New Insights into Plant Extracellular DNA. A Study in Soybean Root Extracellular Trap
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material, PEP-13 Elicitation, and exDNA Separation
2.2. DNA Purification and Sequencing
2.3. Bioinformatic Analyses
3. Results
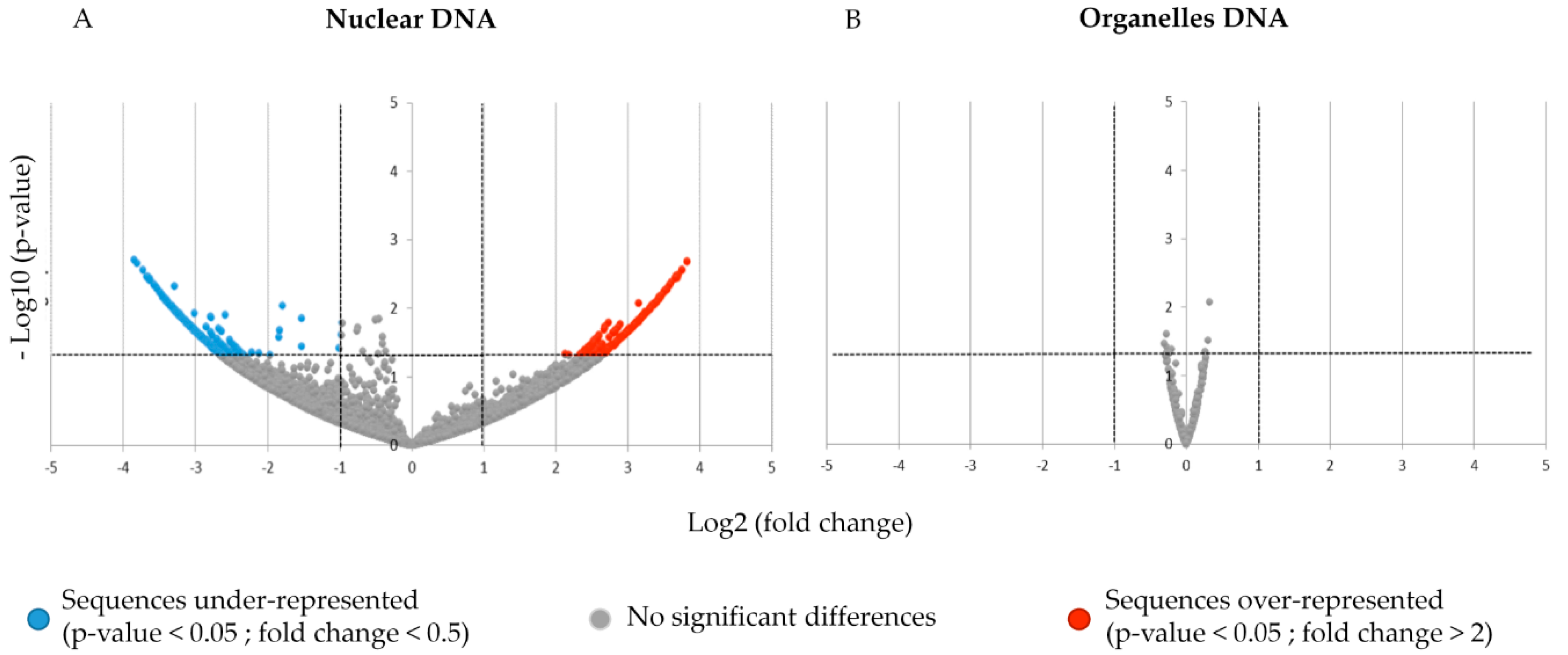
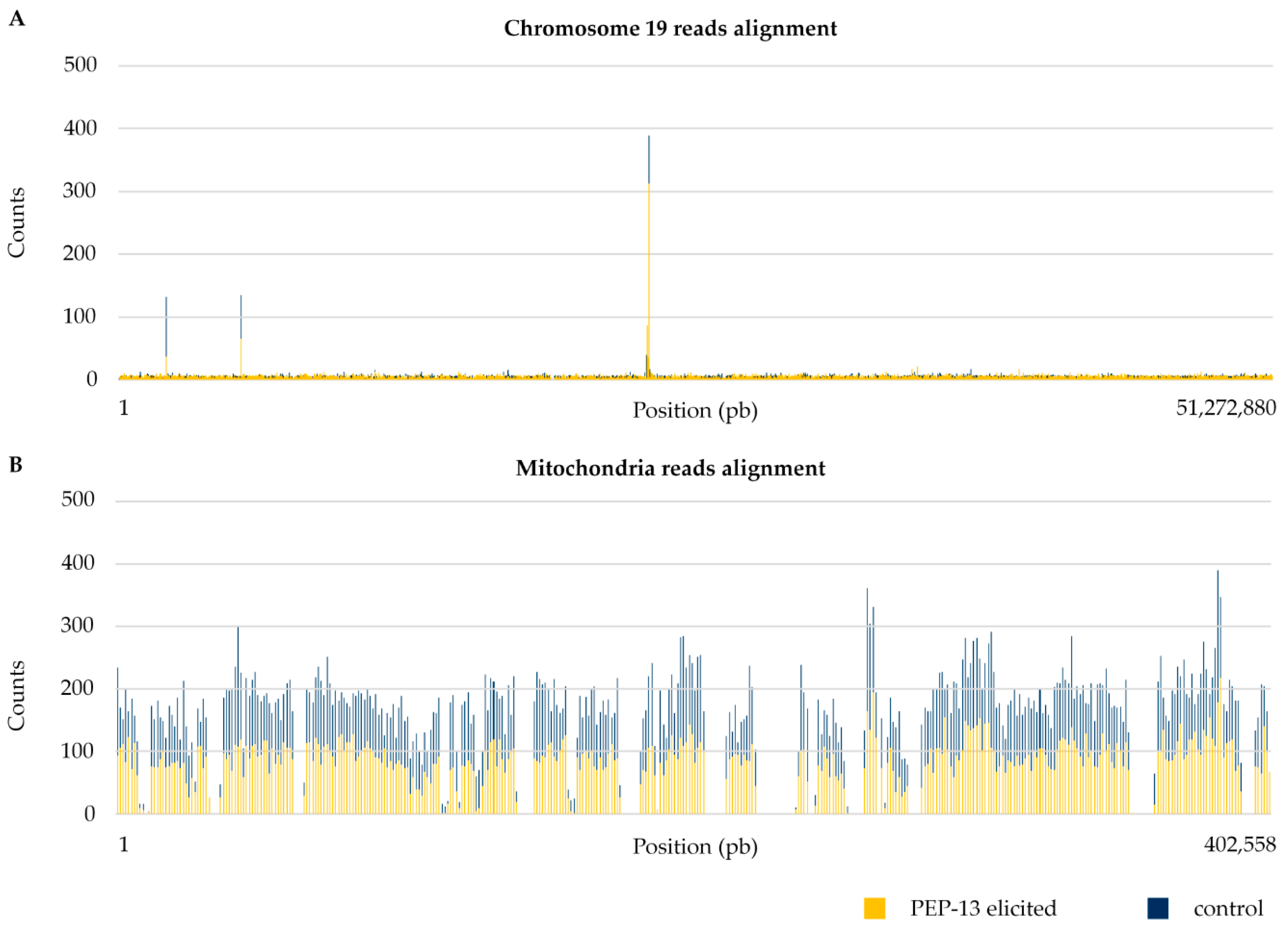
3.1. Differences between Nuclear and Organelle DNA Coverage
3.2. Impact of PEP-13 Elicitation on exDNA Sequence
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kozlowski, H.N.; Lai, E.T.L.; Havugimana, P.C.; White, C.; Emili, A.; Sakac, D.; Binnington, B.; Neschadim, A.; McCarthy, S.D.S.; Branch, D.R. Extracellular histones identified in crocodile blood inhibit in-vitro HIV-1 infection. AIDS 2016, 30, 2043–2052. [Google Scholar] [PubMed]
- Palić, D.; Ostojić, J.; Andreasen, C.B.; Roth, J.A. Fish cast NETs: Neutrophil extracellular traps are released from fish neutrophils. Dev. Comp. Immunol. 2007, 31, 805–816. [Google Scholar] [CrossRef] [PubMed]
- Pieper, J.; Locke, M.; Ruzaike, G.; Voigt, S.; Methner, U.; Berndt, A. In vitro and in vivo generation of heterophil extracellular traps after Salmonella exposure. Vet. Immunol. Immunopathol. 2017, 188, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Brinkmann, V.; Reichard, U.; Goosmann, C.; Fauler, B.; Uhlemann, Y.; Weiss, D.S.; Weinrauch, Y.; Zychlinsky, A. Neutrophil Extracellular Traps Kill Bacteria. Science 2004, 303, 1532–1535. [Google Scholar] [CrossRef] [PubMed]
- von Köckritz-Blickwede, M.; Goldmann, O.; Thulin, P.; Heinemann, K.; Norrby-Teglund, A.; Rohde, M.; Medina, E. Phagocytosis-independent antimicrobial activity of mast cells by means of extracellular trap formation. Blood 2008, 111, 3070–3080. [Google Scholar] [CrossRef] [PubMed]
- Webster, S.J.; Daigneault, M.; Bewley, M.A.; Preston, J.A.; Marriott, H.M.; Walmsley, S.R.; Read, R.C.; Whyte, M.K.B.; Dockrell, D.H. Distinct Cell Death Programs in Monocytes Regulate Innate Responses Following Challenge with Common Causes of Invasive Bacterial Disease. J. Immunol. 2010, 185, 2968–2979. [Google Scholar] [CrossRef] [PubMed]
- Dworski, R.; Simon, H.-U.; Hoskins, A.; Yousefi, S. Eosinophil and neutrophil extracellular DNA traps in human allergic asthmatic airways. J. Allergy Clin. Immunol. 2011, 127, 1260–1266. [Google Scholar] [CrossRef]
- Simon, D.; Hoesli, S.; Roth, N.; Staedler, S.; Yousefi, S.; Simon, H.-U. Eosinophil extracellular DNA traps in skin diseases. J. Allergy Clin. Immunol. 2011, 127, 194–199. [Google Scholar] [CrossRef]
- Aulik, N.A.; Hellenbrand, K.M.; Czuprynski, C.J. Mannheimia haemolytica and Its Leukotoxin Cause Macrophage Extracellular Trap Formation by Bovine Macrophages. Infect. Immun. 2012, 80, 1923–1933. [Google Scholar] [CrossRef]
- Yipp, B.G.; Petri, B.; Salina, D.; Jenne, C.N.; Scott, B.N.V.; Zbytnuik, L.D.; Pittman, K.; Asaduzzaman, M.; Wu, K.; Meijndert, H.C.; et al. Infection-induced NETosis is a dynamic process involving neutrophil multitasking in vivo. Nat. Med. 2012, 18, 1386–1393. [Google Scholar] [CrossRef]
- Simon, D.; Simon, H.-U.; Yousefi, S. Extracellular DNA traps in allergic, infectious, and autoimmune diseases. Allergy 2013, 68, 409–416. [Google Scholar] [CrossRef] [PubMed]
- Delgado-Rizo, V.; Martínez-Guzmán, M.A.; Iñiguez-Gutierrez, L.; García-Orozco, A.; Alvarado-Navarro, A.; Fafutis-Morris, M. Neutrophil Extracellular Traps and Its Implications in Inflammation: An Overview. Front. Immunol. 2017, 8, 81. [Google Scholar] [CrossRef] [PubMed]
- de Bont, C.M.; Koopman, W.J.H.; Boelens, W.C.; Pruijn, G.J.M. Stimulus-dependent chromatin dynamics, citrullination, calcium signalling and ROS production during NET formation. BBA Mol. Cell Res. 2018, 1865, 1621–1629. [Google Scholar] [CrossRef] [PubMed]
- Yipp, B.G.; Kubes, P. NETosis: How vital is it? Blood 2013, 122, 2784–2794. [Google Scholar] [CrossRef]
- Yousefi, S.; Mihalache, C.; Kozlowski, E.; Schmid, I.; Simon, H.U. Viable neutrophils release mitochondrial DNA to form neutrophil extracellular traps. Cell Death Differ. 2009, 16, 1438–1444. [Google Scholar] [CrossRef]
- Allesen-Holm, M.; Barken, K.B.; Yang, L.; Klausen, M.; Webb, J.S.; Kjelleberg, S.; Molin, S.; Givskov, M.; Tolker-Nielsen, T. A characterization of DNA release in Pseudomonas aeruginosa cultures and biofilms. Mol. Microbiol. 2006, 59, 1114–1128. [Google Scholar] [CrossRef]
- Martins, M.; Uppuluri, P.; Thomas, D.P.; Cleary, I.A.; Henriques, M.; Lopez-Ribot, J.L.; Oliveira, R. Presence of Extracellular DNA in the Candida albicans Biofilm Matrix and its Contribution to Biofilms. Mycopathologia 2010, 169, 323–331. [Google Scholar] [CrossRef]
- Tetz, V.V.; Tetz, G.V. Effect of Extracellular DNA Destruction by DNase I on Characteristics of Forming Biofilms. DNA Cell Biol. 2010, 29, 399–405. [Google Scholar] [CrossRef]
- Ibáñez de Aldecoa, A.L.; Zafra, O.; González-Pastor, J.E. Mechanisms and Regulation of Extracellular DNA Release and Its Biological Roles in Microbial Communities. Front. Microbiol. 2017, 8, 1390. [Google Scholar] [CrossRef]
- Pathan, S.I.; Arfaioli, P.; Ceccherini, M.T.; Ascher-Jenull, J.; Pietramellara, G. Preliminary evidences of the presence of extracellular DNA single stranded forms in soil. PLoS ONE 2020, 15, e0227296. [Google Scholar] [CrossRef]
- Lennon, J.T. Diversity and Metabolism of Marine Bacteria Cultivated on Dissolved DNA. AEM 2007, 73, 2799–2805. [Google Scholar] [CrossRef] [PubMed]
- Vuillemin, A.; Horn, F.; Alawi, M.; Henny, C.; Wagner, D.; Crowe, S.A.; Kallmeyer, J. Preservation and Significance of Extracellular DNA in Ferruginous Sediments from Lake Towuti, Indonesia. Front. Microbiol. 2017, 8, 1440. [Google Scholar] [CrossRef]
- Driouich, A.; Follet-Gueye, M.-L.; Vicré-Gibouin, M.; Hawes, M. Root border cells and secretions as critical elements in plant host defense. Curr. Opin. Plant Biol. 2013, 16, 489–495. [Google Scholar] [CrossRef]
- Driouich, A.; Smith, C.; Ropitaux, M.; Chambard, M.; Boulogne, I.; Bernard, S.; Follet-Gueye, M.; Vicré, M.; Moore, J. Root extracellular traps versus neutrophil extracellular traps in host defence, a case of functional convergence? Biol. Rev. 2019, 94, 1685–1700. [Google Scholar] [CrossRef] [PubMed]
- Wen, F.; White, G.J.; VanEtten, H.D.; Xiong, Z.; Hawes, M.C. Extracellular DNA Is Required for Root Tip Resistance to Fungal Infection. Plant Physiol. 2009, 151, 820–829. [Google Scholar] [CrossRef] [PubMed]
- Ropitaux, M.; Bernard, S.; Schapman, D.; Follet-Gueye, M.-L.; Vicré, M.; Boulogne, I.; Driouich, A. Root Border Cells and Mucilage Secretions of Soybean, Glycine Max (Merr) L.: Characterization and Role in Interactions with the Oomycete Phytophthora Parasitica. Cells 2020, 9, 2215. [Google Scholar] [CrossRef] [PubMed]
- Tran, T.M.; MacIntyre, A.; Khokhani, D.; Hawes, M.; Allen, C. Extracellular DNases of Ralstonia solanacearum modulate biofilms and facilitate bacterial wilt virulence: Extracellular DNases modulate R. solanacearum biofilm and virulence. Environ. Microbiol. 2016, 18, 4103–4117. [Google Scholar] [CrossRef] [PubMed]
- Wen, F.; Curlango-Rivera, G.; Huskey, D.A.; Xiong, Z.; Hawes, M.C. Visualization of extracellular DNA released during border cell separation from the root cap. Am. J. Bot. 2017, 104, 970–978. [Google Scholar] [CrossRef]
- Hawes, M.; Allen, C.; Turgeon, B.G.; Curlango-Rivera, G.; Tran, T.M.; Huskey, D.A.; Xiong, Z. Root Border Cells and Their Role in Plant Defense. Annu. Rev. Phytopathol. 2016, 54, 143–161. [Google Scholar] [CrossRef]
- Brunner, F.; Rosahl, S.; Lee, J.; Rudd, J.J.; Geiler, C.; Kauppinen, S.; Rasmussen, G.; Scheel, D.; Nürnberger, T. Pep-13, a plant defense-inducing pathogen-associated pattern from Phytophthora transglutaminases. EMBO J. 2002, 21, 6681–6688. [Google Scholar] [CrossRef]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M.; et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [PubMed]
- Quinlan, A.R.; Hall, I.M. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 2010, 26, 841–842. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550–571. [Google Scholar] [CrossRef] [PubMed]
- Hawes, M.C.; Curlango-Rivera, G.; Wen, F.; White, G.J.; VanEtten, H.D.; Xiong, Z. Extracellular DNA: The tip of root defenses? Plant Sci. 2011, 180, 741–745. [Google Scholar] [CrossRef]
- Parker, J.E.; Hahlbrock, K.; Scheel, D. Different cell-waU components from Phytophthora megasperma f. sp. glycinea elicit phytoalexin production in soybean and parsley. Planta 1988, 176, 75–82. [Google Scholar] [CrossRef]
- Saijo, Y.; Loo, E.P.; Yasuda, S. Pattern recognition receptors and signaling in plant-microbe interactions. Plant J. 2018, 93, 592–613. [Google Scholar] [CrossRef]
- Gao, H.; Bhattacharyya, M.K. The soybean-Phytophthora resistance locus Rps1-k encompasses coiled coil-nucleotide binding-leucine rich repeat-like genes and repetitive sequences. BMC Plant Biol. 2008, 8, 29. [Google Scholar] [CrossRef]
- Song, H.; Sun, W.; Yang, G.; Sun, J. WRKY transcription factors in legumes. BMC Plant Biol. 2018, 18, 243. [Google Scholar] [CrossRef]
- Vicré, M.; Santaella, C.; Blanchet, S.; Gateau, A.; Driouich, A. Root Border-Like Cells of Arabidopsis. Microscopical Characterization and Role in the Interaction with Rhizobacteria. Plant Physiol. 2005, 138, 998–1008. [Google Scholar] [CrossRef]
- Cannesan, M.A.; Gangneux, C.; Lanoue, A.; Giron, D.; Laval, K.; Hawes, M.; Driouich, A.; Vicré-Gibouin, M. Association between border cell responses and localized root infection by pathogenic Aphanomyces euteiches. Ann. Bot. 2011, 108, 459–469. [Google Scholar] [CrossRef] [PubMed]
- Sirois, S.H.; Buckley, D.H. Factors governing extracellular DNA degradation dynamics in soil. Environ. Microbiol. Rep. 2019, 11, 173–184. [Google Scholar] [CrossRef] [PubMed]
- Vinograd, J.; Lebowitz, J. Physical and Topological Properties of Circular DNA. J. Gen. Physiol. 1966, 49, 103–125. [Google Scholar] [CrossRef]
- Yakovchuk, P. Base-stacking and base-pairing contributions into thermal stability of the DNA double helix. Nucleic Acids Res. 2006, 34, 564–574. [Google Scholar] [CrossRef] [PubMed]
- Schmutz, J.; Cannon, S.B.; Schlueter, J.; Ma, J.; Mitros, T.; Nelson, W.; Hyten, D.L.; Song, Q.; Thelen, J.J.; Cheng, J.; et al. Genome sequence of the palaeopolyploid soybean. Nature 2010, 463, 178–183. [Google Scholar] [CrossRef]
- Chang, S.; Wang, Y.; Lu, J.; Gai, J.; Li, J.; Chu, P.; Guan, R.; Zhao, T. The Mitochondrial Genome of Soybean Reveals Complex Genome Structures and Gene Evolution at Intercellular and Phylogenetic Levels. PLoS ONE 2013, 8, e56502. [Google Scholar]
- Saski, C.; Lee, S.-B.; Daniell, H.; Wood, T.C.; Tomkins, J.; Kim, H.-G.; Jansen, R.K. Complete Chloroplast Genome Sequence of Glycine max and Comparative Analyses with other Legume Genomes. Plant Mol. Biol. 2005, 59, 309–322. [Google Scholar] [CrossRef]
- Ogram, A.; Sayler, G.S.; Gustin, D.; Lewis, R.J. DNA adsorption to soils and sediments. Environ. Sci. Technol. 1988, 22, 982–984. [Google Scholar] [CrossRef]
- Douarche, C. Étude de L’adsorption de l’ADN Simple Brin et Double Brin Aux Interfaces; Université de Lille: Lille, France, 2007. [Google Scholar]
- Dangl, M.; Brosch, G.; Haas, H.; Loidl, P.; Lusser, A. Comparative analysis of HD2 type histone deacetylases in higher plants. Planta 2001, 213, 280–285. [Google Scholar] [CrossRef]
- Kochanek, S.; Renz, D.; Doerfler, W. Differences in the accessibility of methylated and unmethylated DNA to DNase I. Nucl. Acids Res. 1993, 21, 5843–5845. [Google Scholar] [CrossRef][Green Version]
- Fulneček, J.; Matyasek, R.; Kovarık, A. Distribution of 5-methylcytosine residues in 5S rRNA genes in Arabidopsis thaliana and Secale cereale. Mol. Genet. Genom. 2002, 268, 510–517. [Google Scholar] [CrossRef] [PubMed]
- Vanyushin, B.F. DNA Methylation in Plants. In DNA Methylation: Basic Mechanisms; Doerfler, W., Böhm, P., Eds.; Current Topics in Microbiology and Immunology; Springer: Berlin/Heidelberg, Germany, 2006; pp. 67–122. [Google Scholar]
- Yuan, D.-H.; Xing, J.-F.; Luan, M.-W.; Ji, K.-K.; Guo, J.; Xie, S.-Q.; Zhang, Y.-M. DNA N6-Methyladenine Modification in Wild and Cultivated Soybeans Reveals Different Patterns in Nucleus and Cytoplasm. Front. Genet. 2020, 11, 736. [Google Scholar] [CrossRef] [PubMed]
- Lood, C.; Blanco, L.P.; Purmalek, M.M.; Carmona-Rivera, C.; De Ravin, S.S.; Smith, C.K.; Malech, H.L.; Ledbetter, J.A.; Elkon, K.B.; Kaplan, M.J. Neutrophil extracellular traps enriched in oxidized mitochondrial DNA are interferogenic and contribute to lupus-like disease. Nat. Med. 2016, 22, 146–153. [Google Scholar] [CrossRef] [PubMed]
- Solymosi, K.; Lethin, J.; Aronsson, H. Diversity and Plasticity of Plastids in Land Plants. In Plastids; Maréchal, E., Ed.; Methods in Molecular Biology; Springer: New York, NY, USA, 2018; Volume 1829, pp. 55–72. [Google Scholar]
- Boffey, S.A.; Lloyd, D. Division and Segregation of Organelles; Cambridge University Press: Cambridge, UK, 1988. [Google Scholar]
- Okazaki, K.; Kabeya, Y.; Suzuki, K.; Mori, T.; Ichikawa, T.; Matsui, M.; Nakanishi, H.; Miyagishima, S. The PLASTID DIVISION1 and 2 Components of the Chloroplast Division Machinery Determine the Rate of Chloroplast Division in Land Plant Cell Differentiation. Plant Cell 2009, 21, 1769–1780. [Google Scholar] [CrossRef]
- Chang, N.; Sun, Q.; Li, Y.; Mu, Y.; Hu, J.; Feng, Y.; Liu, X.; Gao, H. PDV2 has a dosage effect on chloroplast division in Arabidopsis. Plant Cell Rep. 2017, 36, 471–480. [Google Scholar] [CrossRef]
- Wala, J.A.; Bandopadhayay, P.; Greenwald, N.F.; O’Rourke, R.; Sharpe, T.; Stewart, C.; Schumacher, S.; Li, Y.; Weischenfeldt, J.; Yao, X.; et al. SvABA: Genome-wide detection of structural variants and indels by local assembly. Genome Res. 2018, 28, 581–591. [Google Scholar] [CrossRef]
- Hurles, M. Gene Duplication: The Genomic Trade in Spare Parts. PLoS Biol. 2004, 2, e206. [Google Scholar] [CrossRef]
- Lefevre, T.; Raymond, M.; Thomas, F. Biologie Évolutive; Biologie; De Boeck Supérieur: Paris, France, 2016. [Google Scholar]
- Duran-Flores, D.; Heil, M. Extracellular self-DNA as a damage-associated molecular pattern (DAMP) that triggers self-specific immunity induction in plants. Brain Behav. Immun. 2017, 72, 78–88. [Google Scholar] [CrossRef]

| Cellular Structure | exDNA | |
|---|---|---|
| Counts Mean | Coverage (X) | |
| Nuclear DNA | 1,177,342.00 | 0.25 |
| Mitochondrial DNA | 35,765.33 | 17.88 |
| Plastidial DNA | 11,777.67 | 15.70 |
| Chromosome | Position (pb) | Condition | Counts Mean | Log2 (FC) | p-Value | Identification in Soybean or Blast (% Identity) | |
|---|---|---|---|---|---|---|---|
| START | STOP | ||||||
| Gm07 | 26,655,001 | 27,656,000 | E | 8 | 3.8293 | 0.0021 | PREDICTED: Glycine max putative dual specificity protein phosphatase DSP8 |
| NE | 0 | ||||||
| Gm15 | 15,541,001 | 15,542,000 | E | 8 | 3.7595 | 0.0028 | N/A |
| NE | 0 | ||||||
| Gm07 | 7,109,001 | 7,110,000 | E | 7 | 3.7505 | 0.0028 | N/A |
| NE | 0 | ||||||
| Gm09 | 27,858,001 | 27,859,000 | E | 7 | 3.7060 | 0.0033 | 85.88% Glycine max retrotransposon gmw1-45m6-re-2 |
| NE | 0 | ||||||
| Gm16 | 14,656,001 | 14,657,000 | E | 7 | 3.6978 | 0.0034 | 87.13% PREDICTED: Glycine soja organic cation/carnitine transporter 7-like |
| NE | 0 | ||||||
| Gm05 | 2,829,001 | 2,830,000 | E | 7 | 3.6796 | 0.0035 | N/A |
| NE | 0 | ||||||
| Gm08 | 12,008,001 | 12,009,000 | E | 7 | 3.6777 | 0.0036 | Auxin-induced protein 6B-like |
| NE | 0 | ||||||
| Gm01 | 51,034,001 | 51,035,000 | E | 7 | 3.6873 | 0.0036 | Phosphopantethine adenyltransferase |
| NE | 0 | ||||||
| Gm13 | 23,974,001 | 23,975,000 | E | 7 | 3.6657 | 0.0037 | 60S ribosomal protein L23 |
| NE | 0 | ||||||
| Gm14 | 46,813,001 | 46,814,000 | E | 7 | 3.6904 | 0.0020 | N/A |
| NE | 0 | ||||||
| Gm20 | 45,220,001 | 45,221,000 | E | 0 | −3.8522 | 0.0022 | N/A |
| NE | 9 | ||||||
| Gm07 | 22,405,001 | 22,406,000 | E | 0 | −3.8070 | 0.0022 | N/A |
| NE | 8 | ||||||
| Gm13 | 15,882,001 | 15,883,000 | E | 0 | −3.7247 | 0.0029 | N/A |
| NE | 8 | ||||||
| Gm14 | 20,131,001 | 20,132,000 | E | 0 | −3.6824 | 0.0036 | N/A |
| NE | 8 | ||||||
| Gm01 | 29,642,001 | 29,643,000 | E | 0 | −3.6575 | 0.0036 | N/A |
| NE | 7 | ||||||
| Gm19 | 26,749,001 | 26,750,000 | E | 0 | −3.6457 | 0.0038 | N/A |
| NE | 7 | ||||||
| Gm09 | 46,814,001 | 46,815,000 | E | 0 | −3.6416 | 0.0037 | TPR (tetratricopeptide) repeat-containing protein ZIP4-like |
| NE | 7 | ||||||
| Gm15 | 28,346,001 | 28,347,000 | E | 0 | −3.6416 | 0.0037 | N/A |
| NE | 7 | ||||||
| Gm12 | 19,408,001 | 19,409,000 | E | 0 | −3.6362 | 0.0040 | N/A |
| NE | 7 | ||||||
| Gm16 | 127,001 | 128,000 | E | 0 | −3.6279 | 0.0040 | Shaggy-related protein kinase kappa-like |
| NE | 4 | ||||||
| Chromosome | Position (pb) | Identification in Soybean or Blast (% Identity) | Comment | |
|---|---|---|---|---|
| START | STOP | |||
| Gm01 | 15,120,001 | 15,121,000 | N/A | Repeated sequence, identified only in Glycine max |
| Gm02 | 21,073,001 | 21,077,000 | LOC100785390 protein kinase and PP2C-like (protein phosphatase 2C) domain-containing protein [Glycine max (soybean)] | Kinases involved in plant defense response [37] |
| GM04 | 26,981,001 | 26,983,000 | N/A | Repeated sequence, identified only in Glycine max and Glycine soja |
| Gm09 | 25,428,001 | 25,429,000 | 79.66% Pisum sativum clone Ps-phage20 Ogre retrotransposon, partial sequence | Identified only in Fabaceae |
| Gm09 | 23,598,001 | 23,599,000 | 96.05% Glycine max NB-LRR (nucleotide binding—leucine rich repeat) type disease resistance protein Rps1-k-1 (Rps1-k-1) and NB-LRR type disease resistance protein Rps1-k-2 (Rps1-k-2) | Repeated sequence, involved in plant defense against Phytophthora sojae. Might be near or a part of heterochromatic DNA [38]. Identified only in Glycine max |
| Gm10 | 21,046,001 | 21,047,000 | 93.01% PREDICTED: Glycine soja probable WRKY transcription factor 11 (LOC114391634), mRNA | Transcription factor WRKY involved in defense response [39]. Identified only in Glycine max |
| Gm17 | 25,874,001 | 25,877,000 | 97.96% PREDICTED: Glycine soja L-ascorbate oxidase homolog (LOC114411184), mRNA | Identified in Fabaceae and malvaceae (Gossypium) |
| Gm18 | 32,363,001 | 32,364,000 | 72.09% PREDICTED: Glycine soja uncharacterized LOC114387464, ncRNA | Repeated sequence, identified only in Glycine max |
| Gm18 | 32,392,001 | 32,393,000 | N/A | Repeated sequence |
| Gm19 | 24,013,001 | 24,014,000 | Triacyl-glycerol lipase 2 | Identified only in Fabaceae |
| Gm20 | 20,695,001 | 20,697,000 | N/A | Repeated sequence, identified only in Glycine max |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chambard, M.; Plasson, C.; Derambure, C.; Coutant, S.; Tournier, I.; Lefranc, B.; Leprince, J.; Kiefer-Meyer, M.-C.; Driouich, A.; Follet-Gueye, M.-L.; et al. New Insights into Plant Extracellular DNA. A Study in Soybean Root Extracellular Trap. Cells 2021, 10, 69. https://doi.org/10.3390/cells10010069
Chambard M, Plasson C, Derambure C, Coutant S, Tournier I, Lefranc B, Leprince J, Kiefer-Meyer M-C, Driouich A, Follet-Gueye M-L, et al. New Insights into Plant Extracellular DNA. A Study in Soybean Root Extracellular Trap. Cells. 2021; 10(1):69. https://doi.org/10.3390/cells10010069
Chicago/Turabian StyleChambard, Marie, Carole Plasson, Céline Derambure, Sophie Coutant, Isabelle Tournier, Benjamin Lefranc, Jérôme Leprince, Marie-Christine Kiefer-Meyer, Azeddine Driouich, Marie-Laure Follet-Gueye, and et al. 2021. "New Insights into Plant Extracellular DNA. A Study in Soybean Root Extracellular Trap" Cells 10, no. 1: 69. https://doi.org/10.3390/cells10010069
APA StyleChambard, M., Plasson, C., Derambure, C., Coutant, S., Tournier, I., Lefranc, B., Leprince, J., Kiefer-Meyer, M.-C., Driouich, A., Follet-Gueye, M.-L., & Boulogne, I. (2021). New Insights into Plant Extracellular DNA. A Study in Soybean Root Extracellular Trap. Cells, 10(1), 69. https://doi.org/10.3390/cells10010069





