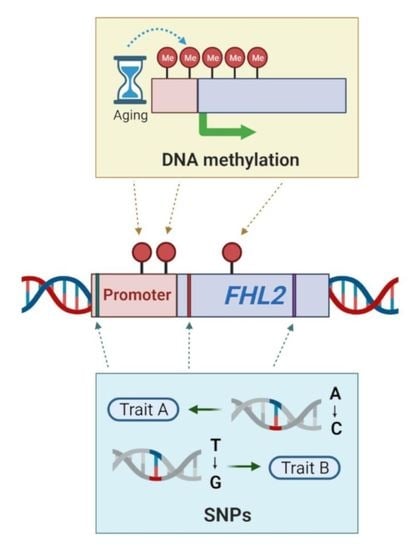
How (Epi)Genetic Regulation of the LIM-Domain Protein FHL2 Impacts Multifactorial Disease
Abstract
1. Introduction
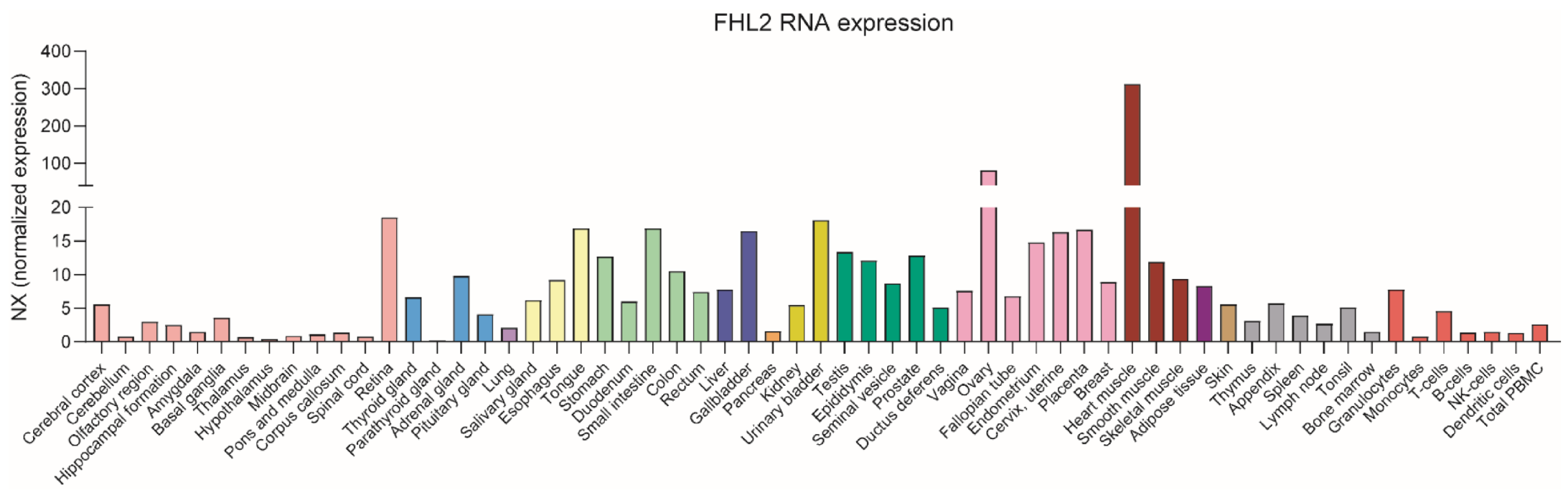
2. FHL2 Tissue Expression
3. Regulation of FHL2 Expression by Specific Transcription Factors
4. FHL2 Genetic Variation
4.1. FHL2 SNPs and Metabolic Phenotypes
4.2. FHL2 SNPs in Cardiovascular Disease and Lung Inflammation
4.3. FHL2 SNPs in Coagulation and Cancer
4.4. FHL2 SNPs and Diverse Traits
5. Methylation of the FHL2 Gene
5.1. Hyper-Methylation of FHL2 in Aging
5.2. Hyper-Methylation of FHL2 in Metabolism
6. Concluding Remarks and Perspective
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bach, I. The LIM domain: Regulation by association. Mech. Dev. 2000, 91, 5–17. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef] [PubMed]
- Orii, N.; Ganapathiraju, M.K. Wiki-Pi: A Web-Server of Annotated Human Protein-Protein Interactions to Aid in Discovery of Protein Function. PLoS ONE 2012, 7, e49029. [Google Scholar] [CrossRef] [PubMed]
- Tran, M.K.; Kurakula, K.; Koenis, D.S.; de Vries, C.J.M. Protein-protein interactions of the LIM-only protein FHL2 and functional implication of the interactions relevant in cardiovascular disease. Biochim. Biophys. Acta Mol. Cell Res. 2016, 1863, 219–228. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.Y.; Tsai, H.Y.; Tsai, S.H.; Chu, P.H.; Huang, P.H.; Chen, J.W.; Lin, S.J. Deletion of the FHL2 gene attenuates intima-media thickening in a partially ligated carotid artery ligated mouse model. J. Cell. Mol. Med. 2020, 24, 160–173. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.; Bradford, W.H.; Zhang, J.; Sheikh, F. Four and a half LIM domain protein signaling and cardiomyopathy. Biophys. Rev. 2018, 10, 1073–1085. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Han, S.; Wang, Y.; Cui, C.; Zhu, Q.; Jiang, X.; Yang, C.; Du, H.; Yu, C.; Li, Q.; et al. The LIM-only protein FHL2 is involved in autophagy to regulate the development of skeletal muscle cell. Int. J. Biol. Sci. 2019, 15, 838–846. [Google Scholar] [CrossRef]
- van de Pol, V.; Vos, M.; DeRuiter, M.C.; Goumans, M.J.; de Vries, C.J.M.; Kurakula, K. LIM-only protein FHL2 attenuates inflammation in vascular smooth muscle cells through inhibition of the NFκB pathway. Vascul. Pharmacol. 2020, 125–126, 106634. [Google Scholar] [CrossRef]
- Jin, X.; Jiao, X.; Jiao, J.; Zhang, T.; Cui, B. Increased expression of FHL2 promotes tumorigenesis in cervical cancer and is correlated with poor prognosis. Gene 2018, 669, 99–106. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Yu, S.; Xu, H.; Zheng, Y.; Lin, J.; Wu, M.; Wang, J.; Wang, A.; Lan, Q.; Furnari, F.; et al. FHL2 interacts with EGFR to promote glioblastoma growth. Oncogene 2018, 37, 1386–1398. [Google Scholar] [CrossRef]
- Verset, L.; Feys, L.; Trépant, A.L.; De Wever, O.; Demetter, P. FHL2: A scaffold protein of carcinogenesis, tumour-stroma interactions and treatment response. Histol. Histopathol. 2016, 31, 469–478. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Lv, X.; He, C.; Davis, J.S.; Wang, C.; Hua, G. Four and a half lim domains 2 (FHL2) contribute to the epithelial ovarian cancer carcinogenesis. Int. J. Mol. Sci. 2020, 21, 7751. [Google Scholar] [CrossRef] [PubMed]
- Chu, P.-H.; Bardwell, W.M.; Gu, Y.; Ross, J.; Chen, J. FHL2 (SLIM3) Is Not Essential for Cardiac Development and Function. Mol. Cell. Biol. 2000, 20, 7460–7462. [Google Scholar] [CrossRef]
- Neuman, N.A.; Ma, S.; Schnitzler, G.R.; Zhu, Y.; Lagna, G.; Hata, A. The four-and-a-half LIM domain protein 2 regulates vascular smooth muscle phenotype and vascular tone. J. Biol. Chem. 2009, 284, 13202–13212. [Google Scholar] [CrossRef]
- Purcell, N.H.; Darwis, D.; Bueno, O.F.; Müller, J.M.; Schüle, R.; Molkentin, J.D. Extracellular Signal-Regulated Kinase 2 Interacts with and Is Negatively Regulated by the LIM-Only Protein FHL2 in Cardiomyocytes. Mol. Cell. Biol. 2004, 24, 1081–1095. [Google Scholar] [CrossRef]
- Clemente-Olivo, M.P.; Habibe, J.J.; Vos, M.; Ottenhoff, R.; Jongejan, A.; Herrema, H.; Zelcer, N.; Kooijman, S.; Rensen, P.C.N.; van Raalte, D.H.; et al. Four-and-a-half LIM domain protein 2 (FHL2) deficiency protects mice from diet-induced obesity and high FHL2 expression marks human obesity. Metabolism 2021, 121, 154815. [Google Scholar] [CrossRef]
- Ng, C.F.; Zhou, W.J.W.; Ng, P.K.S.; Li, M.S.; Ng, Y.K.; Lai, P.B.S.; Tsui, S.K.W. Characterization of human FHL2 transcript variants and gene expression regulation in hepatocellular carcinoma. Gene 2011, 481, 41–47. [Google Scholar] [CrossRef]
- Bai, S.; Zha, J.; Zhao, H.; Ross, F.P.; Teitelbaum, S.L. Tumor necrosis factor receptor-associated factor 6 is an intranuclear transcriptional coactivator in osteoclasts. J. Biol. Chem. 2008, 283, 30861–30867. [Google Scholar] [CrossRef]
- Lv, H.; Zhang, M.; Shang, Z.; Li, J.; Zhang, S.; Lian, D.; Zhang, R. Genome-wide haplotype association study identify the FGFR2 gene as a risk gene for Acute Myeloid Leukemia. Oncotarget 2017, 8, 7891–7899. [Google Scholar] [CrossRef] [PubMed]
- McKay, J.D.; Hung, R.J.; Han, Y.; Zong, X.; Carreras-Torres, R.; Christiani, D.C.; Caporaso, N.E.; Johansson, M.; Xiao, X.; Li, Y.; et al. Large-scale association analysis identifies new lung cancer susceptibility loci and heterogeneity in genetic susceptibility across histological subtypes. Nat. Genet. 2017, 49, 1126–1132. [Google Scholar] [CrossRef] [PubMed]
- Howard, D.M.; Adams, M.J.; Shirali, M.; Clarke, T.K.; Marioni, R.E.; Davies, G.; Coleman, J.R.I.; Alloza, C.; Shen, X.; Barbu, M.C.; et al. Genome-wide association study of depression phenotypes in UK Biobank identifies variants in excitatory synaptic pathways. Nat. Commun. 2018, 9, 1–10. [Google Scholar] [CrossRef]
- Palumbo, D.; Affinito, O.; Monticelli, A.; Cocozza, S. DNA Methylation variability among individuals is related to CpGs cluster density and evolutionary signatures. BMC Genomics 2018, 19, 229. [Google Scholar] [CrossRef] [PubMed]
- Ciccarone, F.; Tagliatesta, S.; Caiafa, P.; Zampieri, M. DNA methylation dynamics in aging: How far are we from understanding the mechanisms? Mech. Ageing Dev. 2018, 174, 3–17. [Google Scholar] [CrossRef]
- Jung, S.E.; Lim, S.M.; Hong, S.R.; Lee, E.H.; Shin, K.J.; Lee, H.Y. DNA methylation of the ELOVL2, FHL2, KLF14, C1orf132/MIR29B2C, and TRIM59 genes for age prediction from blood, saliva, and buccal swab samples. Forensic Sci. Int. Genet. 2019, 38, 1–8. [Google Scholar] [CrossRef]
- Lee, H.Y.; Hong, S.R.; Lee, J.E.; Hwang, I.K.; Kim, N.Y.; Lee, J.M.; Fleckhaus, J.; Jung, S.E.; Lee, Y.H. Epigenetic age signatures in bones. Forensic Sci. Int. Genet. 2020, 46, 102261. [Google Scholar] [CrossRef] [PubMed]
- Zbieć-Piekarska, R.; Spólnicka, M.; Kupiec, T.; Parys-Proszek, A.; Makowska, Z.; Pałeczka, A.; Kucharczyk, K.; Płoski, R.; Branicki, W. Development of a forensically useful age prediction method based on DNA methylation analysis. Forensic Sci. Int. Genet. 2015, 17, 173–179. [Google Scholar] [CrossRef]
- Genini, M. Subtractive cloning and characterization of DRAL, a novel LIM-domain protein down-regulated in Rhabdomyosarcoma. DNA Cell Biol. 1997, 16, 433–442. [Google Scholar] [CrossRef]
- Kong, Y.; Shelton, J.M.; Rothermel, B.; Li, X.; Richardson, J.A.; Bassel-Duby, R.; Williams, R.S. Cardiac-specific LIM protein FHL2 modifies the hypertrophic response to β-adrenergic stimulation. Circulation 2001, 103, 2731–2738. [Google Scholar] [CrossRef]
- Arimura, T.; Hayashi, T.; Matsumoto, Y.; Shibata, H.; Hiroi, S.; Nakamura, T.; Inagaki, N.; Hinohara, K.; Takahashi, M.; Manatsu, S.I.; et al. Structural analysis of four and half LIM protein-2 in dilated cardiomyopathy. Biochem. Biophys. Res. Commun. 2007, 357, 162–167. [Google Scholar] [CrossRef]
- Martin, B.; Schneider, R.; Janetzky, S.; Waibler, Z.; Pandur, P.; Kühl, M.; Behrens, J.; von der Mark, K.; Starzinski-Powitz, A.; Wixler, V. The LIM-only protein FHL2 interacts with β-catenin and promotes differentiation of mouse myoblasts. J. Cell Biol. 2002, 159, 113–122. [Google Scholar] [CrossRef]
- Kim, S.Y.; Volkl, S.; Ludwig, S.; Schneider, H.; Wixler, V.; Park, J. Deficiency of Fhl2 leads to delayed neuronal cell migration and premature astrocyte differentiation. J. Cell Sci. 2019, 132, jcs228940. [Google Scholar] [CrossRef]
- Johannessen, M.; Møler, S.; Hansen, T.; Moens, U.; Van Ghelue, M. The multifunctional roles of the four-and-a-half-LIM only protein FHL2. Cell. Mol. Life Sci. 2006, 63, 268–284. [Google Scholar] [CrossRef] [PubMed]
- Chu, P.H.; Yeh, L.K.; Lin, H.C.; Jung, S.M.; Ma, D.H.K.; Wang, I.J.; Wu, H.H.; Shiu, T.F.; Chen, J. Deletion of the FHL2 gene attenuating neovascularization after corneal injury. Investig. Ophthalmol. Vis. Sci. 2008, 49, 5314–5318. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Huss, S.; Stellmacher, C.; Goltz, D.; Khlistunova, I.; Adam, A.C.; Trebicka, J.; Kirfel, J.; Büttner, R.; Weiskirchen, R. Deficiency in four and one half LIM domain protein 2 (FHL2) aggravates liver fibrosis in mice. BMC Gastroenterol. 2013, 13, 8. [Google Scholar] [CrossRef] [PubMed]
- Nouët, Y.; Dahan, J.; Labalette, C.; Levillayer, F.; Julien, B.; Jouvion, G.; Cairo, S.; Vives, F.L.; Ribeiro, A.; Huerre, M.; et al. The four and a half LIM-only protein 2 regulates liver homeostasis and contributes to carcinogenesis. J. Hepatol. 2012, 57, 1029–1036. [Google Scholar] [CrossRef]
- Sommer, J.; Dorn, C.; Gäbele, E.; Bataille, F.; Freese, K.; Seitz, T.; Thasler, W.E.; Büttner, R.; Weiskirchen, R.; Bosserhoff, A.; et al. Four-And-A-Half LIM-Domain Protein 2 (FHL2) Deficiency Aggravates Cholestatic Liver Injury. Cells 2020, 9, 248. [Google Scholar] [CrossRef] [PubMed]
- Li, S.Y.; Huang, P.H.; Tarng, D.C.; Lin, T.P.; Yang, W.C.; Chang, Y.H.; Yang, A.H.; Lin, C.C.; Yang, M.H.; Chen, J.W.; et al. Four-and-a-half LIM domains protein 2 is a coactivator of Wnt signaling in diabetic kidney disease. J. Am. Soc. Nephrol. 2015, 26, 3072–3084. [Google Scholar] [CrossRef]
- Ebrahimian, T.; Dierick, F.; Simon, D.; Heidari, M.; Orthwein, A.; Mann, K.K.; Lehoux, S. FHL2 Is Essential for Spleen T Cell–Dependent B Cell Activation and Antibody Response. ImmunoHorizons 2020, 4, 259–273. [Google Scholar] [CrossRef]
- Wixler, V. The role of FHL2 in wound healing and inflammation. FASEB J. 2019, 33, 7799–7809. [Google Scholar] [CrossRef]
- Chu, P.H.; Yeh, H.I.; Wu, H.H.; Hong, R.C.; Shiu, T.F.; Yang, C.M. Deletion of the FHL2 gene attenuates the formation of atherosclerotic lesions after a cholesterol-enriched diet. Life Sci. 2010, 86, 365–371. [Google Scholar] [CrossRef]
- Ebrahimian, T.; Simon, D.; Lemarié, C.A.; Simeone, S.; Heidari, M.; Mann, K.K.; Wassmann, S.; Lehoux, S. Absence of Four-and-a-Half LIM domain protein 2 decreases atherosclerosis in ApoE-/- Mice. Arterioscler. Thromb. Vasc. Biol. 2015, 35, 1190–1197. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kurakula, K.; Sommer, D.; Sokolovic, M.; Moerland, P.D.; Scheij, S.; van Loenen, P.B.; Koenis, D.S.; Zelcer, N.; van Tiel, C.M.; de Vries, C.J.M. LIM-Only Protein FHL2 Is a Positive Regulator of Liver X Receptors in Smooth Muscle Cells Involved in Lipid Homeostasis. Mol. Cell. Biol. 2015, 35, 52–62. [Google Scholar] [CrossRef]
- Rönn, T.; Volkov, P.; Gillberg, L.; Kokosar, M.; Perfilyev, A.; Jacobsen, A.L.; Jørgensen, S.W.; Brøns, C.; Jansson, P.A.; Eriksson, K.F.; et al. Impact of age, BMI and HbA1c levels on the genome-wide DNA methylation and mRNA expression patterns in human adipose tissue and identification of epigenetic biomarkers in blood. Hum. Mol. Genet. 2015, 24, 3792–3813. [Google Scholar] [CrossRef] [PubMed]
- Cao, C.Y.; Mok, S.W.F.; Cheng, V.W.S.; Tsui, S.K.W. The FHL2 regulation in the transcriptional circuitry of human cancers. Gene 2015, 572, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Kleiber, K.; Strebhardt, K.; Martin, B.T. The biological relevance of FHL2 in tumour cells and its role as a putative cancer target. Anticancer Res. 2007, 27, 55–61. [Google Scholar] [PubMed]
- Hua, G.; He, C.; Lv, X.; Fan, L.; Wang, C.; Remmenga, S.W.; Rodabaugh, K.J.; Yang, L.; Lele, S.M.; Yang, P.; et al. The four and a half LIM domains 2 (FHL2) regulates ovarian granulosa cell tumor progression via controlling AKT1 transcription. Cell Death Dis. 2016, 7, e2297. [Google Scholar] [CrossRef] [PubMed]
- Martin, B.T.; Kleiber, K.; Wixler, V.; Raab, M.; Zimmer, B.; Kaufmann, M.; Strebhardt, K. FHL2 regulates cell cycle-dependent and doxorubicin-induced p21 Cip1/Waf1 expression in breast cancer cells. Cell Cycle 2007, 6, 1779–1788. [Google Scholar] [CrossRef]
- McGrath, M.J.; Binge, L.C.; Sriratana, A.; Wang, H.; Robinson, P.A.; Pook, D.; Fedele, C.G.; Brown, S.; Dyson, J.M.; Cottle, D.L.; et al. Regulation of the transcriptional coactivator FHL2 licenses activation of the androgen receptor in castrate-resistant prostate cancer. Cancer Res. 2013, 73, 5066–5079. [Google Scholar] [CrossRef]
- Kahl, P.; Gullotti, L.; Heukamp, L.C.; Wolf, S.; Friedrichs, N.; Vorreuther, R.; Solleder, G.; Bastian, P.J.; Ellinger, J.; Metzger, E.; et al. Androgen receptor coactivators lysine-specific histone demethylase 1 and four and a half LIM domain protein 2 predict risk of prostate cancer recurrence. Cancer Res. 2006, 66, 11341–11347. [Google Scholar] [CrossRef]
- Xu, J.; Zhou, J.; Li, M.S.; Ng, C.F.; Ng, Y.K.; Lai, P.B.S.; Tsui, S.K.W. Transcriptional regulation of the tumor suppressor FHL2 by p53 in human kidney and liver cells. PLoS ONE 2014, 9, e99359. [Google Scholar] [CrossRef]
- Wang, Q.; Wang, X.; Tian, X.; Tang, R.; Xu, X. Four and a half LIM domains 2 contributes to the development of human tongue squamous cell carcinoma. J. Mol. Histol. 2016, 47, 105–116. [Google Scholar] [CrossRef]
- Qiao, L.; Wang, Y.; Pang, R.; Wang, J.; Dai, Y.; Ma, J.; Gu, Q.; Li, Z.; Zhang, Y.; Zou, B.; et al. Oncogene functions of FHL2 are independent from NF-κBIα in gastrointestinal cancer. Pathol. Oncol. Res. 2009, 15, 31–36. [Google Scholar] [CrossRef] [PubMed]
- Labalette, C.; Nouët, Y.; Levillayer, F.; Colnot, S.; Chen, J.; Claude, V.; Huerre, M.; Perret, C.; Buendia, M.A.; Wei, Y. Deficiency of the LIM-only protein FHL2 reduces intestinal tumorigenesis in Apc Mutant Mice. PLoS ONE 2010, 5, e10371. [Google Scholar] [CrossRef] [PubMed]
- Guo, Z.; Zhang, W.; Xia, G.; Niu, L.; Zhang, Y.; Wang, X.; Zhang, Y.; Jiang, B.; Wang, J. Sp1 upregulates the Four and Half Lim 2 (FHL2) expression in gastrointestinal cancers through transcription regulation. Mol. Carcinog. 2010, 49, 826–836. [Google Scholar] [CrossRef]
- Philippar, U.; Schratt, G.; Dieterich, C.; Müller, J.M.; Galgóczy, P.; Engel, F.B.; Keating, M.T.; Gertler, F.; Schüle, R.; Vingron, M.; et al. The SRF target gene Fhl2 antagonizes RhoA/MAL-dependent activation of SRF. Mol. Cell 2004, 16, 867–880. [Google Scholar] [CrossRef] [PubMed]
- Morlon, A.; Sassone-Corsi, P. The LIM-only protein FHL2 is a serum-inducible transcriptional coactivator of AP-1. Proc. Natl. Acad. Sci. USA 2003, 100, 3977–3982. [Google Scholar] [CrossRef]
- Kurakula, K.; Vos, M.; Logiantara, A.; Roelofs, J.J.T.H.; Nieuwenhuis, M.A.; Koppelman, G.H.; Postma, D.S.; Brandsma, C.A.; Sin, D.D.; Bossé, Y.; et al. Deficiency of FHL2 attenuates airway inflammation in mice and genetic variation associates with human bronchial hyper-responsiveness. Allergy Eur. J. Allergy Clin. Immunol. 2015, 70, 1531–1544. [Google Scholar] [CrossRef]
- Bae, H.; Lunetta, K.L.; Murabito, J.M.; Andersen, S.L.; Schupf, N.; Perls, T.; Sebastiani, P. Genetic associations with age of menopause in familial longevity. Menopause 2019, 26, 1204–1212. [Google Scholar] [CrossRef]
- Kroone, C.; Vos, M.; Rademakers, T.; Kuijpers, M.; Hoogenboezem, M.; Van Buul, J.; Heemskerk, J.W.M.; Ruf, W.; Van Hylckama Vlieg, A.; Versteeg, H.H.; et al. LIM-only protein FHL2 attenuates vascular tissue factor activity, inhibits thrombus formation in mice and FHL2 genetic variation associates with human venous thrombosis. Haematologica 2020, 105, 1677–1685. [Google Scholar] [CrossRef]
- Tachmazidou, I.; Süveges, D.; Min, J.L.; Ritchie, G.R.S.; Steinberg, J.; Walter, K.; Iotchkova, V.; Schwartzentruber, J.; Huang, J.; Memari, Y.; et al. Whole-Genome Sequencing Coupled to Imputation Discovers Genetic Signals for Anthropometric Traits. Am. J. Hum. Genet. 2017, 100, 865–884. [Google Scholar] [CrossRef]
- Zhu, Z.; Guo, Y.; Shi, H.; Liu, C.L.; Panganiban, R.A.; Chung, W.; O’Connor, L.J.; Himes, B.E.; Gazal, S.; Hasegawa, K.; et al. Shared genetic and experimental links between obesity-related traits and asthma subtypes in UK Biobank. J. Allergy Clin. Immunol. 2020, 145, 537–549. [Google Scholar] [CrossRef]
- Kichaev, G.; Bhatia, G.; Loh, P.R.; Gazal, S.; Burch, K.; Freund, M.K.; Schoech, A.; Pasaniuc, B.; Price, A.L. Leveraging Polygenic Functional Enrichment to Improve GWAS Power. Am. J. Hum. Genet. 2019, 104, 65–75. [Google Scholar] [CrossRef]
- Friedrich, F.W.; Reischmann, S.; Schwalm, A.; Unger, A.; Ramanujam, D.; Münch, J.; Müller, O.J.; Hengstenberg, C.; Galve, E.; Charron, P.; et al. FHL2 expression and variants in hypertrophic cardiomyopathy. Basic Res. Cardiol. 2014, 109, 451. [Google Scholar] [CrossRef]
- Ntalla, I.; Weng, L.C.; Cartwright, J.H.; Hall, A.W.; Sveinbjornsson, G.; Tucker, N.R.; Choi, S.H.; Chaffin, M.D.; Roselli, C.; Barnes, M.R.; et al. Multi-ancestry GWAS of the electrocardiographic PR interval identifies 202 loci underlying cardiac conduction. Nat. Commun. 2020, 11, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.H.; Raffield, L.M.; Mousas, A.; Sakaue, S.; Huffman, J.E.; Moscati, A.; Trivedi, B.; Jiang, T.; Akbari, P.; Vuckovic, D.; et al. Trans-ethnic and Ancestry-Specific Blood-Cell Genetics in 746,667 Individuals from 5 Global Populations. Cell 2020, 182, 1198–1213. [Google Scholar] [CrossRef] [PubMed]
- Vuckovic, D.; Bao, E.L.; Akbari, P.; Lareau, C.A.; Mousas, A.; Jiang, T.; Chen, M.H.; Raffield, L.M.; Tardaguila, M.; Huffman, J.E.; et al. The Polygenic and Monogenic Basis of Blood Traits and Diseases. Cell 2020, 182, 1214–1231.e11. [Google Scholar] [CrossRef]
- Evangelou, E.; Warren, H.R.; Mosen-Ansorena, D.; Mifsud, B.; Pazoki, R.; Gao, H.; Ntritsos, G.; Dimou, N.; Cabrera, C.P.; Karaman, I.; et al. Genetic analysis of over 1 million people identifies 535 new loci associated with blood pressure traits. Nat. Genet. 2018, 50, 1412–1425. [Google Scholar] [CrossRef] [PubMed]
- Bacos, K.; Gillberg, L.; Volkov, P.; Olsson, A.H.; Hansen, T.; Pedersen, O.; Gjesing, A.P.; Eiberg, H.; Tuomi, T.; Almgren, P.; et al. Blood-based biomarkers of age-associated epigenetic changes in human islets associate with insulin secretion and diabetes. Nat. Commun. 2016, 7, 11089. [Google Scholar] [CrossRef]
- Qian, Z.; Mao, L.; Fernald, A.A.; Yu, H.; Luo, R.; Jiang, Y.; Anastasi, J.; Valk, P.J.; Delwel, R.; Le Beau, M.M. Enhanced expression of FHL2 leads to abnormal myelopoiesis in vivo. Leukemia 2009, 23, 1650–1657. [Google Scholar] [CrossRef][Green Version]
- Cheng, Z.; Dai, Y.; Pang, Y.; Jiao, Y.; Zhao, H.; Zhang, Z.; Qin, T.; Hu, N.; Zhang, Y.; Ke, X.; et al. Enhanced expressions of FHL2 and iASPP predict poor prognosis in acute myeloid leukemia. Cancer Gene Ther. 2019, 26, 17–25. [Google Scholar] [CrossRef]
- Lu, W.; Yu, T.; Liu, S.; Li, S.; Li, S.; Liu, J.; Xu, Y.; Xing, H.; Tian, Z.; Tang, K.; et al. FHL2 interacts with iASPP and impacts the biological functions of leukemia cells. Oncotarget 2017, 8, 40885–40895. [Google Scholar] [CrossRef] [PubMed][Green Version]
- McLoughlin, P.; Ehler, E.; Carlile, G.; Licht, J.D.; Schäfer, B.W. The LIM-only protein DRAL/FHL2 interacts with and is a corepressor for the promyelocytic leukemia zinc finger protein. J. Biol. Chem. 2002, 277, 37045–37053. [Google Scholar] [CrossRef]
- Pasšliç, Z.; Greif, P.A.; Jurinoviç, V.; Mulaw, M.; Kakadia, P.M.; Tizazu, B.; Archangelo, L.F.; Krause, A.; Bohlander, S.K. FHL2 interacts with CALM and is highly expressed in acute erythroid leukemia. Blood Cancer J. 2011, 1, e42. [Google Scholar] [CrossRef]
- Jones, P.A. Functions of DNA methylation: Islands, start sites, gene bodies and beyond. Nat. Rev. Genet. 2012, 13, 484–492. [Google Scholar] [CrossRef]
- Smith, J.; Sen, S.; Weeks, R.J.; Eccles, M.R.; Chatterjee, A. Promoter DNA Hypermethylation and Paradoxical Gene Activation. Trends Cancer 2020, 6, 392–406. [Google Scholar] [CrossRef] [PubMed]
- Horvath, S. DNA methylation age of human tissues and cell types. Genome Biol. 2013, 14, R115. [Google Scholar] [CrossRef] [PubMed]
- Hannum, G.; Guinney, J.; Zhao, L.; Zhang, L.; Hughes, G.; Sadda, S.V.; Klotzle, B.; Bibikova, M.; Fan, J.B.; Gao, Y.; et al. Genome-wide Methylation Profiles Reveal Quantitative Views of Human Aging Rates. Mol. Cell 2013, 49, 359–367. [Google Scholar] [CrossRef] [PubMed]
- Garagnani, P.; Bacalini, M.G.; Pirazzini, C.; Gori, D.; Giuliani, C.; Mari, D.; Di Blasio, A.M.; Gentilini, D.; Vitale, G.; Collino, S.; et al. Methylation of ELOVL2 gene as a new epigenetic marker of age. Aging Cell 2012, 11, 1132–1134. [Google Scholar] [CrossRef] [PubMed]
- Correia Dias, H.; Cunha, E.; Corte Real, F.; Manco, L. Age prediction in living: Forensic epigenetic age estimation based on blood samples. Leg. Med. 2020, 47, 101763. [Google Scholar] [CrossRef]
- Steegenga, W.T.; Boekschoten, M.V.; Lute, C.; Hooiveld, G.J.; De Groot, P.J.; Morris, T.J.; Teschendorff, A.E.; Butcher, L.M.; Beck, S.; Müller, M. Genome-wide age-related changes in DNA methylation and gene expression in human PBMCs. Age (Omaha) 2014, 36, 1523–1540. [Google Scholar] [CrossRef]
- Kananen, L.; Marttila, S.; Nevalainen, T.; Jylhävä, J.; Mononen, N.; Kähönen, M.; Raitakari, O.T.; Lehtimäki, T.; Hurme, M. Aging-associated DNA methylation changes in middle-aged individuals: The Young Finns study. BMC Genom. 2016, 17, 1–12. [Google Scholar] [CrossRef]
- Correia Dias, H.; Cordeiro, C.; Corte Real, F.; Cunha, E.; Manco, L. Age Estimation Based on DNA Methylation Using Blood Samples From Deceased Individuals. J. Forensic Sci. 2020, 65, 465–470. [Google Scholar] [CrossRef]
- Freire-Aradas, A.; Phillips, C.; Mosquera-Miguel, A.; Girón-Santamaría, L.; Gómez-Tato, A.; Casares De Cal, M.; Álvarez-Dios, J.; Ansede-Bermejo, J.; Torres-Español, M.; Schneider, P.M.; et al. Development of a methylation marker set for forensic age estimation using analysis of public methylation data and the Agena Bioscience EpiTYPER system. Forensic Sci. Int. Genet. 2016, 24, 65–74. [Google Scholar] [CrossRef]
- Hamano, Y.; Manabe, S.; Morimoto, C.; Fujimoto, S.; Ozeki, M.; Tamaki, K. Forensic age prediction for dead or living samples by use of methylation-sensitive high resolution melting. Leg. Med. 2016, 21, 5–10. [Google Scholar] [CrossRef] [PubMed]
- Cho, S.; Jung, S.E.; Hong, S.R.; Lee, E.H.; Lee, J.H.; Lee, S.D.; Lee, H.Y. Independent validation of DNA-based approaches for age prediction in blood. Forensic Sci. Int. Genet. 2017, 29, 250–256. [Google Scholar] [CrossRef] [PubMed]
- Dias, H.C.; Cordeiro, C.; Pereira, J.; Pinto, C.; Real, F.C.; Cunha, E.; Manco, L. DNA methylation age estimation in blood samples of living and deceased individuals using a multiplex SNaPshot assay. Forensic Sci. Int. 2020, 311, 110267. [Google Scholar] [CrossRef] [PubMed]
- Woźniak, A.; Heidegger, A.; Piniewska-Róg, D.; Pośpiech, E.; Xavier, C.; Pisarek, A.; Kartasińska, E.; Boroń, M.; Freire-Aradas, A.; Wojtas, M.; et al. Development of the VISAGE enhanced tool and statistical models for epigenetic age estimation in blood, buccal cells and bones. Aging (Albany. N. Y.) 2021, 13, 6459–6484. [Google Scholar] [CrossRef]
- Giuliani, C.; Cilli, E.; Bacalini, M.G.; Pirazzini, C.; Sazzini, M.; Gruppioni, G.; Franceschi, C.; Garagnani, P.; Luiselli, D. Inferring chronological age from DNA methylation patterns of human teeth. Am. J. Phys. Anthropol. 2016, 159, 585–595. [Google Scholar] [CrossRef]
- Turner, D.C.; Gorski, P.P.; Maasar, M.F.; Seaborne, R.A.; Baumert, P.; Brown, A.D.; Kitchen, M.O.; Erskine, R.M.; Dos-Remedios, I.; Voisin, S.; et al. DNA methylation across the genome in aged human skeletal muscle tissue and muscle-derived cells: The role of HOX genes and physical activity. Sci. Rep. 2020, 10, 1–19. [Google Scholar] [CrossRef]
- Spólnicka, M.; Pośpiech, E.; Pepłońska, B.; Zbieć-Piekarska, R.; Makowska, Ż.; Pięta, A.; Karłowska-Pik, J.; Ziemkiewicz, B.; Wężyk, M.; Gasperowicz, P.; et al. DNA methylation in ELOVL2 and C1orf132 correctly predicted chronological age of individuals from three disease groups. Int. J. Legal Med. 2018, 132, 1–11. [Google Scholar] [CrossRef]
- Li, J.; Zhu, X.; Yu, K.; Jiang, H.; Zhang, Y.; Wang, B.; Liu, X.; Deng, S.; Hu, J.; Deng, Q.; et al. Exposure to polycyclic aromatic hydrocarbons and accelerated DNA methylation aging. Environ. Health Perspect. 2018, 126, 067005. [Google Scholar] [CrossRef] [PubMed]
- Bacalini, M.G.; Deelen, J.; Pirazzini, C.; De Cecco, M.; Giuliani, C.; Lanzarini, C.; Ravaioli, F.; Marasco, E.; Van Heemst, D.; Suchiman, H.E.D.; et al. Systemic Age-Associated DNA Hypermethylation of ELOVL2 Gene: In Vivo and in Vitro Evidences of a Cell Replication Process. J. Gerontol. Ser. A Biol. Sci. Med. Sci. 2017, 72, 1015–1023. [Google Scholar] [CrossRef] [PubMed]
- Bysani, M.; Perfilyev, A.; De Mello, V.D.; Rönn, T.; Nilsson, E.; Pihlajamäki, J.; Ling, C. Epigenetic alterations in blood mirror age-associated DNA methylation and gene expression changes in human liver. Epigenomics 2017, 9, 105–122. [Google Scholar] [CrossRef] [PubMed]
- Johansson, Å.; Enroth, S.; Gyllensten, U. Continuous Aging of the Human DNA Methylome Throughout the Human Lifespan. PLoS ONE 2013, 8, e67378. [Google Scholar] [CrossRef] [PubMed]
- Taniguchi, I.; Iwaya, C.; Ohnaka, K.; Shibata, H.; Yamamoto, K. Genome-wide DNA methylation analysis reveals hypomethylation in the low-CpG promoter regions in lymphoblastoid cell lines. Hum. Genom. 2017, 11, 8. [Google Scholar] [CrossRef]
- Wan, J.; Oliver, V.F.; Wang, G.; Zhu, H.; Zack, D.J.; Merbs, S.L.; Qian, J. Characterization of tissue-specific differential DNA methylation suggests distinct modes of positive and negative gene expression regulation. BMC Genom. 2015, 16, 49. [Google Scholar] [CrossRef]

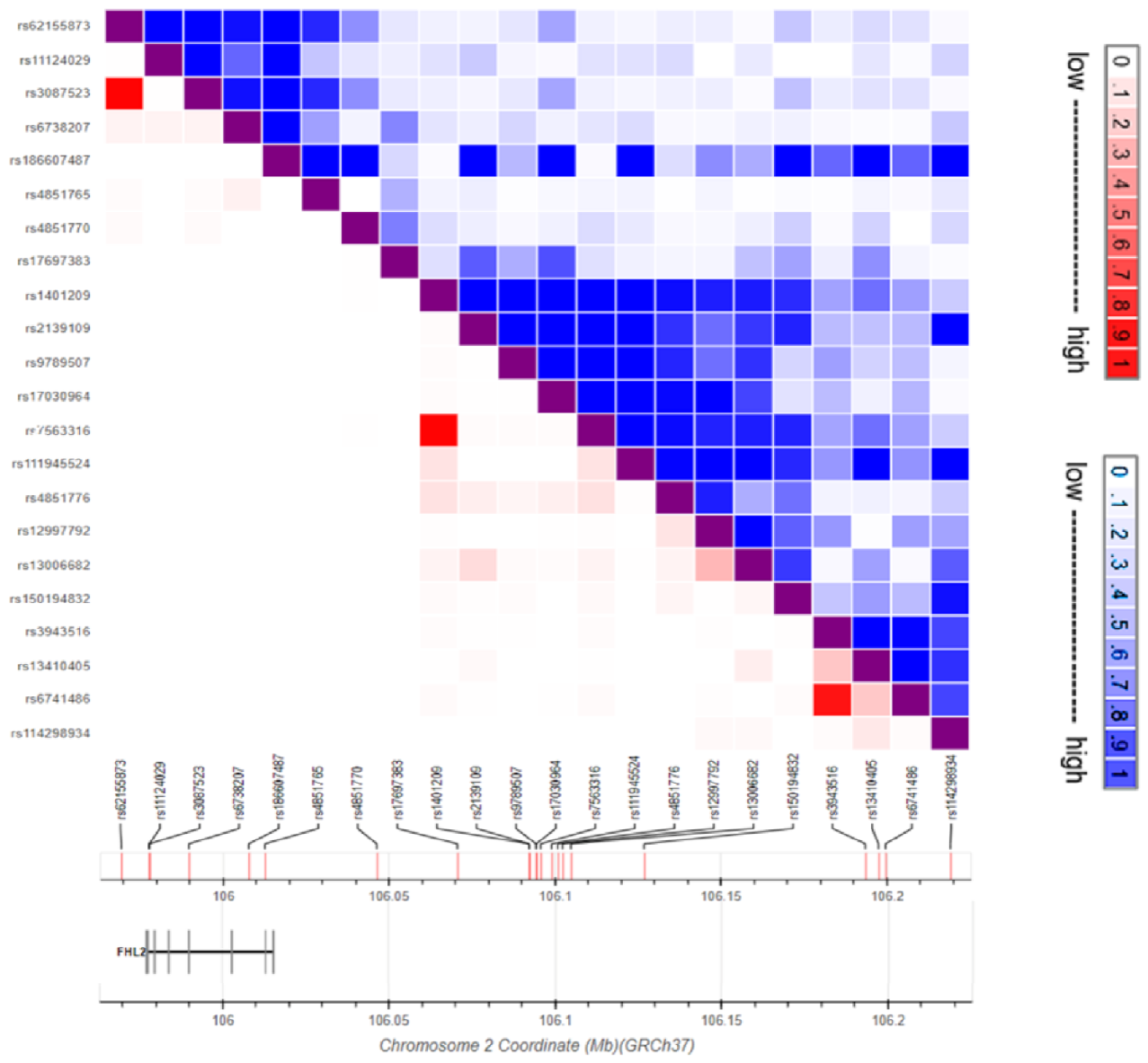

| SNP ID | Ref | Alt | Risk Allele | Location | Associated Trait | SNP Type | Reference |
|---|---|---|---|---|---|---|---|
| rs4851765 | C | T/A | NA | 2:105396175 | Severity of bronchial hyper-responsiveness | non-coding | [57] |
| rs114298934 | C | A | NA | 2:105602735 | Age of menopause | non-coding | [58] |
| rs4851770 | T | C | NA | 2:105429876 | Venous thrombosis | non-coding | [59] |
| rs186607487 | A | G | A | 2:105391292 | Fat body mass | non-coding | [60] |
| rs3087523 a | G | A | NA | 2:105361319 | BMI | coding | [61] |
| [62] | |||||||
| Cardiac myopathy | [63] | ||||||
| rs11124029 | G | A | NA | 2:105361304 | Cardiac myopathy | coding | [63] |
| rs1401209 b | T | G | T | 2:105475760 | Acute myeloid leukemia | intergenic | [19] |
| rs9789507 | T | C | C | 2:105477863 | |||
| rs7563316 b | C | T | C | 2:105479257 | |||
| rs17030964 | C | A | C | 2:105477930 | |||
| rs4851776 | G | A | A | 2:105484430 | |||
| rs2139109 | C | T | C | 2:105475835 | |||
| rs12997792 | C | T | T | 2:105485871 | |||
| rs13006682 | T | C | C | 2:105488399 | PR interval | intergenic | [64] |
| rs17697383 | C | A/T | C | 2:105454164 | Lung cancer in ever smokers | intergenic | [20] |
| rs62155873 a | C | T | NA | 2:105352905 | Smoking status | intergenic | [62] |
| rs13410405 | T | G | G | 2:105581084 | Myeloid white cell count | intergenic | [65] |
| rs3943516 c | A | G | G | 2:105577095 | Platelet count | intergenic | [65] |
| [66] | |||||||
| rs6741486 c | A | G | G | 2:105583173 | Platelet count | intergenic | [65] |
| rs111945524 | C | T | T | 2:105482561 | Unipolar depression | intergenic | [21] |
| rs6738207 | G | A | NA | 2:105373259 | Height | non-coding | [62] |
| rs150194832 | G | C | G | 2:105510423 | Pulse pressure | intergenic | [67] |
| CpG ID | Position in Chromosome | Tissue | References |
|---|---|---|---|
| cg17129821 | chr2:105986385 a | Liver | [93] |
| cg19850931 | chr2:105993347 a | Whole blood | [43] |
| Adipose tissue | [43] | ||
| cg17291435 | chr2:106015527 a | Liver | [93] |
| cg02054792 | chr2:106014950 a | Liver | [93] |
| cg10349436 | chr2:106015079 a | Adipose tissue | [43] |
| cg02445447 | chr2:106015595 a | Liver | [93] |
| cg06639320 | chr2:106015740 a | Whole blood | [68,78,81,83,85] |
| Pancreatic islets | [68] | ||
| Leucocytes | [94] | ||
| Granulocytes | [92] | ||
| Liver | [93] | ||
| Lymphoblastoid line | [95] | ||
| Saliva | [24] | ||
| cg22454769 | chr2:106015768 a | Whole blood | [68,78,81,83,85] |
| Adipose tissue | [43] | ||
| Pancreatic islets | [68] | ||
| Leucocytes | [94] | ||
| Granulocytes | [92] | ||
| Liver | [93] | ||
| Skeletal muscle | [89] | ||
| cg24079702 | chr2:106015772 a | Whole blood | [68,78,81,83,85] |
| Pancreatic islets | [68] | ||
| Leucocytes | [94] | ||
| Granulocytes | [92] | ||
| Liver | [93] | ||
| cg26344233 | chr2:106015818 a | Whole blood | [43] |
| Adipose tissue | [43] | ||
| cg06907053 | chr2:106015870 a | Whole blood | [43] |
| Adipose tissue | [43] | ||
| Liver | [93] | ||
| Not specified (8 CpG sites) | chr2: 106015678–106016008 a | Teeth | [88] |
| Not specified (12 CpG sites) | chr2:105399282–105399340 b | Whole blood | [82] |
| Not specified | chr2:105399282 b | Whole blood | [86] |
| Not specified | chr2:105399291 b | Whole blood | [79] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Habibe, J.J.; Clemente-Olivo, M.P.; de Vries, C.J. How (Epi)Genetic Regulation of the LIM-Domain Protein FHL2 Impacts Multifactorial Disease. Cells 2021, 10, 2611. https://doi.org/10.3390/cells10102611
Habibe JJ, Clemente-Olivo MP, de Vries CJ. How (Epi)Genetic Regulation of the LIM-Domain Protein FHL2 Impacts Multifactorial Disease. Cells. 2021; 10(10):2611. https://doi.org/10.3390/cells10102611
Chicago/Turabian StyleHabibe, Jayron J., Maria P. Clemente-Olivo, and Carlie J. de Vries. 2021. "How (Epi)Genetic Regulation of the LIM-Domain Protein FHL2 Impacts Multifactorial Disease" Cells 10, no. 10: 2611. https://doi.org/10.3390/cells10102611
APA StyleHabibe, J. J., Clemente-Olivo, M. P., & de Vries, C. J. (2021). How (Epi)Genetic Regulation of the LIM-Domain Protein FHL2 Impacts Multifactorial Disease. Cells, 10(10), 2611. https://doi.org/10.3390/cells10102611