Identification of Targets from LRRK2 Rescue Phenotypes
Abstract
:1. Introduction
2. Materials and Methods
2.1. Fly Stocks
2.2. Generation of Transgenic Strains
2.3. Western Blot
2.4. Immunofluorescence and Confocal Microscopy
2.5. Climbing and Lifespan Assays
2.6. TU Tagging
2.7. Library Preparation and RNA-Sequencing
2.8. Alignment Coverage Analysis
2.9. Differential Transcript Analysis
2.10. Pathway Analysis
2.11. Statistical Analysis
2.12. Real-Time PCR
3. Results
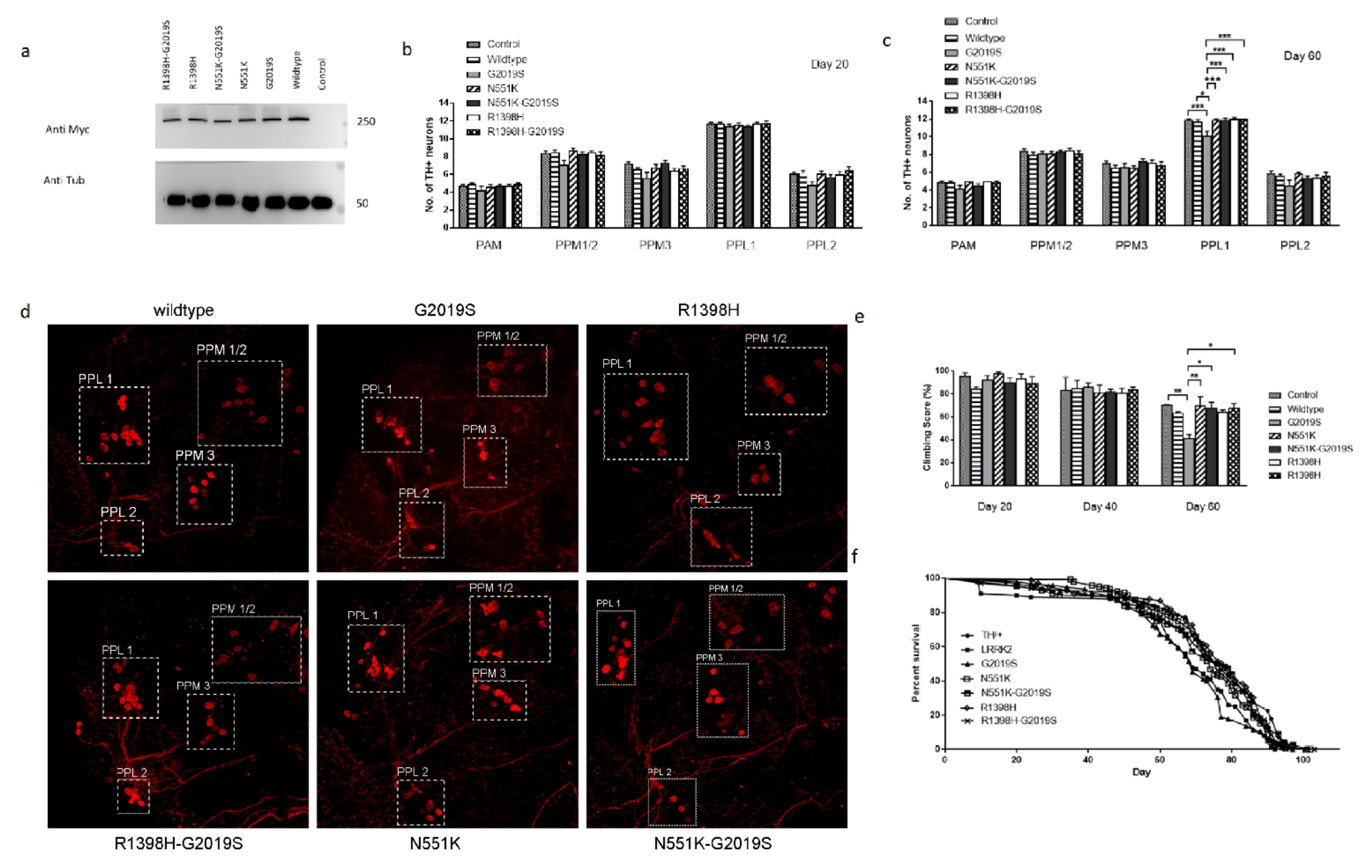
3.1. N551K and R1398H Variants Protected DA Integrity In Vivo
3.2. Molecular Analyses Using RNA-Seq Data Acquired from DA Neurons of Transgenic Fly Mutants Identify New Pathways and Targets
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Spillantini, M.G.; Crowther, R.A.; Jakes, R.; Hasegawa, M.; Goedert, M. α-Synuclein in filamentous inclusions of Lewy bodies from Parkinson’s disease and dementia with Lewy bodies. Proc. Natl. Acad. Sci. USA 1998, 95, 6469–6473. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kondo, K.; Obitsu, S.; Teshima, R. α-Synuclein aggregation and transmission are enhanced by leucine-rich repeat kinase 2 in human neuroblastoma SH-SY5Y cells. Biol. Pharm. Bull. 2011. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guerreiro, P.S.; Huang, Y.; Gysbers, A.; Cheng, D.; Gai, W.P.; Outeiro, T.F.; Halliday, G.M. LRRK2 interactions with α-synuclein in Parkinson’s disease brains and in cell models. J. Mol. Med. 2013. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bosgraaf, L.; Van Haastert, P.J.M. Roc, a Ras/GTPase domain in complex proteins. Biochim. Biophys. Acta (BBA) Mol. Cell Res. 2003, 1643, 5–10. [Google Scholar] [CrossRef] [Green Version]
- Mills, R.D.; Mulhern, T.D.; Liu, F.; Culvenor, J.G.; Cheng, H.-C. Prediction of the Repeat Domain Structures and Impact of Parkinsonism-Associated Variations on Structure and Function of all Functional Domains of Leucine-Rich Repeat Kinase 2 (LRRK2). Hum. Mutat. 2014, 35, 395–412. [Google Scholar] [CrossRef]
- Di Fonzo, A.; Rohé, C.F.; Ferreira, J.; Chien, H.F.; Vacca, L.; Stocchi, F.; Guedes, L.; Fabrizio, E.; Manfredi, M.; Vanacore, N.; et al. A frequent LRRK2 gene mutation associated with autosomal dominant Parkinson’s disease. Lancet 2005, 365, 412–415. [Google Scholar] [CrossRef]
- Gilks, W.P.; Abou-Sleiman, P.M.; Gandhi, S.; Jain, S.; Singleton, A.; Lees, A.J.; Shaw, K.; Bhatia, K.P.; Bonifati, V.; Quinn, N.P.; et al. A common LRRK2 mutation in idiopathic Parkinson’s disease. Lancet 2005, 365, 415–416. [Google Scholar] [CrossRef]
- Gloeckner, C.J.; Kinkl, N.; Schumacher, A.; Braun, R.J.; O’Neill, E.; Meitinger, T.; Kolch, W.; Prokisch, H.; Ueffing, M. The Parkinson disease causing LRRK2 mutation I2020T is associated with increased kinase activity. Hum. Mol. Genet. 2006. [Google Scholar] [CrossRef]
- Li, N.N.; Tan, E.K.; Chang, X.L.; Mao, X.Y.; Zhang, J.H.; Zhao, D.M.; Liao, Q.; Peng, R. Genetic analysis of LRRK2 A419V variant in ethnic Chinese. Neurobiol. Aging 2012. [Google Scholar] [CrossRef]
- Li, K.; Tang, B.S.; Liu, Z.H.; Kang, J.F.; Zhang, Y.; Shen, L.; Li, N.; Yan, X.X.; Xia, K.; Guo, J.F. LRRK2 A419V variant is a risk factor for Parkinson’s disease in Asian population. Neurobiol. Aging 2015. [Google Scholar] [CrossRef]
- Wang, C.; Cai, Y.; Zheng, Z.; Tang, B.S.; Xu, Y.; Wang, T.; Ma, J.; Chen, S.D.; Langston, J.W.; Tanner, C.M.; et al. Penetrance of LRRK2 G2385R and R1628P is modified by common PD-associated genetic variants. Parkinsonism Relat. Disord. 2012. [Google Scholar] [CrossRef] [PubMed]
- Ross, O.A.; Wu, Y.R.; Lee, M.C.; Funayama, M.; Chen, M.L.; Soto, A.I.; Mata, I.F.; Lee-Chen, G.J.; Chiung, M.C.; Tang, M.; et al. Analysis of Lrrk2 R1628P as a risk factor for Parkinson’s disease. Ann. Neurol. 2008. [Google Scholar] [CrossRef] [PubMed]
- Tan, E.K.; Zhao, Y.; Tan, L.; Lim, H.Q.; Lee, J.; Yuen, Y.; Pavanni, R.; Wong, M.C.; Fook-Chong, S.; Liu, J.J. Analysis of LRRK2 Gly2385Arg genetic variant in non-Chinese Asians. Mov. Disord. 2007. [Google Scholar] [CrossRef] [PubMed]
- Zimprich, A.; Biskup, S.; Leitner, P.; Lichtner, P.; Farrer, M.; Lincoln, S.; Kachergus, J.; Hulihan, M.; Uitti, R.J.; Calne, D.B.; et al. Mutations in LRRK2 cause autosomal-dominant parkinsonism with pleomorphic pathology. Neuron 2004. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singh, A.; Zhi, L.; Zhang, H. LRRK2 and mitochondria: Recent advances and current views. Brain Res. 2019. [Google Scholar] [CrossRef]
- Saha, S.; Guillily, M.D.; Ferree, A.; Lanceta, J.; Chan, D.; Ghosh, J.; Hsu, C.H.; Segal, L.; Raghavan, K.; Matsumoto, K.; et al. LRRK2 modulates vulnerability to mitochondrial dysfunction in Caenorhabditis elegans. J. Neurosci. 2009, 29, 9210–9218. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Yan, M.H.; Fujioka, H.; Liu, J.; Wilson-Delfosse, A.; Chen, S.G.; Perry, G.; Casadesus, G.; Zhu, X. LRRK2 regulates mitochondrial dynamics and function through direct interaction with DLP1. Hum. Mol. Genet. 2012, 21, 1931–1944. [Google Scholar] [CrossRef] [Green Version]
- Gillardon, F. Interaction of elongation factor 1-alpha with leucine-rich repeat kinase 2 impairs kinase activity and microtubule bundling in vitro. Neuroscience 2009, 163, 533–539. [Google Scholar] [CrossRef]
- Parisiadou, L.; Xie, C.; Cho, H.J.; Lin, X.; Gu, X.-L.; Long, C.-X.; Lobbestael, E.; Baekelandt, V.; Taymans, J.-M.; Sun, L.; et al. Phosphorylation of Ezrin/Radixin/Moesin Proteins by LRRK2 Promotes the Rearrangement of Actin Cytoskeleton in Neuronal Morphogenesis. J. Neurosci. 2009, 29, 13971–13980. [Google Scholar] [CrossRef]
- Civiero, L.; Cogo, S.; Biosa, A.; Greggio, E. The role of LRRK2 in cytoskeletal dynamics. Biochem. Soc. Trans. 2018, 46, 1653–1663. [Google Scholar] [CrossRef]
- Bravo-San Pedro, J.M.; Niso-Santano, M.; Gómez-Sánchez, R.; Pizarro-Estrella, E.; Aiastui-Pujana, A.; Gorostidi, A.; Climent, V.; López De Maturana, R.; Sanchez-Pernaute, R.; López De Munain, A.; et al. The LRRK2 G2019S mutant exacerbates basal autophagy through activation of the MEK/ERK pathway. Cell. Mol. Life Sci. 2013, 70, 121–136. [Google Scholar] [CrossRef]
- Orenstein, S.J.; Kuo, S.H.; Tasset, I.; Arias, E.; Koga, H.; Fernandez-Carasa, I.; Cortes, E.; Honig, L.S.; Dauer, W.; Consiglio, A.; et al. Interplay of LRRK2 with chaperone-mediated autophagy. Nat. Neurosci. 2013. [Google Scholar] [CrossRef] [Green Version]
- Berwick, D.C.; Harvey, K. LRRK2 functions as a Wnt signaling scaffold, bridging cytosolic proteins and membrane-localized LRP6. Hum. Mol. Genet. 2012, 21, 4966–4979. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sancho, R.M.; Law, B.M.H.; Harvey, K. Mutations in the LRRK2 Roc-COR tandem domain link Parkinson’s disease to Wnt signalling pathways. Hum. Mol. Genet. 2009. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wallings, R.; Manzoni, C.; Bandopadhyay, R. Cellular processes associated with LRRK2 function and dysfunction. FEBS J. 2015, 282, 2806–2826. [Google Scholar] [CrossRef]
- Angeles, D.C.; Gan, B.H.; Onstead, L.; Zhao, Y.; Lim, K.L.; Dachsel, J.; Melrose, H.; Farrer, M.; Wszolek, Z.K.; Dickson, D.W.; et al. Mutations in LRRK2 increase phosphorylation of peroxiredoxin 3 exacerbating oxidative stress-induced neuronal death. Hum. Mutat. 2011, 32, 1390–1397. [Google Scholar] [CrossRef]
- Chua, L.L.; Ho, P.; Toh, J.; Tan, E.K. Chetomin rescues pathogenic phenotype of LRRK2 mutation in drosophila. Aging 2020. [Google Scholar] [CrossRef] [PubMed]
- West, A.B.; Moore, D.J.; Biskup, S.; Bugayenko, A.; Smith, W.W.; Ross, C.A.; Dawson, V.L.; Dawson, T.M. Parkinson’s disease-associated mutations in leucine-rich repeat kinase 2 augment kinase activity. Proc. Natl. Acad. Sci. USA 2005, 102, 16842–16847. [Google Scholar] [CrossRef] [Green Version]
- Russo, I.; Di Benedetto, G.; Kaganovich, A.; Ding, J.; Mercatelli, D.; Morari, M.; Cookson, M.R.; Bubacco, L.; Greggio, E. Leucine-rich repeat kinase 2 controls protein kinase A activation state through phosphodiesterase 4. J. Neuroinflamm. 2018. [Google Scholar] [CrossRef]
- Anand, V.S.; Reichling, L.J.; Lipinski, K.; Stochaj, W.; Duan, W.; Kelleher, K.; Pungaliya, P.; Brown, E.L.; Reinhart, P.H.; Somberg, R.; et al. Investigation of leucine-rich repeat kinase 2: Enzymological properties and novel assays. FEBS J. 2009. [Google Scholar] [CrossRef]
- Jaleel, M.; Nichols, R.J.; Deak, M.; Campbell, D.G.; Gillardon, F.; Knebel, A.; Alessi, D.R. LRRK2 phosphorylates moesin at threonine-558: Characterization of how Parkinson’s disease mutants affect kinase activity. Biochem. J. 2007. [Google Scholar] [CrossRef] [PubMed]
- Rudenko, I.N.; Chia, R.; Cookson, M.R. Is inhibition of kinase activity the only therapeutic strategy for LRRK2-associated Parkinson’s disease? BMC Med. 2012, 10, 20. [Google Scholar] [CrossRef] [PubMed]
- Volpicelli-Daley, L.A.; Abdelmotilib, H.; Liu, Z.; Stoyka, L.; Daher, J.P.L.; Milnerwood, A.J.; Unni, V.K.; Hirst, W.D.; Yue, Z.; Zhao, H.T.; et al. G2019s-LRRK2 expression augments α-synuclein sequestration into inclusions in neurons. J. Neurosci. 2016, 36, 7415–7427. [Google Scholar] [CrossRef] [PubMed]
- Healy, D.G.; Falchi, M.; O’Sullivan, S.S.; Bonifati, V.; Durr, A.; Bressman, S.; Brice, A.; Aasly, J.; Zabetian, C.P.; Goldwurm, S.; et al. Phenotype, genotype, and worldwide genetic penetrance of LRRK2-associated Parkinson’s disease: A case-control study. Lancet Neurol. 2008. [Google Scholar] [CrossRef] [Green Version]
- Nguyen, A.P.T.; Moore, D.J. Understanding the GTPase activity of LRRK2: Regulation, function, and neurotoxicity. In Advances in Neurobiology; Springer: Cham, Switzerland, 2017. [Google Scholar]
- Tan, E.K.; Peng, R.; Teo, Y.Y.; Tan, L.C.; Angeles, D.; Ho, P.; Chen, M.L.; Lin, C.H.; Mao, X.Y.; Chang, X.L.; et al. Multiple LRRK2 variants modulate risk of Parkinson disease: A Chinese multicenter study. Hum. Mutat. 2010. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.R.; Chang, K.H.; Chang, W.T.; Hsiao, Y.C.; Hsu, H.C.; Jiang, P.R.; Chen, Y.C.; Chao, C.Y.; Chang, Y.C.; Lee, B.H.; et al. Genetic variants of LRRK2 in Taiwanese Parkinson’s disease. PLoS ONE 2013. [Google Scholar] [CrossRef] [Green Version]
- Gopalai, A.A.; Lim, J.L.; Li, H.H.; Zhao, Y.; Lim, T.T.; Eow, G.B.; Puvanarajah, S.; Viswanathan, S.; Norlinah, M.I.; Abdul Aziz, Z.; et al. LRRK2 N551K and R1398H variants are protective in Malays and Chinese in Malaysia: A case–control association study for Parkinson’s disease. Mol. Genet. Genom. Med. 2019. [Google Scholar] [CrossRef]
- Hui, K.Y.; Fernandez-Hernandez, H.; Hu, J.; Schaffner, A.; Pankratz, N.; Hsu, N.Y.; Chuang, L.S.; Carmi, S.; Villaverde, N.; Li, X.; et al. Functional variants in the LRRK2 gene confer shared effects on risk for Crohn’s disease and Parkinson’s disease. Sci. Transl. Med. 2018. [Google Scholar] [CrossRef] [Green Version]
- Nixon-Abell, J.; Berwick, D.C.; Grannó, S.; Spain, V.A.; Blackstone, C.; Harvey, K. Protective LRRK2 R1398H variant enhances GTPase and Wnt signaling activity. Front. Mol. Neurosci. 2016. [Google Scholar] [CrossRef] [Green Version]
- Berwick, D.C.; Javaheri, B.; Wetzel, A.; Hopkinson, M.; Nixon-Abell, J.; Grannò, S.; Pitsillides, A.A.; Harvey, K. Pathogenic LRRK2 variants are gain-of-function mutations that enhance LRRK2-mediated repression of β-catenin signaling. Mol. Neurodegener. 2017. [Google Scholar] [CrossRef] [Green Version]
- Angeles, D.C.; Ho, P.; Chua, L.L.; Wang, C.; Yap, Y.W.; Ng, C.; Zhou, Z.D.; Lim, K.-L.; Wszolek, Z.K.; Wang, H.Y.; et al. Thiol peroxidases ameliorate LRRK2 mutant-induced mitochondrial and dopaminergic neuronal degeneration in Drosophila. Hum. Mol. Genet. 2014, 23, 3157–3165. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miller, M.R.; Robinson, K.J.; Cleary, M.D.; Doe, C.Q. TU-tagging: Cell type-specific RNA isolation from intact complex tissues. Nat. Methods 2009. [Google Scholar] [CrossRef] [PubMed]
- Trapnell, C.; Pachter, L.; Salzberg, S.L. TopHat: Discovering splice junctions with RNA-Seq. Bioinformatics 2009. [Google Scholar] [CrossRef] [PubMed]
- Trapnell, C.; Roberts, A.; Goff, L.; Pertea, G.; Kim, D.; Kelley, D.R.; Pimentel, H.; Salzberg, S.L.; Rinn, J.L.; Pachter, L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goff, L.A.; Trapnell, C.; Kelley, D. CummeRbund: Visualization and Exploration of Cufflinks High-Throughput Sequencing Data; R Package Version; R Foundation: Vienna, Austria, 2012. [Google Scholar]
- Ross, O.A.; Soto-Ortolaza, A.I.; Heckman, M.G.; Aasly, J.O.; Abahuni, N.; Annesi, G.; Bacon, J.A.; Bardien, S.; Bozi, M.; Brice, A.; et al. Association of LRRK2 exonic variants with susceptibility to Parkinson’s disease: A case-control study. Lancet Neurol. 2011. [Google Scholar] [CrossRef] [Green Version]
- Infante, J.; Prieto, C.; Sierra, M.; Sánchez-Juan, P.; González-Aramburu, I.; Sánchez-Quintana, C.; Berciano, J.; Combarros, O.; Sainz, J. Comparative blood transcriptome analysis in idiopathic and LRRK2 G2019S-associated Parkinson’s disease. Neurobiol. Aging 2015. [Google Scholar] [CrossRef]
- Infante, J.; Prieto, C.; Sierra, M.; Sánchez-Juan, P.; González-Aramburu, I.; Sánchez-Quintana, C.; Berciano, J.; Combarros, O.; Sainz, J. Identification of candidate genes for Parkinson’s disease through blood transcriptome analysis in LRRK2-G2019S carriers, idiopathic cases, and controls. Neurobiol. Aging 2015. [Google Scholar] [CrossRef]
- Dhekne, H.S.; Yanatori, I.; Gomez, R.C.; Tonelli, F.; Diez, F.; Schüle, B.; Steger, M.; Alessi, D.R.; Pfeffer, S.R. A pathway for parkinson’s disease LRRK2 kinase to block primary cilia and sonic hedgehog signaling in the brain. eLife 2018. [Google Scholar] [CrossRef]
- Schulz, C.; Paus, M.; Frey, K.; Schmid, R.; Kohl, Z.; Mennerich, D.; Winkler, J.; Gillardon, F. Leucine-rich repeat kinase 2 modulates retinoic acid-induced neuronal differentiation of murine embryonic stem cells. PLoS ONE 2011. [Google Scholar] [CrossRef]
- Rideout, H.J. Neuronal death signaling pathways triggered by mutant LRRK2. Biochem. Soc. Trans. 2017, 45, 123–129. [Google Scholar] [CrossRef]
- Cho, H.J.; Xie, C.; Cai, H. AGE-induced neuronal cell death is enhanced in G2019S LRRK2 mutation with increased RAGE expression. Transl. Neurodegener. 2018. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Melachroinou, K.; Leandrou, E.; Valkimadi, P.E.; Memou, A.; Hadjigeorgiou, G.; Stefanis, L.; Rideout, H.J. Activation of FADD-Dependent neuronal death pathways as a predictor of pathogenicity for LRRK2 mutations. PLoS ONE 2016. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Salašová, A.; Yokota, C.; Potěšil, D.; Zdráhal, Z.; Bryja, V.; Arenas, E. A proteomic analysis of LRRK2 binding partners reveals interactions with multiple signaling components of the WNT/PCP pathway. Mol. Neurodegener. 2017. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ramonet, D.; Dietz, G.P.H. Novel Cell-Based Assay for Identification of LRRK2 Inhibitors Using Its Aberrant Regulation of a Pluripotency Gene. Slas Discov. 2020. [Google Scholar] [CrossRef] [PubMed]
- Russo, I.; Kaganovich, A.; Ding, J.; Landeck, N.; Mamais, A.; Varanita, T.; Biosa, A.; Tessari, I.; Bubacco, L.; Greggio, E.; et al. Transcriptome analysis of LRRK2 knock-out microglia cells reveals alterations of inflammatory- and oxidative stress-related pathways upon treatment with α-synuclein fibrils. Neurobiol. Dis. 2019. [Google Scholar] [CrossRef]
- Häbig, K.; Walter, M.; Poths, S.; Riess, O.; Bonin, M. RNA interference of LRRK2-microarray expression analysis of a Parkinson’s disease key player. Neurogenetics 2008. [Google Scholar] [CrossRef]
- Botta-Orfila, T.; Tolosa, E.; Gelpi, E.; Sànchez-Pla, A.; Martí, M.J.; Valldeoriola, F.; Fernández, M.; Carmona, F.; Ezquerra, M. Microarray expression analysis in idiopathic and LRRK2-associated Parkinson’s disease. Neurobiol. Dis. 2012. [Google Scholar] [CrossRef]
- Parisiadou, L.; Yu, J.; Sgobio, C.; Xie, C.; Liu, G.; Sun, L.; Gu, X.L.; Lin, X.; Crowley, N.A.; Lovinger, D.M.; et al. LRRK2 regulates synaptogenesis and dopamine receptor activation through modulation of PKA activity. Nat. Neurosci. 2014. [Google Scholar] [CrossRef] [Green Version]
- Pandey, N.; Fahey, M.T.; Jong, Y.J.I.; O’Malley, K.L. Sequences Located within the N-Terminus of the PD-Linked LRRK2 Lead to Increased Aggregation and Attenuation of 6-Hydroxydopamine-Induced Cell Death. PLoS ONE 2012. [Google Scholar] [CrossRef] [Green Version]
- Xiong, Y.; Dawson, V.L.; Dawson, T.M. LRRK2 GTPase dysfunction in the pathogenesis of Parkinson’s disease. Biochem. Soc. Trans. 2012, 40, 1074–1079. [Google Scholar] [CrossRef] [Green Version]
- Yang, D.; Li, T.; Liu, Z.; Arbez, N.; Yan, J.; Moran, T.H.; Ross, C.A.; Smith, W.W. LRRK2 kinase activity mediates toxic interactions between genetic mutation and oxidative stress in a Drosophila model: Suppression by curcumin. Neurobiol. Dis. 2012. [Google Scholar] [CrossRef] [PubMed]
- Tsika, E.; Moore, D.J. Contribution of GTPase activity to LRRK2-associated Parkinson disease. Small GTPases 2013, 4, 164–170. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, Y.M.; Park, S.H.; Shin, D.-I.; Hwang, J.-Y.; Park, B.; Park, Y.-J.; Lee, T.H.; Chae, H.Z.; Jin, B.K.; Oh, T.H.; et al. Oxidative Modification of Peroxiredoxin Is Associated with Drug-induced Apoptotic Signaling in Experimental Models of Parkinson Disease. J. Biol. Chem. 2008, 283, 9986–9998. [Google Scholar] [CrossRef] [PubMed] [Green Version]



| Category | Term | Count | % | p-Value | Ensembl ID | Gene Symbol | List Total | Fold Enrichment | FDR p-Value |
|---|---|---|---|---|---|---|---|---|---|
| SP_PIR_KEYWORDS | oxidoreductase | 11 | 9.166666667 | 0.002426169 | ENSG00000104808, ENSG00000090013, ENSG00000167815, ENSG00000101365, ENSG00000137992, ENSG00000197601, ENSG00000122787, ENSG00000151376, ENSG00000129151, ENSG00000196177, ENSG00000163738 | DHDH, BLVRB, PRDX2, IDH3B, DBT, FAR1, AKR1D1, ME3, BBOX1, ACADSB, MTHFD2L | 119 | 3.163745925 | 0.035624767 |
| GOTERM_BP_FAT | GO:0055114~oxidation reduction | 11 | 9.166666667 | 0.022170929 | ENSG00000104808, ENSG00000090013, ENSG00000167815, ENSG00000101365, ENSG00000197601, ENSG00000122787, ENSG00000151376, ENSG00000129151, ENSG00000196177, ENSG00000163738, ENSG00000023909 | DHDH, BLVRB, PRDX2, IDH3B, FAR1, AKR1D1, ME3, BBOX1, ACADSB, MTHFD2L, GCLM | 103 | 2.260935625 | 0.467306665 |
| INTERPRO | IPR016040:NAD(P)-binding domain | 7 | 5.833333333 | 0.000634 | ENSG00000104808, ENSG00000090013, ENSG00000101444, ENSG00000197601, ENSG00000151376, ENSG00000124217, ENSG00000163738 | DHDH, BLVRB, AHCY, FAR1, ME3, MOCS3, MTHFD2L | 117 | 6.644615385 | 0.169621933 |
| SP_PIR_KEYWORDS | nadp | 6 | 5 | 0.002759564 | ENSG00000104808, ENSG00000090013, ENSG00000008130, ENSG00000197601, ENSG00000122787, ENSG00000151376 | DHDH, BLVRB, NADK, FAR1, AKR1D1, ME3 | 119 | 6.216871364 | 0.037948511 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Toh, J.; Chua, L.L.; Ho, P.; Sandanaraj, E.; Tang, C.; Wang, H.; Tan, E.K. Identification of Targets from LRRK2 Rescue Phenotypes. Cells 2021, 10, 76. https://doi.org/10.3390/cells10010076
Toh J, Chua LL, Ho P, Sandanaraj E, Tang C, Wang H, Tan EK. Identification of Targets from LRRK2 Rescue Phenotypes. Cells. 2021; 10(1):76. https://doi.org/10.3390/cells10010076
Chicago/Turabian StyleToh, Joanne, Ling Ling Chua, Patrick Ho, Edwin Sandanaraj, Carol Tang, Hongyan Wang, and Eng King Tan. 2021. "Identification of Targets from LRRK2 Rescue Phenotypes" Cells 10, no. 1: 76. https://doi.org/10.3390/cells10010076





