Böttinger, L.; Klay, D.; Weeber, F.; Huelsz-Prince, G.; Iakoba-chvili, N.; Amatngalim, G.D.; et al. Long-term expanding human airway organoids for disease modeling. EMBO J. 2019, 38. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, H.; Sugano, M.; Miyashita, T.; Hashimoto, H.; Ochiai, A.; Suzuki, K.; Tsuboi, M.; Ishii, G. Organoid culture containing cancer cells and stromal cells reveals that podoplanin-positive cancer-associated fibroblasts enhance prolifera-tion of lung cancer cells. Lung Cancer 2019, 134, 100–107. [Google Scholar] [CrossRef] [PubMed]
- Stratmann, A.T.; Fecher, D.; Wangorsch, G.; Göttlich, C.; Walles, T.; Walles, H.; Dandekar, T.; Dandekar, G.; Nietzer, S.L. Establishment of a human 3D lung cancer model based on a biological tissue matrix combined with a Boolean in silico model. Mol. Oncol. 2013, 8, 351–365. [Google Scholar] [CrossRef] [PubMed]
- Wallstabe, L.; Göttlich, C.; Nelke, L.C.; Kühnemundt, J.; Schwarz, T.; Nerreter, T.; Einsele, H.; Walles, H.; Dandekar, G.; Nietzer, S.L.; et al. ROR1-CAR T cells are effective against lung and breast cancer in advanced microphysiologic 3D tu-mor models. JCI Insight 2019, 4. [Google Scholar] [CrossRef] [Green Version]
- Saforo, D.; Omer, L.; Smolenkov, A.; Barve, A.; Casson, L.; Boyd, N.L.; Clark, G.; Siskind, L.; Beverly, L.J. Primary lung cancer samples cultured under microenvironment-mimetic conditions enrich for mesenchymal stem-like cells that pro-mote metastasis. Sci. Rep. 2019, 9, 4177. [Google Scholar] [CrossRef]
- Kuriakose, A.E.; Hu, W.; Nguyen, K.T.; Menon, J.U. Scaffold-based lung tumor culture on porous PLGA microparticle substrates. PLoS ONE 2019, 14, e0217640. [Google Scholar] [CrossRef]
- Zhang, M.; Boughton, P.; Rose, B.; Lee, C.-S.; Hong, A.M. The Use of Porous Scaffold as a Tumor Model. Int. J. Biomater. 2013, 2013, 396056. [Google Scholar] [CrossRef] [Green Version]
- Mondal, A.; Gebeyehu, A.; Miranda, M.; Bahadur, D.; Patel, N.; Ramakrishnan, S.; Rishi, A.K.; Singh, M. Characteriza-tion and printability of Sodium alginate -Gelatin hydrogel for bioprinting NSCLC co-culture. Sci. Rep. 2019, 9, 1–12. [Google Scholar] [CrossRef]
- Simon, K.A.; Mosadegh, B.; Minn, K.T.; Lockett, M.R.; Lockett, M.R.; Boucher, D.M.; Hall, A.B.; Hillier, S.M.; Udagawa, T.; Eustace, B.K.; et al. Metabolic response of lung cancer cells to radiation in a paper-based 3D cell culture system. Biomaterials 2016, 95, 47–59. [Google Scholar] [CrossRef] [Green Version]
- Chang, C.-W.; Cheng, Y.-J.; Tu, M.; Chen, Y.-H.; Peng, C.-C.; Liao, W.-H.; Tung, Y.-C. A polydimethylsiloxane-polycarbonate hybrid microfluidic device capable of generating perpendicular chemical and oxygen gradients for cell culture studies. Lab. Chip 2014, 14, 3762–3772. [Google Scholar] [CrossRef] [PubMed]
- Jin, X.; Luan, H.; Chai, H.; Yan, L.; Zhang, J.; Wang, Q.; Cao, L. Netrin 1 interference potentiates epithelial-to-mesenchymal transition through the PI3K/AKT pathway under the hypoxic microenvironment conditions of non-small cell lung cancer. Int. J. Oncol. 2019, 54, 1457–1465. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Gao, Y.; Hao, Y.; Li, E.; Wang, Y.; Zhang, J.; Wang, W.; Gao, Z.; Wang, Q. Application of a microfluidic chip-based 3D co-culture to test drug sensitivity for individualized treatment of lung cancer. Biomaterials 2013, 34, 4109–4117. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Kulasinghe, A.; Bogseth, A.; O’Byrne, K.; Punyadeera, C.; Papautsky, I. Isolation of circulating tumor cells in non-small-cell-lung-cancer patients using a multi-flow microfluidic channel. Microsyst. Nanoeng. 2019, 5, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Zhang, X.; Dai, X.; Wang, X.; Li, X.; Diao, J.; Xu, T. Tumor-like lung cancer model based on 3D bioprinting. 3 Biotech 2018, 8, 501. [Google Scholar] [CrossRef]
- Utama, R.H.; Atapattu, L.; O’Mahony, A.P.; Fife, C.M.; Baek, J.; Allard, T.; O’Mahony, K.J.; Ribeiro, J.C.; Gaus, K.; Kavallaris, M. A 3D Bioprinter Specifically Designed for the High-Throughput Production of Matrix-Embedded Multicellar Spheroids. Iscience 2020, 23, 101621. [Google Scholar] [CrossRef]
- Däster, S.; Amatruda, N.; Calabrese, D.; Ivanek, R.; Turrini, E.; Droeser, R.A.; Zajac, P.; Fimognari, C.; Spagnoli, G.C.; Iezzi, G.; et al. Induction of hypoxia and necrosis in multicellular tumor spheroids is associated with resistance to chemo-therapy treatment. Oncotarget 2016, 8, 1725–1736. [Google Scholar] [CrossRef] [Green Version]
- Lv, D.; Hu, Z.; Lu, L.; Lu, H.; Xu, X. Three dimensional cell culture: A powerful tool in tumor research and drug discovery (Review). Oncol. Lett. 2017, 14, 6999–7010. [Google Scholar] [CrossRef] [Green Version]
- Gamerith, G.; Rainer, J.; Huber, J.M.; Hackl, H.; Trajanoski, Z.; Koeck, S.; Lorenz, E.; Kern, J.; Kofler, R.; Kelm, J.M.; et al. 3D-cultivation of NSCLC cell lines induce gene expression alterations of key cancer-associated pathways and mimic in-vivo conditions. Oncotarget 2017, 8, 112647–112661. [Google Scholar] [CrossRef] [Green Version]
- Bhattacharya, S.; Calar, K.; De La Puente, P. Mimicking tumor hypoxia and tumor-immune interactions employing three-dimensional in vitro models. J. Exp. Clin. Cancer Res. 2020, 39, 1–16. [Google Scholar] [CrossRef]
- Barkauskas, C.E.; Chung, M.-I.; Fioret, B.; Gao, X.; Katsura, H.; Hogan, B.L.M. Lung organoids: Current uses and future promise. Development 2017, 144, 986–997. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, M.; Mun, H.; Sung, C.O.; Cho, E.J.; Jeon, H.-J.; Chun, S.-M.; Jung, D.J.; Shin, T.H.; Jeong, G.S.; Kim, D.K.; et al. Patient-derived lung cancer organoids as in vitro cancer models for therapeutic screening. Nat. Commun. 2019, 10, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Chaicharoenaudomrung, N.; Kunhorm, P.; Noisa, P. Three-dimensional cell culture systems as an in vitro platform for cancer and stem cell modeling. World J. Stem Cells 2019, 11, 1065–1083. [Google Scholar] [CrossRef] [PubMed]
- Grist, S.M.; Nasseri, S.S.; Laplatine, L.; Schmok, J.C.; Yao, D.; Hua, J.; Chrostowski, L.; Cheung, K.C. Long-term monitor-ing in a microfluidic system to study tumour spheroid response to chronic and cycling hypoxia. Sci. Rep. 2019, 9, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Xu, Z.; Li, E.; Guo, Z.; Yu, R.; Hao, H.; Xu, Y.; Sun, Z.; Li, X.; Lyu, J.; Wang, Q. Design and Construction of a Multi-Organ Microfluidic Chip Mimicking the in vivo Microenvironment of Lung Cancer Metastasis. ACS Appl. Mater. Interfaces 2016, 8, 25840–25847. [Google Scholar] [CrossRef]
- Rebelo, S.P.; Pinto, C.; Martins, T.R.; Harrer, N.; Estrada, M.F.; Loza-Alvarez, P.; Cabeçadas, J.; Alves, P.C.; Gualda, E.J.; Sommergruber, W.; et al. 3D-3-culture: A tool to unveil macrophage plasticity in the tumour microenvironment. Biomaterials 2018, 163, 185–197. [Google Scholar] [CrossRef] [PubMed]
- Tam, R.Y.; Yockell-Lelièvre, J.; Smith, L.J.; Julian, L.M.; Baker, A.E.G.; Choey, C.; Hasim, M.S.; Dimitroulakos, J.; Stanford, W.L.; Shoichet, M.S. Rationally Designed 3D Hydrogels Model Invasive Lung Diseases Enabling High-Content Drug Screening. Adv. Mater. 2019, 31, e1806214. [Google Scholar] [CrossRef] [PubMed]
- Glunde, K.; Shah, T.; Winnard, P.T.; Raman, V.; Takagi, T.; Vesuna, F.; Artemov, D.; Bhujwalla, Z.M. Hypoxia Regulates Choline Kinase Expression through Hypoxia-Inducible Factor-1α Signaling in a Human Prostate Cancer Model. Cancer Res. 2008, 68, 172–180. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Godet, I.; Shin, Y.J.; Ju, J.A.; Ye, I.C.; Wang, G.; Gilkes, D.M. Fate-mapping post-hypoxic tumor cells reveals a ROS-resistant phenotype that promotes metastasis. Nat. Commun. 2019, 10, 1–18. [Google Scholar] [CrossRef] [PubMed] [Green Version]



| Feature | 2D | 3D |
|---|---|---|
| Cell morphology | Shape changed, flat | Real shape, aggregates |
| Cell polarity | Partial | Replicate |
| Cell proliferation | High proliferation | Replication of proliferation rate in vivo |
| Cell differentiation | Nonspontaneous | Spontaneous may occur via cellular interactions |
| Cell stage | The same for all cells in culture | Heterogenous cell cycle stage (proliferating, hypoxic, quiescent, necrotic) |
| Cell interactions | Limited | Replicate in vivo |
| Stiffness | High | Low |
| Culture formation | Quick | Slow |
| Culture duration | Long | Short |
| Cell culture | High reproductivity | Low reproductivity |
| In vivo like | No | Mimics in vivo tissue and interactions: cell to cell, cell to extracellular matrix (ECM), cell to growth factor |
| Tumoral heterogeneity | Basic | Approximation to in vivo |
| Exposure to nutrients, oxygen, drugs | Equally | Variable access |
| Drug response | Rather lack of correlation with human tumors | Similar response pattern as in human tumors |
| Costs | Average | More expensive |
| Model | Cells | Results | Reference |
|---|---|---|---|
| Spheroids | |||
| 3D high-throughput screening system | A549 | Hypoxia level of A549 spheroid was declined with TGF-β2 and elevated with TGF-β receptor I inhibitor (SB431542). | [78] |
| Spherical microtissues (hanging drop technology) | A549, Colo699 in combination with a fibroblast cell line (SV80) and two endothelial cell lines | Hypoxia marker (CA IX) was significantly expressed in microtissues that consisted of A549 cancer cells co-cultured with fibroblasts or endothelial cells. | [79] |
| 3D cell co-culture collagen gel model | Human lung: adenocarcinoma cells (HCC), fibroblast cells (MRC-5) and macrophages | Hypoxia and/or serum starvation conditions induced elevated secretion of VEGF in the 3D co-culture model in vitro, but not MMP-1. | [80] |
| 3D patient-derived tumor spheroids (PDS) | I/II stage NSCLC tumors | Long term 3D in vitro NSCLC model is useful for drug screening. | [81] |
| Organoids | |||
| Primary lung cancer organoids | Primary lung cancer tissues and paired non-neoplastic airway tissues (epithelial cells) | Cultured for long-term expansion over 6 months without any change in spherical organoid morphology and maintained proliferation capacity. | [82] |
| Hybrid cancer organoids | Podoplanin-positive cancer-associated fibroblasts (CAFs) and NSCLC PC-9 cells | The proliferation of PC-9 cells in hybrid cancer organoids containing podoplanin-overexpressing CAFs was significantly higher. | [83] |
| 3D scaffolds | |||
| Decellularized scaffolds | HCC827, A549 | Quantitative read-outs for proliferation, apoptosis and invasion were established in the complex 3D tumor model. | [84] |
| Microphysiologic 3D lung model (SISmuc platform) | A549 | Antitumor activity of ROR1-CAR T was specific and potent against A549 lung cancer. | [85] |
| 3D on human embryonic stem cell-qualified Matrigel-coated plates | Resections derived from NSCLC patients | 3D system allowed for the isolation and expansion of stromal progenitors from tumor resections. | [86] |
| Synthetic scaffolds on porous PLGA | A549 | Microparticles were used for A549 lung cancer cell culture. | [87] |
| Variotis tissue scaffold | NCI-H460 | NSCLC cells showed enhanced expression of CA IX hypoxia marker. | [88] |
| Hydrogels | |||
| Sodium alginate -gelatin (SA-GL) | NSCLC patient xenograft cells and lung CAFs co-cultures. | SA-GL hydrogel enhances printability and viability of NSCLC cells and CAF co-culture which allows 3D co-culture spheroid formation within the printed scaffold. | [89] |
| 3D tissue-like constructCells-in-Gels-in-Paper (CiGiP) | A549 | A549 cells showed increased levels of HIF1-α, decreased proliferation and reduced sensitivity to ionizing radiation. | [90] |
| Microfluidic devices | |||
| PDMS–PC hybrid microfluidic device | A549 | Drug testing results showed an increase in A549 cell apoptosis due to the hypoxia-activated cytotoxicity of tirapazamine. | [91] |
| 3D microfluidic chip | A549 and PC9 cells in vitro | Netrin-1 mediated epithelial–mesenchymal transition (EMT) of A549 and PC9 cells in vitro was associated with the phosphoinositide 3 kinase/AKT pathway, but only in hypoxia. | [92] |
| Spheroids in device-assisted culture | Primary lung cancer cells, SPCA-1 | Developed a high-throughput model for assessing drug sensitivities in vitro. There was a large discrepancy between drug sensitivity levels in 2D versus 3D. | [93] |
| Multi-flow microfluidic (MFM) system | Blood derived from NSCLC patients; NSCLC cell lines: HCC827, H460 | Effective separation of circulating tumor cells (CTCs) from 6 out of 8 NSCLC patients. | [94] |
| 3D bioprinting | |||
| 3D bioprinting using gelatin–sodium alginate-lung cancer cells suspension as the bio-ink | A549 and 95-D | Cell viability remained over 90%. Cell invasion and migration capabilities were improved in 3D printed cells compared to 2D cultured cells. | [95] |
| 3D bioprinter for high-throughput printing of spheroids | NSCLC (H460), neuroblastoma (SK-N-BE(2), glioblastoma (U87vIII) cells | Organization of the printed spheroids, presence of apoptotic and hypoxic cells was comparable to manually prepared spheroids. | [96] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ziółkowska-Suchanek, I. Mimicking Tumor Hypoxia in Non-Small Cell Lung Cancer Employing Three-Dimensional In Vitro Models. Cells 2021, 10, 141. https://doi.org/10.3390/cells10010141
Ziółkowska-Suchanek I. Mimicking Tumor Hypoxia in Non-Small Cell Lung Cancer Employing Three-Dimensional In Vitro Models. Cells. 2021; 10(1):141. https://doi.org/10.3390/cells10010141
Chicago/Turabian StyleZiółkowska-Suchanek, Iwona. 2021. "Mimicking Tumor Hypoxia in Non-Small Cell Lung Cancer Employing Three-Dimensional In Vitro Models" Cells 10, no. 1: 141. https://doi.org/10.3390/cells10010141
APA StyleZiółkowska-Suchanek, I. (2021). Mimicking Tumor Hypoxia in Non-Small Cell Lung Cancer Employing Three-Dimensional In Vitro Models. Cells, 10(1), 141. https://doi.org/10.3390/cells10010141





