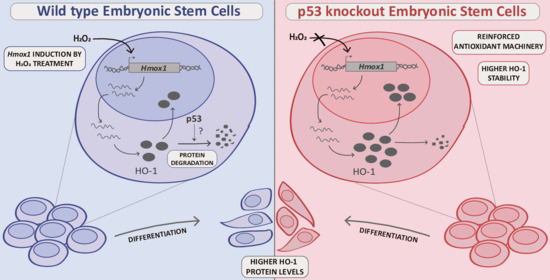
Novel Interplay between p53 and HO-1 in Embryonic Stem Cells
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture and Treatments
2.2. Gene Expression Analysis
2.3. Complementation Assay
2.4. Nuclear-Cytoplasmic Ratio Estimation
2.5. Bioinformatics Analysis
2.6. Flow Cytometry
2.7. Statistical Analysis
3. Results
3.1. HO-1 Protein Levels Are Boosted in p53 Knockout ES Cells
3.2. HO-1 Half-Life Is Longer in p53 KO ES Cells
3.3. p53 KO ES Cells Exhibit Higher HO-1 Cytoplasmic Levels
3.4. HO-1 Increases during Differentiation in WT and p53 KO ES Cells
3.5. HO-1 Is Not Induced in Oxidant Conditions in p53 KO ES Cells Consistent with an Adapted Stress Response
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Abdulla, E.M.; Campbell, I.C. Studies of Neurotoxicity in Cellular Models. In In Vitro Methods in Pharmaceutical Research; Elsevier: Amsterdam, The Netherlands, 1997; pp. 155–180. [Google Scholar]
- Tower, J. Stress and stem cells. Wiley Interdiscip. Rev. Dev. Biol. 2012, 1, 789–802. [Google Scholar] [CrossRef]
- Circu, M.L.; Aw, T.Y. Reactive oxygen species, cellular redox systems, and apoptosis. Free Radic. Biol. Med. 2010, 48, 749–762. [Google Scholar] [CrossRef] [PubMed]
- Sauer, H.; Wartenberg, M.; Hescheler, J. Reactive oxygen species as intracellular messengers during cell growth and differentiation. Cell. Physiol. Biochem. 2001, 11, 173–186. [Google Scholar] [CrossRef] [PubMed]
- Sart, S.; Song, L.; Li, Y. Controlling redox status for stem cell survival, expansion, and differentiation. Oxid. Med. Cell. Longev. 2015. [Google Scholar] [CrossRef]
- Snezhkina, A.V.; Kudryavtseva, A.V.; Kardymon, O.L.; Savvateeva, M.V.; Melnikova, N.V.; Krasnov, G.S.; Dmitriev, A.A. ROS Generation and Antioxidant Defense Systems in Normal and Malignant Cells. Oxid. Med. Cell. Longev. 2019, 2019, 6175804. [Google Scholar] [CrossRef]
- Martin, G.R. Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proc. Natl. Acad. Sci. USA 1981, 78, 7634–7638. [Google Scholar] [CrossRef]
- Binetruy, B.; Heasley, L.; Bost, F.; Caron, L.; Aouadi, M. Regulation of ES cell lineage commitment by mitogen activated protein kinases. Stem Cells 2007. [Google Scholar] [CrossRef]
- Saretzki, G.; Walter, T.; Atkinson, S.; Passos, J.F.; Bareth, B.; Keith, W.N.; Stewart, R.; Hoare, S.; Stojkovic, M.; Armstrong, L.; et al. Downregulation of Multiple Stress Defense Mechanisms During Differentiation of Human Embryonic Stem Cells. Stem Cells 2008, 26, 455–464. [Google Scholar] [CrossRef]
- Armstrong, L.; Tilgner, K.; Saretzki, G.; Atkinson, S.P.; Stojkovic, M.; Moreno, R.; Przyborski, S.; Lako, M. Human Induced Pluripotent Stem Cell Lines Show Stress Defense Mechanisms and Mitochondrial Regulation Similar to Those of Human Embryonic Stem Cells. Stem Cells 2010, 28, 661–673. [Google Scholar] [CrossRef]
- Cieślar-Pobuda, A.; Yue, J.; Lee, H.C.; Skonieczna, M.; Wei, Y.H. ROS and Oxidative Stress in Stem Cells. Oxid. Med. Cell. Longev. 2017. [Google Scholar] [CrossRef]
- Tsatmali, M.; Walcott, E.C.; Crossin, K.L. Newborn neurons acquire high levels of reactive oxygen species and increased mitochondrial proteins upon differentiation from progenitors. Brain Res. 2005, 1040, 137–150. [Google Scholar] [CrossRef] [PubMed]
- Jang, J.; Wang, Y.; Kim, H.-S.; Lalli, M.A.; Kosik, K.S. Nrf2, a Regulator of the Proteasome, Controls Self-Renewal and Pluripotency in Human Embryonic Stem Cells. Stem Cells 2014, 32, 2616–2625. [Google Scholar] [CrossRef] [PubMed]
- Ito, K.; Hirao, A.; Arai, F.; Takubo, K.; Matsuoka, S.; Miyamoto, K.; Ohmura, M.; Naka, K.; Hosokawa, K.; Ikeda, Y.; et al. Reactive oxygen species act through p38 MAPK to limit the lifespan of hematopoietic stem cells. Nat. Med. 2006, 12, 446–451. [Google Scholar] [CrossRef]
- Schmelter, M.; Ateghang, B.; Helmig, S.; Wartenberg, M.; Sauer, H. Embryonic stem cells utilize reactive oxygen species as transducers of mechanical strain-induced cardiovascular differentiation. FASEB J. 2006, 20. [Google Scholar] [CrossRef]
- Smith, J.; Ladi, E.; Mayer-Pröschel, M.; Noble, M. Redox state is a central modulator of the balance between self-renewal and differentiation in a dividing glial precursor cell. Proc. Natl. Acad. Sci. USA 2000, 97, 10032–10037. [Google Scholar] [CrossRef]
- Marión, R.M.; Strati, K.; Li, H.; Murga, M.; Blanco, R.; Ortega, S.; Fernandez-Capetillo, O.; Serrano, M.; Blasco, M.A. A p53-mediated DNA damage response limits reprogramming to ensure iPS cell genomic integrity Europe PMC Funders Group. Nature 2009, 460, 1149–1153. [Google Scholar] [CrossRef]
- Chen, Y.; Liu, K.; Shi, Y.; Shao, C. The tango of ROS and p53 in tissue stem cells. Cell Death Differ. 2018, 25, 637–639. [Google Scholar] [CrossRef]
- Kastenhuber, E.R.; Lowe, S.W. Putting p53 in Context. Cell 2017, 170, 1062–1078. [Google Scholar] [CrossRef]
- Wang, C.; Liang, C.C.; Bian, Z.C.; Zhu, Y.; Guan, J.L. FIP200 is required for maintenance and differentiation of postnatal neural stem cells. Nat. Neurosci. 2013, 16, 532–542. [Google Scholar] [CrossRef]
- Jung, H.; Kim, M.J.; Kim, D.O.; Kim, W.S.; Yoon, S.-J.; Park, Y.-J.; Yoon, S.R.; Kim, T.-D.; Suh, H.-W.; Yun, S.; et al. Article TXNIP Maintains the Hematopoietic Cell Pool by Switching the Function of p53 under Oxidative Stress. Cell Metab. 2013, 18, 75–85. [Google Scholar] [CrossRef]
- Liu, D.; Xu, Y. P53, oxidative stress, and aging. Antioxid. Redox Signal. 2011, 15, 1669–1678. [Google Scholar] [CrossRef] [PubMed]
- Jaiswal, S.K.; Oh, J.J.; DePamphilis, M.L. Cell cycle arrest and apoptosis are not dependent on p53 prior to p53-dependent embryonic stem cell differentiation. Stem Cells 2020. [Google Scholar] [CrossRef] [PubMed]
- Lin, T.; Lin, Y. p53 switches off pluripotency on differentiation. Stem Cell Res. Ther. 2017, 8, 44. [Google Scholar] [CrossRef] [PubMed]
- Abdelalim, E.M.; Tooyama, I. The p53 inhibitor, pifithrin-α, suppresses self-renewal of embryonic stem cells. Biochem. Biophys. Res. Commun. 2012, 420, 605–610. [Google Scholar] [CrossRef]
- Gozzelino, R.; Jeney, V.; Soares, M.P. Mechanisms of cell protection by heme oxygenase-1. Annu. Rev. Pharmacol. Toxicol. 2010. [Google Scholar] [CrossRef] [PubMed]
- Kozakowska, M.; Szade, K.; Dulak, J.; Jozkowicz, A. Role of heme oxygenase-1 in postnatal differentiation of stem cells: A possible cross-talk with MicroRNAs. Antioxid. Redox Signal. 2014. [Google Scholar] [CrossRef]
- Meiller, A.; Alvarez, S.; Drané, P.; Lallemand, C.; Blanchard, B.; Tovey, M.; May, E. p53-dependent stimulation of redox-related genes in the lymphoid organs of gamma-irradiated--mice identification of Haeme-oxygenase 1 as a direct p53 target gene. Nucleic Acids Res. 2007, 35, 6924–6934. [Google Scholar] [CrossRef]
- Nam, S.Y.; Sabapathy, K. p53 promotes cellular survival in a context-dependent manner by directly inducing the expression of haeme-oxygenase-1. Oncogene 2011, 30, 4476–4486. [Google Scholar] [CrossRef]
- Tonelli, C.; Chio, I.I.C.; Tuveson, D.A. Transcriptional Regulation by Nrf2. Antioxid. Redox Signal. 2018, 29, 1727–1745. [Google Scholar] [CrossRef]
- Rotblat, B.; Melino, G.; Knight, R.A. NRF2 and p53: Januses in cancer? Oncotarget 2012, 3, 1272–1283. [Google Scholar] [CrossRef]
- Pereira, E.J.; Burns, J.S.; Lee, C.Y.; Marohl, T.; Calderon, D.; Wang, L.; Atkins, K.A.; Wang, C.-C.; Janes, K.A. Sporadic activation of an oxidative stress-dependent NRF2–p53 signaling network in breast epithelial spheroids and premalignancies. Sci. Signal. 2020, 13. [Google Scholar] [CrossRef] [PubMed]
- Tung, M.C.; Lin, P.L.; Wang, Y.C.; He, T.Y.; Lee, M.C.; Der Yeh, S.; Chen, C.Y.; Lee, H. Mutant p53 confers chemoresistance in non-small cell lung cancer by upregulating Nrf2. Oncotarget 2015, 6, 41692–41705. [Google Scholar] [CrossRef] [PubMed]
- Lisek, K.; Campaner, E.; Ciani, Y.; Walerych, D.; Del Sal, G. Mutant p53 tunes the NRF2-dependent antioxidant response to support survival of cancer cells. Oncotarget 2018, 9, 20508–20523. [Google Scholar] [CrossRef] [PubMed]
- Mohammadzadeh, M.; Halabian, R.; Gharehbaghian, A.; Amirizadeh, N.; Jahanian-Najafabadi, A.; Mohammadi, A.; Mehryar, R.; Roudkenar, H. Nrf-2 overexpression in mesenchymal stem cells reduces oxidative stress-induced apoptosis and cytotoxicity. Cell Stress Chaperones 2012, 17, 553–565. [Google Scholar] [CrossRef]
- Yoon, D.S.; Choi, Y.; Lee, J.W. Cellular localization of NRF2 determines the self-renewal and osteogenic differentiation potential of human MSCs via the P53-SIRT1 axis. Cell Death Dis. 2016, 7, e2093. [Google Scholar] [CrossRef]
- Stepniewski, J.; Pacholczak, T.; Skrzypczyk, A.; Ciesla, M.; Szade, A.; Szade, K.; Bidanel, R.; Langrzyk, A.; Grochowski, R.; Vandermeeren, F.; et al. Heme oxygenase-1 affects generation and spontaneous cardiac differentiation of induced pluripotent stem cells. IUBMB Life 2018. [Google Scholar] [CrossRef]
- Waisman, A.; Vazquez Echegaray, C.; Solari, C.; Cosentino, M.S.; Martyn, I.; Deglincerti, A.; Ozair, M.Z.; Ruzo, A.; Barañao, L.; Miriuka, S.; et al. Inhibition of Cell Division and DNA Replication Impair Mouse-Naïve Pluripotency Exit. J. Mol. Biol. 2017. [Google Scholar] [CrossRef]
- Cosentino, M.S.; Oses, C.; Vázquez Echegaray, C.; Solari, C.; Waisman, A.; Álvarez, Y.; Petrone, M.V.; Francia, M.; Schultz, M.; Sevlever, G.; et al. Kat6b Modulates Oct4 and Nanog Binding to Chromatin in Embryonic Stem Cells and Is Required for Efficient Neural Differentiation. J. Mol. Biol. 2019, 431, 1148–1159. [Google Scholar] [CrossRef]
- Losino, N.; Waisman, A.; Solari, C.; Luzzani, C.; Espinosa, D.F.; Sassone, A.; Muro, A.F.; Miriuka, S.; Sevlever, G.; Barañao, L.; et al. EDA-containing fibronectin increases proliferation of embryonic stem cells. PLoS ONE 2013. [Google Scholar] [CrossRef]
- Wells, C.A.; Mosbergen, R.; Korn, O.; Choi, J.; Seidenman, N.; Matigian, N.A.; Vitale, A.M.; Shepherd, J. Stemformatics: Visualisation and sharing of stem cell gene expression. Stem Cell Res. 2013, 10, 387–395. [Google Scholar] [CrossRef]
- Enver, T.; Soneji, S.; Joshi, C.; Brown, J.; Iborra, F.; Orntoft, T.; Thykjaer, T.; Maltby, E.; Smith, K.; Dawud, R.A.; et al. Cellular differentiation hierarchies in normal and culture-adapted human embryonic stem cells. Hum. Mol. Genet. 2005, 14, 3129–3140. [Google Scholar] [CrossRef] [PubMed]
- Kyrkou, A.; Stellas, D.; Syrrou, M.; Klinakis, A.; Fotsis, T.; Murphy, C. Generation of human induced pluripotent stem cells in defined, feeder-free conditions. Stem Cell Res. 2016, 17, 458–460. [Google Scholar] [CrossRef] [PubMed]
- Yi, H.; Xue, L.; Guo, M.; Ma, J.; Zeng, Y.; Wang, W.; Cai, J.; Hu, H.; Shu, H.; Shi, Y.; et al. Gene expression atlas for human embryogenesis. FASEB J. 2010, 24, 3341–3350. [Google Scholar] [CrossRef] [PubMed]
- Pandya, H.; Shen, M.J.; Ichikawa, D.M.; Sedlock, A.B.; Choi, Y.; Johnson, K.R.; Kim, G.; Brown, M.A.; Elkahloun, A.G.; Maric, D.; et al. Differentiation of human and murine induced pluripotent stem cells to microglia-like cells. Nat. Neurosci. 2017, 20, 753–759. [Google Scholar] [CrossRef] [PubMed]
- Mariani, J.; Coppola, G.; Zhang, P.; Abyzov, A.; Provini, L.; Tomasini, L.; Amenduni, M.; Szekely, A.; Palejev, D.; Wilson, M.; et al. FOXG1-Dependent Dysregulation of GABA/Glutamate Neuron Differentiation in Autism Spectrum Disorders. Cell 2015, 162, 375–390. [Google Scholar] [CrossRef]
- An, X.; Schulz, V.P.; Li, J.; Wu, K.; Liu, J.; Xue, F.; Hu, J.; Mohandas, N.; Gallagher, P.G. Global transcriptome analyses of human and murine terminal erythroid differentiation. Blood 2014, 123, 3466–3477. [Google Scholar] [CrossRef]
- Granchi, D.; Ochoa, G.; Leonardi, E.; Devescovi, V.; Baglìo, S.R.; Osaba, L.; Baldini, N.; Ciapetti, G. Gene expression patterns related to osteogenic differentiation of bone marrow-derived mesenchymal stem cells during Ex vivo expansion. Tissue Eng. Part C Methods 2010, 16, 511–524. [Google Scholar] [CrossRef]
- Si-Tayeb, K.; Noto, F.K.; Nagaoka, M.; Li, J.; Battle, M.A.; Duris, C.; North, P.E.; Dalton, S.; Duncan, S.A. Highly efficient generation of human hepatocyte-like cells from induced pluripotent stem cells. Hepatology 2010, 51, 297–305. [Google Scholar] [CrossRef]
- Teo, A.K.K.; Arnold, S.J.; Trotter, M.W.B.; Brown, S.; Ang, L.T.; Chng, Z.; Robertson, E.J.; Dunn, N.R.; Vallier, L. Pluripotency factors regulate definitive endoderm specification through eomesodermin. Genes Dev. 2011, 25, 238–250. [Google Scholar] [CrossRef]
- Yeow, J.; Kaur, A.; Anscomb, M.D.; New, E.J. A novel flavin derivative reveals the impact of glucose on oxidative stress in adipocytes. Chem. Commun. 2014, 50, 8181–8184. [Google Scholar] [CrossRef]
- Kaur, A.; Brigden, K.W.L.; Cashman, T.F.; Fraser, S.T.; New, E.J. Mitochondrially targeted redox probe reveals the variations in oxidative capacity of the haematopoietic cells. Org. Biomol. Chem. 2015, 13, 4343–4349. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.C.; Nguyen, T.; Pickett, C.B. Phosphorylation of Nrf2 at Ser-40 by protein kinase C regulates antioxidant response element-mediated transcription. J. Biol. Chem. 2002, 277, 42769–42774. [Google Scholar] [CrossRef] [PubMed]
- Inobe, T.; Matouschek, A. Paradigms of protein degradation by the proteasome. Curr. Opin. Struct. Biol. 2014, 24, 156–164. [Google Scholar] [CrossRef] [PubMed]
- Lecker, S.H.; Goldberg, A.L.; Mitch, W.E. Protein degradation by the ubiquitin-proteasome pathway in normal and disease states. J. Am. Soc. Nephrol. 2006, 17, 1807–1819. [Google Scholar] [CrossRef] [PubMed]
- Sacca, P.; Meiss, R.; Casas, G.; Mazza, O.; Calvo, J.C.; Navone, N.; Vazquez, E. Nuclear translocation of haeme oxygenase-1 is associated to prostate cancer. Br. J. Cancer 2007, 97, 1683–1689. [Google Scholar] [CrossRef] [PubMed]
- Lin, Q.; Weis, S.; Yang, G.; Weng, Y.H.; Helston, R.; Rish, K.; Smith, A.; Bordner, J.; Polte, T.; Gaunitz, F.; et al. Heme oxygenase-1 protein localizes to the nucleus and activates transcription factors important in oxidative stress. J. Biol. Chem. 2007. [Google Scholar] [CrossRef]
- Suliman, H.B.; Zobi, F.; Piantadosi, C.A. Heme Oxygenase-1/Carbon Monoxide System and Embryonic Stem Cell Differentiation and Maturation into Cardiomyocytes. Antioxid. Redox Signal. 2016. [Google Scholar] [CrossRef]
- Lin, C.-Y.; Peng, C.-Y.; Huang, T.-T.; Wu, M.-L.; Lai, Y.-L.; Peng, D.H.; Chen, P.-F.; Chen, H.-F.; Yen, B.L.; Wu, K.K.; et al. Exacerbation of Oxidative Stress-Induced Cell Death and Differentiation in Induced Pluripotent Stem Cells Lacking Heme Oxygenase-1. Stem Cells Dev. 2012, 21, 1675–1687. [Google Scholar] [CrossRef]
- Lai, Y.-L.; Lin, C.-Y.; Jiang, W.-C.; Ho, Y.-C.; Chen, C.-H.; Yet, S.-F. Loss of heme oxygenase-1 accelerates mesodermal gene expressions during embryoid body development from mouse embryonic stem cells. Redox Biol. 2018, 15, 51–61. [Google Scholar] [CrossRef]
- Oki, S.; Ohta, T.; Shioi, G.; Hatanaka, H.; Ogasawara, O.; Okuda, Y.; Kawaji, H.; Nakaki, R.; Sese, J.; Meno, C. Ch IP -Atlas: A data-mining suite powered by full integration of public Ch IP -seq data. EMBO Rep. 2018, 19. [Google Scholar] [CrossRef]
- Solari, C.; Vázquez Echegaray, C.; Cosentino, M.S.M.S.; Petrone, M.V.M.V.; Waisman, A.; Luzzani, C.; Francia, M.; Villodre, E.; Lenz, G.; Miriuka, S.; et al. Manganese Superoxide Dismutase Gene Expression Is Induced by Nanog and Oct4, Essential Pluripotent Stem Cells’ Transcription Factors. PLoS ONE 2015. [Google Scholar] [CrossRef] [PubMed]
- Corbett, A.H. Post-transcriptional regulation of gene expression and human disease. Curr. Opin. Cell Biol. 2018, 52, 96–104. [Google Scholar] [CrossRef] [PubMed]
- Ray, P.D.; Huang, B.W.; Tsuji, Y. Reactive oxygen species (ROS) homeostasis and redox regulation in cellular signaling. Cell. Signal. 2012, 24, 981–990. [Google Scholar] [CrossRef] [PubMed]
- Amadio, M.; Scapagnini, G.; Laforenza, U.; Intrieri, M.; Romeo, L.; Govoni, S.; Pascale, A. Post-Transcriptional Regulation of HSP70 Expression Following Oxidative Stress in SH-SY5Y Cells: The Potential Involvement of the RNA-Binding Protein HuR. Curr. Pharm. Des. 2008, 14, 2651–2658. [Google Scholar] [CrossRef] [PubMed]
- Je, G.; Kim, Y.S. Mitochondrial ROS-mediated post-transcriptional regulation of α-synuclein through miR-7 and miR-153. Neurosci. Lett. 2017, 661, 132–136. [Google Scholar] [CrossRef] [PubMed]
- Tsushima, K.; Bugger, H.; Wende, A.R.; Soto, J.; Jenson, G.A.; Tor, A.R.; McGlauflin, R.; Kenny, H.C.; Zhang, Y.; Souvenir, R.; et al. Mitochondrial Reactive Oxygen Species in Lipotoxic Hearts Induce Post-Translational Modifications of AKAP121, DRP1, and OPA1 That Promote Mitochondrial Fission. Circ. Res. 2018, 122, 58–73. [Google Scholar] [CrossRef]
- Bigarella, C.L.; Liang, R.; Ghaffari, S. Stem cells and the impact of ROS signaling. Development 2014, 141, 4206–4218. [Google Scholar] [CrossRef]
- Vomund, S.; Schäfer, A.; Parnham, M.J.; Brüne, B.; Von Knethen, A. Molecular Sciences Nrf2, the Master Regulator of Anti-Oxidative Responses. J. Mol. Sci. 2017, 18, 2772. [Google Scholar] [CrossRef]
- Oren, M. Decision making by p53: Life, death and cancer. Cell Death Differ. 2003, 10, 431–442. [Google Scholar] [CrossRef]
- Polyak, K.; Xia, Y.; Zweier, J.L.; Kinzler, K.W.; Vogelstein, B. A model for p53-induced apoptosis. Nature 1997, 389, 300–305. [Google Scholar] [CrossRef]
- Faraonio, R.; Vergara, P.; di Marzo, D.; Pierantoni, M.G.; Napolitano, M.; Russo, T.; Cimino, F. p53 Suppresses the Nrf2-dependent Transcription of Antioxidant Response Genes. J. Biol. Chem. 2006, 281, 39776–39784. [Google Scholar] [CrossRef]
- You, A.; Nam, C.W.; Wakabayashi, N.; Yamamoto, M.; Kensler, T.W.; Kwak, M.K. Transcription factor Nrf2 maintains the basal expression of Mdm2: An implication of the regulation of p53 signaling by Nrf2. Arch. Biochem. Biophys. 2011, 507, 356–364. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.T.; Gu, W. The multiple levels of regulation by p53 ubiquitination. Cell Death Differ. 2010, 17, 86–92. [Google Scholar] [CrossRef] [PubMed]
- Sablina, A.A.; Budanov, A.V.; Ilyinskaya, G.V.; Agapova, L.S.; Kravchenko, J.E.; Chumakov, P.M. The antioxidant function of the p53 tumor suppressor. Nat. Med. 2005, 11, 1306–1313. [Google Scholar] [CrossRef] [PubMed]
- Jain, A.K.; Barton, M.C. P53: Emerging roles in stem cells, development and beyond. Development 2018, 145. [Google Scholar] [CrossRef] [PubMed]
- Fu, X.; Wu, S.; Li, B.; Xu, Y.; Liu, J. Functions of p53 in pluripotent stem cells. Protein Cell 2020, 11, 71–78. [Google Scholar] [CrossRef]
- Hwang, H.W.; Lee, J.R.; Chou, K.Y.; Suen, C.S.; Hwang, M.J.; Chen, C.; Shieh, R.C.; Chau, L.Y. Oligomerization is crucial for the stability and function of heme oxygenase-1 in the endoplasmic reticulum. J. Biol. Chem. 2009. [Google Scholar] [CrossRef]
- Lin, P.H.; Chiang, M.T.; Chau, L.Y. Ubiquitin-proteasome system mediates heme oxygenase-1 degradation through endoplasmic reticulum-associated degradation pathway. Biochim. Biophys. Acta Mol. Cell Res. 2008, 1783, 1826–1834. [Google Scholar] [CrossRef]
- Song, J.; Zhang, X.; Liao, Z.; Liang, H.; Chu, L.; Dong, W.; Zhang, X.; Ge, Q.; Liu, Q.; Fan, P.; et al. 14-3-3ζ inhibits heme oxygenase-1 (HO-1) degradation and promotes hepatocellular carcinoma proliferation: Involvement of STAT3 signaling. J. Exp. Clin. Cancer Res. 2019, 38. [Google Scholar] [CrossRef]
- Biswas, C.; Shah, N.; Muthu, M.; La, P.; Fernando, A.P.; Sengupta, S.; Yang, G.; Dennery, P.A. Nuclear heme oxygenase-1 (HO-1) modulates subcellular distribution and activation of Nrf2, impacting metabolic and anti-oxidant defenses. J. Biol. Chem. 2014, 289, 26882–26894. [Google Scholar] [CrossRef]
- Mora-Castilla, S.; Tejedo, J.R.; Hmadcha, A.; Cahuana, G.M.; Martín, F.; Soria, B.; Bedoya, F.J. Nitric oxide repression of Nanog promotes mouse embryonic stem cell differentiation. Cell Death Differ. 2010. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.H.; Suh, H.N.; Lee, Y.J.; Seo, B.N.; Ha, J.W.; Han, H.J. Midkine prevented hypoxic injury of mouse embryonic stem cells through activation of Akt and HIF-1α via low-density lipoprotein receptor-related protein-1. J. Cell. Physiol. 2011. [Google Scholar] [CrossRef]
- Trigona, W.L.; Porter, C.M.; Horvath-Arcidiacono, J.A.; Majumdar, A.S.; Bloom, E.T. Could heme-oxygenase-1 have a role in modulating the recipient immune response to embryonic stem cells? Antioxid. Redox Signal. 2007. [Google Scholar] [CrossRef] [PubMed]
- Spike, B.T.; Wahl, G.M. P53, stem cells, and reprogramming: Tumor suppression beyond guarding the genome. Genes Cancer 2011, 2, 404–419. [Google Scholar] [CrossRef] [PubMed]






| Reference | Cell Line | Platform | Accession No | |
|---|---|---|---|---|
| Non-directed diff | Enver et al., 2005 [42] | H7 | Affymetrix U133A | N/A |
| Reprograming | Kyrkou et al., 2016 [43] | Fibroblast and iPS cells | Affymetrix HuGene-1_0-ST V1 | GSE58932 |
| Embryo development | Yi et al., 2010 [44] | Human embryos | Affymetrix HG-U133_Plus_2 | GSE15744 |
| Ectodermal diff | Pandya et al., 2017 [45] | iPS cells and iPS derived microglia | Affymetrix MoGene-1_0-ST V1 | GSE47605 |
| Ectodermal diff | Mariani et al., 2015 [46] | iPS cells | Illumina HiSeq 2000 | GSE61476 |
| Mesodermal diff | An et al., 2014 [47] | Erythroblasts | Illumina HiSeq 2000 | GSE53983 |
| Mesodermal diff | Granchi et al., 2010 [48] | Mesenchymal stem cells | Affymetrix HG-U133_Plus_2 | GSE12267 |
| Endodermal diff | Si-Tayeb et al., 2010 [49] | iPS cells | Affymetrix HG-U133_Plus_2 | GSE14897 |
| Endodermal diff | Teo et al., 2011 [50] | ES cells | Illumina HumanRef-8 V3 | E-MTAB-467 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Toro, A.; Anselmino, N.; Solari, C.; Francia, M.; Oses, C.; Sanchis, P.; Bizzotto, J.; Vazquez Echegaray, C.; Petrone, M.V.; Levi, V.; et al. Novel Interplay between p53 and HO-1 in Embryonic Stem Cells. Cells 2021, 10, 35. https://doi.org/10.3390/cells10010035
Toro A, Anselmino N, Solari C, Francia M, Oses C, Sanchis P, Bizzotto J, Vazquez Echegaray C, Petrone MV, Levi V, et al. Novel Interplay between p53 and HO-1 in Embryonic Stem Cells. Cells. 2021; 10(1):35. https://doi.org/10.3390/cells10010035
Chicago/Turabian StyleToro, Ayelén, Nicolás Anselmino, Claudia Solari, Marcos Francia, Camila Oses, Pablo Sanchis, Juan Bizzotto, Camila Vazquez Echegaray, María Victoria Petrone, Valeria Levi, and et al. 2021. "Novel Interplay between p53 and HO-1 in Embryonic Stem Cells" Cells 10, no. 1: 35. https://doi.org/10.3390/cells10010035
APA StyleToro, A., Anselmino, N., Solari, C., Francia, M., Oses, C., Sanchis, P., Bizzotto, J., Vazquez Echegaray, C., Petrone, M. V., Levi, V., Vazquez, E., & Guberman, A. (2021). Novel Interplay between p53 and HO-1 in Embryonic Stem Cells. Cells, 10(1), 35. https://doi.org/10.3390/cells10010035