Nematostella vectensis, an Emerging Model for Deciphering the Molecular and Cellular Mechanisms Underlying Whole-Body Regeneration
Abstract
:1. Introduction
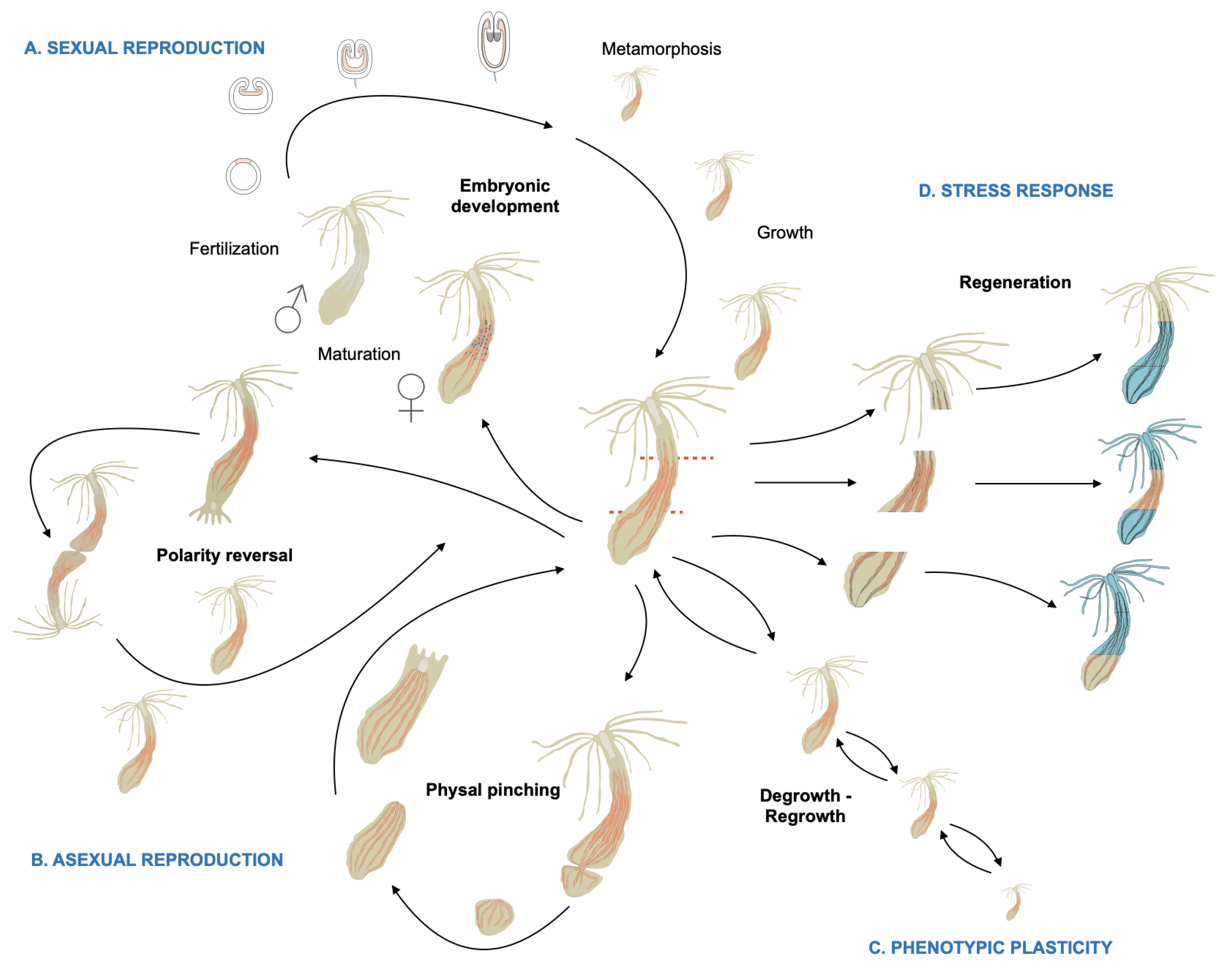
2. Phylogeny, Morphology, Reproduction, and Homeostatic Plasticity of Nematostella vectensis
3. Available Resources, Techniques, and Tools to Study Nematostella vectensis
4. The Morphological, Cellular, and Molecular Basis of Regeneration in Nematostella
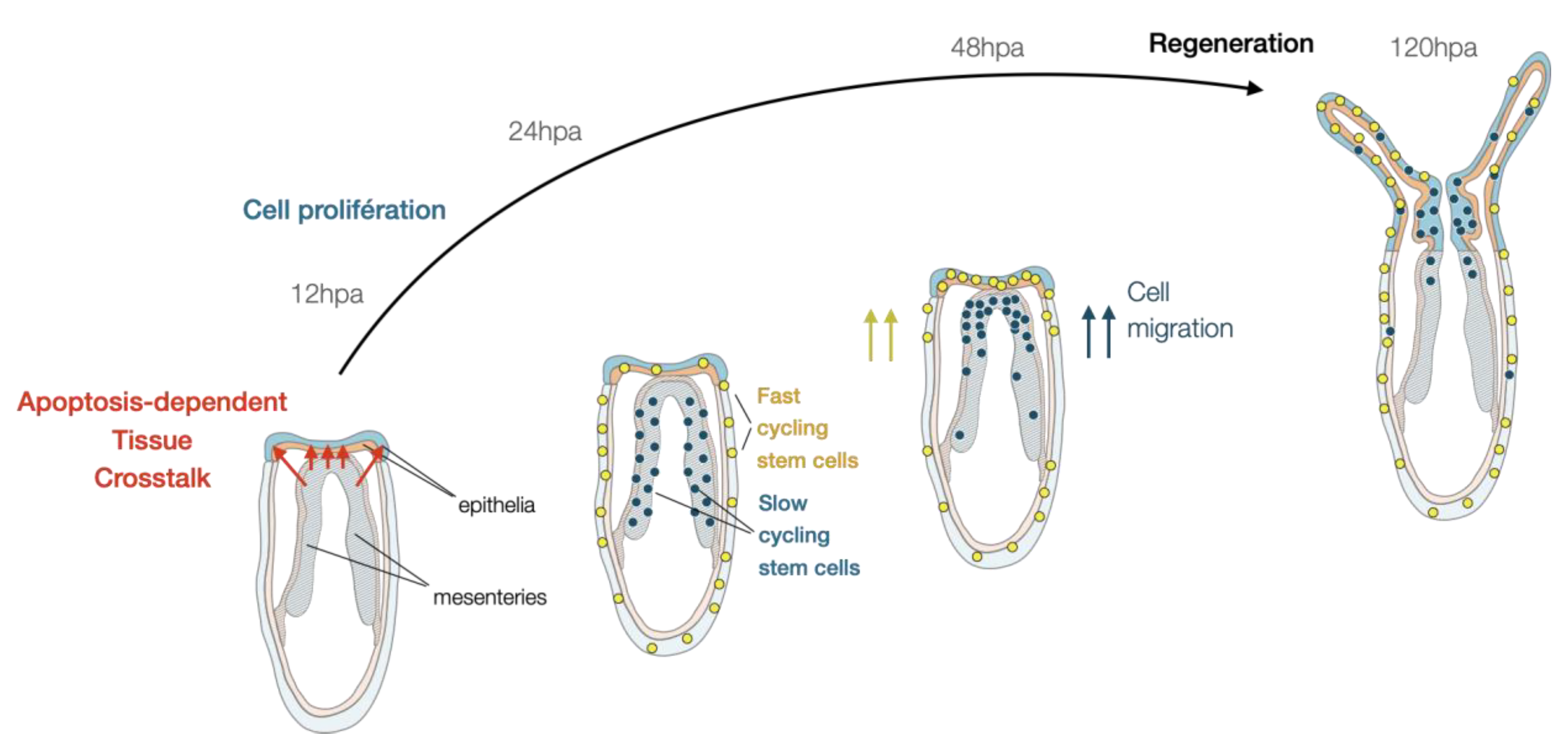
4.1. Morphological and Tissular Dynamics
4.2. Cellular Dynamics Underlying Nematostella Regeneration
4.3. Molecular Wound-Healing Response and Patterning during Nematostella Regeneration
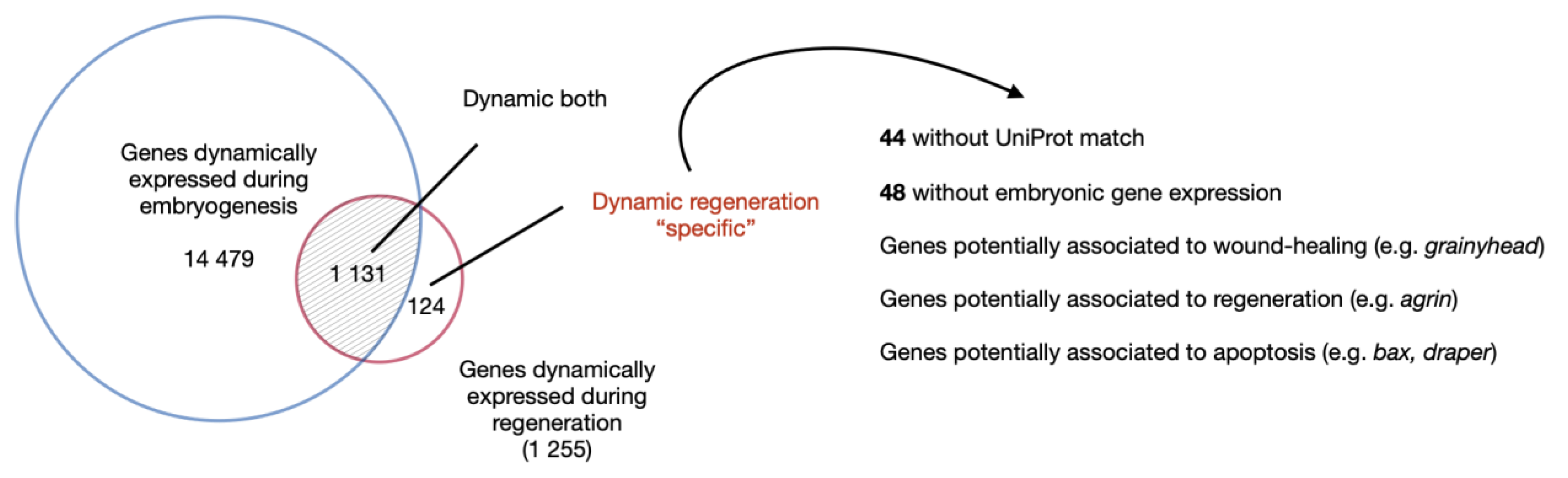
4.4. Regeneration-Specific Genes and Gene Modules
5. Conclusions and Future Directions
Funding
Acknowledgments
Conflicts of Interest
References
- Reaumur, D. Sur les diverses reproductions qui se font dans les ecrivisses, les omars, les crabes, etc. et entre autres sur celles de leurs jambes et de leurs ecailles. Mem Acad. R. Des. Sci. 1712, 226–245. [Google Scholar]
- Bely, A.E.; Nyberg, K.G. Evolution of animal regeneration: Re-emergence of a field. Trends Ecol. Evol. 2010, 25, 161–170. [Google Scholar] [CrossRef] [PubMed]
- Jopling, C.; Boue, S.; Belmonte, J.C.I. Dedifferentiation, transdifferentiation and reprogramming: Three routes to regeneration. Nat. Rev. Mol. Cell Biol. 2011, 12, 79–89. [Google Scholar] [CrossRef] [PubMed]
- Yun, M.H. Changes in Regenerative Capacity through Lifespan. Int. J. Mol. Sci. 2015, 16, 25392–25432. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grillo, M.; Konstantinides, N.; Averof, M. Old questions, new models: Unraveling complex organ regeneration with new experimental approaches. Curr. Opin. Genet. Dev. 2016, 40, 23–31. [Google Scholar] [CrossRef]
- Reddien, P.W. The Cellular and Molecular Basis for Planarian Regeneration. Cell 2018, 175, 327–345. [Google Scholar] [CrossRef] [Green Version]
- Vogg, M.C.; Galliot, B.; Tsiairis, C.D. Model systems for regeneration: Hydra. Development 2019, 146. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Planques, A.; Malem, J.; Parapar, J.; Vervoort, M.; Gazave, E. Morphological, cellular and molecular characterization of posterior regeneration in the marine annelid Platynereis dumerilii. Dev. Biol. 2019, 445, 189–210. [Google Scholar] [CrossRef]
- Giani, V.C.; Yamaguchi, E.; Boyle, M.J.; Seaver, E.C. Somatic and germline expression of piwi during development and regeneration in the marine polychaete annelid Capitella teleta. EvoDevo 2011, 2, 10. [Google Scholar] [CrossRef] [Green Version]
- Gehrke, A.R.; Neverett, E.; Luo, Y.; Brandt, A.; Ricci, L.; Hulett, R.E.; Gompers, A.; Ruby, G.J.; Rokhsar, D.S.; Reddien, P.W.; et al. Acoel genome reveals the regulatory landscape of whole-body regeneration. Science 2019, 363, eaau6173. [Google Scholar] [CrossRef]
- Grudniewska, M.; Mouton, S.; Simanov, D.; Beltman, F.; Grelling, M.; de Mulder, K.; Arindrarto, W.; Weissert, P.M.; van der Elst, S.; Berezikov, E. Transcriptional signatures of somatic neoblasts and germline cells in Macrostomum lignano. Elife 2016, 5, 3389. [Google Scholar] [CrossRef] [Green Version]
- Sprecher, S.G.; Bernardo-Garcia, F.J.; van Giesen, L.; Hartenstein, V.; Reichert, H.; Neves, R.; Bailly, X.; Martinez, P.; Brauchle, M. Functional brain regeneration in the acoel worm Symsagittifera roscoffensis. Biol. Open 2015, 4, 1688–1695. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- DuBuc, T.Q.; Traylor-Knowles, N.; Martindale, M.Q. Initiating a regenerative response; cellular and molecular features of wound healing in the cnidarian Nematostella vectensis. BMC Biol. 2014, 12, 24. [Google Scholar] [CrossRef] [Green Version]
- Kassmer, S.H.; Langenbacher, A.D.; De Tomaso, A.W. Integrin-alpha-6+ Candidate stem cells are responsible for whole body regeneration in the invertebrate chordate Botrylloides diegensis. Nat. Commun. 2020, 11, 4435. [Google Scholar] [CrossRef] [PubMed]
- Prünster, M.M.; Ricci, L.; Brown, F.D.; Tiozzo, S. Modular co-option of cardiopharyngeal genes during non-embryonic myogenesis. EvoDevo 2019, 10, 3. [Google Scholar] [CrossRef] [Green Version]
- van der Burg, C.A.; Pavasovic, A.; Gilding, E.K.; Pelzer, E.S.; Surm, J.M.; Smith, H.L.; Walsh, T.P.; Prentis, P.J. The rapid regenerative response of a model sea anemone species Exaiptasia pallidais characterised by tissue plasticity and highly coordinated cell communication. Mar. Biotechnol. 2020, 22, 285–307. [Google Scholar] [CrossRef] [PubMed]
- Zapata, F.; Goetz, F.E.; Smith, S.A.; Howison, M.; Siebert, S.; Church, S.H.; Sanders, S.M.; Ames, C.L.; McFadden, C.S.; France, S.C.; et al. Phylogenomic Analyses Support Traditional Relationships within Cnidaria. PLoS ONE 2015, 10, e0139068. [Google Scholar] [CrossRef] [Green Version]
- Putnam, N.H.; Srivastava, M.; Hellsten, U.; Dirks, B.; Chapman, J.; Salamov, A.; Terry, A.; Shapiro, H.; Lindquist, E.; Kapitonov, V.V. Sea anemone genome reveals ancestral eumetazoan gene repertoire and genomic organization. Science 2007, 317, 86–94. [Google Scholar] [CrossRef] [Green Version]
- Layden, M.J.; Rentzsch, F.; Röttinger, E. The rise of the starlet sea anemone Nematostella vectensis as a model system to investigate development and regeneration. Wiley Interdiscip. Rev. Dev. Biol. 2016, 5, 408–428. [Google Scholar] [CrossRef] [Green Version]
- Warner, J.F.; Guerlais, V.; Amiel, A.R.; Johnston, H.; Nedoncelle, K.; Röttinger, E. NvERTx: A gene expression database to compare embryogenesis and regeneration in the sea anemone Nematostella vectensis. Development 2018, 145, 162867. [Google Scholar] [CrossRef] [Green Version]
- Warner, J.F.; Amiel, A.R.; Johnston, H.; Röttinger, E. Regeneration is a partial redeployment of the embryonic gene network. bioRxiv 2021. [Google Scholar] [CrossRef] [Green Version]
- Technau, U.; Steele, R.E. Evolutionary crossroads in developmental biology: Cnidaria. J. Embryol. Exp. Morphol. 2011, 138, 1447–1458. [Google Scholar] [CrossRef] [Green Version]
- Technau, U.; Scholz, C.B. Origin and evolution of endoderm and mesoderm. Int. J. Dev. Biol. 2003, 47, 531–539. [Google Scholar]
- Martindale, M.Q.; Pang, K.; Finnerty, J.R. Investigating the origins of triploblasty: Mesodermal gene expression in a diploblastic animal, the sea anemone Nematostella vectensis (phylum, Cnidaria; class, Anthozoa). Development 2004, 131, 2463–2474. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hall, B.K. Evolutionary Developmental Biology (Evo-Devo): Past, Present, and Future. Evol. Educ. Outreach 2012, 5, 184–193. [Google Scholar] [CrossRef] [Green Version]
- Trembley, A. Mémoires Pour Servir à l’Histoire d’un Genre de Polypes d’Eau Douce, à Bras en Forme de Cornes; Wentworth Press: Sydney, Australia, 1744; pp. 1–404. [Google Scholar]
- Galliot, B. Hydra, a fruitful model system for 270 years. Int. J. Dev. Biol. 2012, 56, 411–423. [Google Scholar] [CrossRef] [PubMed]
- Hand, C.; Uhlinger, K.R. The Culture, Sexual and Asexual Reproduction, and Growth of the Sea Anemone Nematostella vectensis. Biol. Bull. 1992, 182, 169–176. [Google Scholar] [CrossRef] [Green Version]
- Rentzsch, F.; Technau, U. Genomics and development of Nematostella vectensis and other anthozoans. Curr. Opin. Genet. Dev. 2016, 39, 63–70. [Google Scholar] [CrossRef]
- Technau, U. Gastrulation and germ layer formation in the sea anemone Nematostella vectensis and other cnidarians. Mech. Dev. 2020, 163, 103628. [Google Scholar] [CrossRef]
- Fritzenwanker, J.; Technau, U. Induction of gametogenesis in the basal cnidarian Nematostella vectensis (Anthozoa). Dev. Genes Evol. 2002, 212, 99–103. [Google Scholar] [CrossRef]
- Genikhovich, G.; Technau, U. The Starlet Sea Anemone Nematostella vectensis: An Anthozoan Model Organism for Studies in Comparative Genomics and Functional Evolutionary Developmental Biology. Cold Spring Harb. Protoc. 2009, 2009, 129. [Google Scholar] [CrossRef]
- Stefanik, D.J.; E Friedman, L.; Finnerty, J.R. Collecting, rearing, spawning and inducing regeneration of the starlet sea anemone, Nematostella vectensis. Nat. Protoc. 2013, 8, 916–923. [Google Scholar] [CrossRef] [PubMed]
- Darling, J.; Reitzel, A.R.; Burton, P.M.; Mazza, M.E.; Ryan, J.; Sullivan, J.C.; Finnerty, J.R. Rising starlet: The starlet sea anemone Nematostella vectensis. BioEssays 2005, 27, 211–221. [Google Scholar] [CrossRef] [PubMed]
- Amiel, A.R.; Röttinger, E. Experimental Tools to Study Regeneration in the Sea Anemone Nematostella vectensis. In Developmental Biology of the Sea Urchin and Other Marine Invertebrates; Humana: New York, NY, USA, 2020; Volume 2219, pp. 69–80. [Google Scholar] [CrossRef]
- Genikhovich, G.; Fried, P.; Prünster, M.M.; Schinko, J.B.; Gilles, A.F.; Fredman, D.; Meier, K.; Iber, D.; Technau, U. Axis Patterning by BMPs: Cnidarian Network Reveals Evolutionary Constraints. Cell Rep. 2015, 10, 1646–1654. [Google Scholar] [CrossRef] [PubMed]
- Leclere, L.; Rentzsch, F. RGM Regulates BMP-Mediated Secondary Axis Formation in the Sea Anemone Nematostella vectensis. Cell Rep. 2014, 9, 1921–1930. [Google Scholar] [CrossRef] [Green Version]
- Lee, P.N.; Kumburegama, S.; Marlow, H.Q.; Martindale, M.Q.; Wikramanayake, A.H. Asymmetric developmental potential along the animal–vegetal axis in the anthozoan cnidarian, Nematostella vectensis, is mediated by Dishevelled. Dev. Biol. 2007, 310, 169–186. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rentzsch, F.; Fritzenwanker, J.; Scholz, C.B.; Technau, U. FGF signalling controls formation of the apical sensory organ in the cnidarian Nematostella vectensis. Development 2008, 135, 1761–1769. [Google Scholar] [CrossRef] [Green Version]
- Wikramanayake, A.H.; Hong, M.; Lee, P.N.; Pang, K.; Byrum, C.A.; Bince, J.M.; Xu, R.; Martindale, M.Q. An ancient role for nuclear β-catenin in the evolution of axial polarity and germ layer segregation. Nature 2003, 426, 446–450. [Google Scholar] [CrossRef]
- Magie, C.R.; Daly, M.; Martindale, M.Q. Gastrulation in the cnidarian Nematostella vectensis occurs via invagination not ingression. Dev. Biol. 2007, 305, 483–497. [Google Scholar] [CrossRef] [Green Version]
- Kirillova, A.; Genikhovich, G.; Pukhlyakova, E.; Demilly, A.; Kraus, Y.; Technau, U. Germ-layer commitment and axis formation in sea anemone embryonic cell aggregates. Proc. Natl. Acad. Sci. USA 2018, 115, 1813–1818. [Google Scholar] [CrossRef] [Green Version]
- Kraus, Y.; Aman, A.; Technau, U.; Genikhovich, G. Pre-bilaterian origin of the blastoporal axial organizer. Nat. Commun. 2016, 7, 11694. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kraus, Y.; Fritzenwanker, J.H.; Genikhovich, G.; Technau, U. The blastoporal organiser of a sea anemone. Curr. Biol. 2007, 17, R874–R876. [Google Scholar] [CrossRef] [Green Version]
- Sinigaglia, C.; Busengdal, H.; Leclère, L.; Technau, U.; Rentzsch, F. The Bilaterian Head Patterning Gene six3/6 Controls Aboral Domain Development in a Cnidarian. PLoS Biol. 2013, 11, e1001488. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leclère, L.; Bause, M.; Sinigaglia, C.; Steger, J.; Rentzsch, F. Development of the aboral domain in Nematostella requires β-catenin and the opposing activities of six3/6 and frizzled5/8. Development 2016, 143, 1766–1777. [Google Scholar] [CrossRef] [Green Version]
- Layden, M.J.; Röttinger, E.; Wolenski, F.S.; Gilmore, T.D.; Martindale, M.Q. Microinjection of mRNA or morpholinos for reverse genetic analysis in the starlet sea anemone, Nematostella vectensis. Nat. Protoc. 2013, 8, 924–934. [Google Scholar] [CrossRef]
- Zimmermann, B.; Robb, S.M.C.; Genikhovich, G.; Fropf, W.J.; Weilguny, L.; He, S.; Chen, S.; Lovegrove-Walsh, J.; Hill, E.M.; Ragkousi, K.; et al. Sea anemone genomes reveal ancestral metazoan chromosomal macrosynteny. bioRxiv 2020. [Google Scholar] [CrossRef]
- Layden, M.J.; Boekhout, M.; Martindale, M.Q. Nematostella vectensis achaete-scute homolog NvashA regulates embryonic ectodermal neurogenesis and represents an ancient component of the metazoan neural specification pathway. Development 2012, 139, 1013–1022. [Google Scholar] [CrossRef] [Green Version]
- Sinigaglia, C.; Busengdal, H.; Lerner, A.; Oliveri, P.; Rentzsch, F. Molecular characterization of the apical organ of the anthozoan Nematostella vectensis. Dev. Biol. 2014, 398, 120–133. [Google Scholar] [CrossRef] [Green Version]
- Röttinger, E.; Dahlin, P.; Martindale, M.Q. A Framework for the Establishment of a Cnidarian Gene Regulatory Network for “Endomesoderm” Specification: The Inputs of ß-Catenin/TCF Signaling. PLoS Genet. 2012, 8, e1003164. [Google Scholar] [CrossRef] [PubMed]
- Fritz, A.E.; Ikmi, A.; Seidel, C.; Paulson, A.; Gibson, M. Mechanisms of tentacle morphogenesis in the sea anemone Nematostella vectensis. Development 2013, 140, 2212–2223. [Google Scholar] [CrossRef] [Green Version]
- Tulin, S.; Aguiar, D.; Istrail, S.; Smith, J. A quantitative reference transcriptome for Nematostella vectensis earlyembryonic development: A pipeline for de novo assembly in emergingmodel systems. EvoDevo 2013, 4, 16. [Google Scholar] [CrossRef] [Green Version]
- Helm, R.R.; Siebert, S.; Tulin, S.; Smith, J.; Dunn, C.W. Characterization of differential transcript abundance through time during Nematostella vectensis development. BMC Genom. 2013, 14, 266. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sebé-Pedrós, A.; Saudemont, B.; Chomsky, E.; Plessier, F.; Mailhé, M.P.; Renno, J.; Loe-Mie, Y.; Lifshitz, A.; Mukamel, Z.; Schmutz, S.; et al. Cnidarian Cell Type Diversity and Regulation Revealed by Whole-Organism Single-Cell RNA-Seq. Cell 2018, 173, 1520–1534.e20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schwaiger, M.; Schönauer, A.; Rendeiro, A.F.; Pribitzer, C.; Schauer, A.; Gilles, A.F.; Schinko, J.B.; Renfer, E.; Fredman, D.; Technau, U. Evolutionary conservation of the eumetazoan gene regulatory landscape. Genome Res. 2014, 24, 639–650. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kusserow, A.; Pang, K.; Sturm, C.; Hrouda, M.; Lentfer, J.; Schmidt, H.A.; Technau, U.; von Haeseler, A.; Hobmayer, B.; Martindale, M.Q.; et al. Unexpected complexity of the Wnt gene family in a sea anemone. Nature 2005, 433, 156–160. [Google Scholar] [CrossRef] [PubMed]
- Finnerty, J.R.; Pang, K.; Burton, P.; Paulson, D.; Martindale, M.Q. Origins of Bilateral Symmetry: Hox and Dpp Expression in a Sea Anemone. Science 2004, 304, 1335–1337. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scholz, C.B.; Technau, U. The ancestral role of Brachyury: Expression of NemBra1 in the basal cnidarian Nematostella vectensis (Anthozoa). Dev. Genes Evol. 2003, 212, 563–570. [Google Scholar] [CrossRef]
- Wolenski, F.S.; Layden, M.J.; Martindale, M.Q.; Gilmore, T.D.; Finnerty, J.R. Characterizing the spatiotemporal expression of RNAs and proteins in the starlet sea anemone, Nematostella vectensis. Nat. Protoc. 2013, 8, 900–915. [Google Scholar] [CrossRef] [Green Version]
- Extavour, C.G.; Pang, K.; Matus, D.Q.; Martindale, M.Q. vasaandnanosexpression patterns in a sea anemone and the evolution of bilaterian germ cell specification mechanisms. Evol. Dev. 2005, 7, 201–215. [Google Scholar] [CrossRef]
- Matus, D.Q.; Thomsen, G.H.; Martindale, M.Q. Dorso/Ventral Genes Are Asymmetrically Expressed and Involved in Germ-Layer Demarcation during Cnidarian Gastrulation. Curr. Biol. 2006, 16, 499–505. [Google Scholar] [CrossRef] [Green Version]
- Saina, M.; Genikhovich, G.; Renfer, E.; Technau, U. BMPs and Chordin regulate patterning of the directive axis in a sea anemone. Proc. Natl. Acad. Sci. USA 2009, 106, 18592–18597. [Google Scholar] [CrossRef] [Green Version]
- Praher, D.; Zimmermann, B.; Genikhovich, G.; Columbus-Shenkar, Y.; Modepalli, V.; Aharoni, R.; Moran, Y.; Technau, U. Characterization of the piRNA pathway during development of the sea anemone Nematostella vectensis. RNA Biol. 2017, 14, 1727–1741. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Amiel, A.R.; Johnston, H.; Chock, T.; Dahlin, P.; Iglesias, M.; Layden, M.; Röttinger, E.; Martindale, M.Q. A bipolar role of the transcription factor ERG for cnidarian germ layer formation and apical domain patterning. Dev. Biol. 2017, 430, 346–361. [Google Scholar] [CrossRef] [PubMed]
- Genikhovich, G.; Technau, U. Anti-acetylated Tubulin Antibody Staining and Phalloidin Staining in the Starlet Sea Anemone Nematostella vectensis. Cold Spring Harb. Protoc. 2009, 2009. [Google Scholar] [CrossRef] [PubMed]
- Gilmore, T.; Wolenski, F.; Finnerty, J. Preparation of antiserum and detection of proteins by Western blotting using the starlet sea anemone, Nematostella vectensis. Protoc. Exch. 2012. [Google Scholar] [CrossRef]
- Renfer, E.; Amon-Hassenzahl, A.; Steinmetz, P.; Technau, U. A muscle-specific transgenic reporter line of the sea anemone, Nematostella vectensis. Proc. Natl. Acad. Sci. USA 2009, 107, 104–108. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ikmi, A.; McKinney, S.A.; Delventhal, K.M.; Gibson, M. TALEN and CRISPR/Cas9-mediated genome editing in the early-branching metazoan Nematostella vectensis. Nat. Commun. 2014, 5, 5486. [Google Scholar] [CrossRef] [PubMed]
- Tournière, O.; Dolan, D.; Richards, G.S.; Sunagar, K.; Columbus-Shenkar, Y.Y.; Moran, Y.; Rentzsch, F. NvPOU4/Brain3 Functions as a Terminal Selector Gene in the Nervous System of the Cnidarian Nematostella vectensis. Cell Rep. 2020, 30, 4473–4489.e5. [Google Scholar] [CrossRef] [PubMed]
- Ikmi, A.; Steenbergen, P.J.; Anzo, M.; McMullen, M.R.; Stokkermans, A.; Ellington, L.R.; Gibson, M.C. Feeding-dependent tentacle development in the sea anemone Nematostella vectensis. Nat. Commun. 2020, 11, 4399. [Google Scholar] [CrossRef]
- Richards, G.S.; Rentzsch, F. Transgenic analysis of a SoxB gene reveals neural progenitor cells in the cnidarian Nematostella vectensis. Development 2014, 141, 4681–4689. [Google Scholar] [CrossRef] [Green Version]
- Havrilak, J.A.; Faltine-Gonzalez, D.; Wen, Y.; Fodera, D.; Simpson, A.C.; Magie, C.R.; Layden, M.J. Characterization of NvLWamide-like neurons reveals stereotypy in Nematostella nerve net development. Dev. Biol. 2017, 431, 336–346. [Google Scholar] [CrossRef] [PubMed]
- He, S.; Viso, F.D.; Chen, C.-Y.; Ikmi, A.; Kroesen, A.E.; Gibson, M.C. An axial Hox code controls tissue segmentation and body patterning in Nematostella vectensis. Science 2018, 361, 1377–1380. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karabulut, A.; He, S.; Chen, C.-Y.; McKinney, S.A.; Gibson, M.C. Electroporation of short hairpin RNAs for rapid and efficient gene knockdown in the starlet sea anemone, Nematostella vectensis. Dev. Biol. 2019, 448, 7–15. [Google Scholar] [CrossRef]
- Layden, M.J.; Johnston, H.; Amiel, A.R.; Havrilak, J.; Steinworth, B.; Chock, T.; Röttinger, E.; Martindale, M.Q. MAPK signaling is necessary for neurogenesis in Nematostella vectensis. BMC Biol. 2016, 14, 61. [Google Scholar] [CrossRef] [Green Version]
- Amiel, A.R.; Johnston, H.T.; Nedoncelle, K.; Warner, J.F.; Ferreira, S.; Röttinger, E. Characterization of Morphological and Cellular Events Underlying Oral Regeneration in the Sea Anemone, Nematostella vectensis. Int. J. Mol. Sci. 2015, 16, 28449–28471. [Google Scholar] [CrossRef] [PubMed]
- Trevino, M.; Stefanik, D.J.; Rodriguez, R.; Harmon, S.; Burton, P.M. Induction of canonical Wnt signaling by alsterpaullone is sufficient for oral tissue fate during regeneration and embryogenesis in Nematostella vectensis. Dev. Dyn. 2011, 240, 2673–2679. [Google Scholar] [CrossRef] [Green Version]
- Marlow, H.; Matus, D.; Martindale, M.Q. Ectopic activation of the canonical wnt signaling pathway affects ectodermal patterning along the primary axis during larval development in the anthozoan Nematostella vectensis. Dev. Biol. 2013, 380, 324–334. [Google Scholar] [CrossRef] [Green Version]
- Faltine-Gonzalez, D.Z.; Layden, M.J. Characterization of nAChRs in Nematostella vectensis supports neuronal and non-neuronal roles in the cnidarian-bilaterian common ancestor. EvoDevo 2019, 10, 27. [Google Scholar] [CrossRef] [PubMed]
- Nakanishi, N.; Renfer, E.; Technau, U.; Rentzsch, F. Nervous systems of the sea anemone Nematostella vectensis are generated by ectoderm and endoderm and shaped by distinct mechanisms. Development 2012, 139, 347–357. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Busengdal, H.; Rentzsch, F. Unipotent progenitors contribute to the generation of sensory cell types in the nervous system of the cnidarian Nematostella vectensis. Dev. Biol. 2017, 431, 59–68. [Google Scholar] [CrossRef] [PubMed]
- Steinmetz, P.R.H.; Aman, A.; Kraus, J.E.M.; Technau, U. Gut—Like ectodermal tissue in a sea anemone challenges germ layer homology. Nat. Ecol. Evol. 2017, 1, 1535–1542. [Google Scholar] [CrossRef]
- Sunagar, K.; Columbus-Shenkar, Y.Y.; Fridrich, A.; Gutkovich, N.; Aharoni, R.; Moran, Y. Cell type-specific expression profiling unravels the development and evolution of stinging cells in sea anemone. BMC Biol. 2018, 16, 108. [Google Scholar] [CrossRef] [Green Version]
- Admoni, Y.; Kozlovski, I.; Lewandowska, M.; Moran, Y. TATA Binding Protein (TBP) Promoter Drives Ubiquitous Expression of Marker Transgene in the Adult Sea Anemone Nematostella vectensis. Genes 2020, 11, 1081. [Google Scholar] [CrossRef] [PubMed]
- Tucker, R.P.; Shibata, B.; Blankenship, T.N. Ultrastructure of the mesoglea of the sea anemone Nematostella vectensis (Edwardsiidae). Invertebr. Biol. 2011, 130, 11–24. [Google Scholar] [CrossRef]
- Jahnel, S.M.; Walzl, M.; Technau, U. Development and epithelial organisation of muscle cells in the sea anemone Nematostella vectensis. Front. Zool. 2014, 11, 44. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marlow, H.Q.; Srivastava, M.; Matus, D.Q.; Rokhsar, D.; Martindale, M.Q. Anatomy and development of the nervous system of Nematostella vectensis, an anthozoan cnidarian. Dev. Neurobiol. 2009, 69, 235–254. [Google Scholar] [CrossRef]
- Chang, E.S.; Neuhof, M.; Rubinstein, N.D.; Diamant, A.; Philippe, H.; Huchon, D.; Cartwright, P. Genomic insights into the evolutionary origin of Myxozoa within Cnidaria. Proc. Natl. Acad. Sci. USA 2015, 112, 14912–14917. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kayal, E.; Bentlage, B.; Pankey, M.S.; Ohdera, A.H.; Medina, M.; Plachetzki, D.C.; Collins, A.G.; Ryan, J.F. Phylogenomics provides a robust topology of the major cnidarian lineages and insights on the origins of key organismal traits. BMC Evol. Biol. 2018, 18, 68. [Google Scholar] [CrossRef] [Green Version]
- Leclère, L.; Röttinger, E. Diversity of Cnidarian Muscles: Function, Anatomy, Development and Regeneration. Front. Cell Dev. Biol. 2017, 4, 157. [Google Scholar] [CrossRef]
- Amiel, A.R.; Michel, V.; Carvalho, J.E.; Shkreli, M.; Petit, C.; Röttinger, E. L’anémone de mer Nematostella vectensis. Med. Sci. 2021, 37, 167–177. [Google Scholar] [CrossRef]
- Steinmetz, P.R.H. A non-bilaterian perspective on the development and evolution of animal digestive systems. Cell Tissue Res. 2019, 377, 321–339. [Google Scholar] [CrossRef] [Green Version]
- Ormestad, M.; Martindale, M.Q.; Röttinger, E. A comparative gene expression database for invertebrates. EvoDevo 2011, 2, 17. [Google Scholar] [CrossRef] [Green Version]
- Hand, C.; Uhlinger, K.R. Asexual Reproduction by Transverse Fission and Some Anomalies in the Sea Anemone Nematostella vectensis Invertebr. Biol. 1995, 114. [Google Scholar] [CrossRef]
- Reitzel, A.; Burton, P.; Krone, C.; Finnerty, J. Comparison of developmental trajectories in the starlet sea anemone Nematostella vectensis: Embryogenesis, regeneration, and two forms of asexual fission. Invertebr. Biol. 2007, 126, 99–112. [Google Scholar] [CrossRef]
- Passamaneck, Y.J.; Martindale, M.Q. Cell proliferation is necessary for the regeneration of oral structures in the anthozoan cnidarian Nematostella vectensis. BMC Dev. Biol. 2012, 12, 34. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Havrilak, J.A.; Al-Shaer, L.; Baban, N.; Akinci, N.; Layden, M.J. Characterization of the dynamics and variability of neuronal subtype responses during growth, degrowth, and regeneration of Nematostella vectensis. BMC Biol. 2021, 19, 104. [Google Scholar] [CrossRef]
- Amiel, A.; Foucher, K.; Ferreira, S.; Röttinger, E. Synergic coordination of stem cells is required to induce a regenerative response in anthozoan cnidarians. bioRxiv 2019. [Google Scholar] [CrossRef]
- Moran, Y.; Fredman, D.; Praher, D.; Li, X.Z.; Wee, L.M.; Rentzsch, F.; Zamore, P.D.; Technau, U.; Seitz, H. Cnidarian microRNAs frequently regulate targets by cleavage. Genome Res. 2014, 24, 651–663. [Google Scholar] [CrossRef] [Green Version]
- Chourrout, D.; Delsuc, F.; Chourrout, P.; Edvardsen, R.B.; Rentzsch, F.; Renfer, E.; Jensen, M.F.; Zhu, B.; de Jong, P.; Steele, E.E.; et al. Minimal ProtoHox cluster inferred from bilaterian and cnidarian Hox complements. Nature 2006, 442, 684–687. [Google Scholar] [CrossRef] [PubMed]
- Bossert, P.E.; Dunn, M.; Thomsen, G.H. A staging system for the regeneration of a polyp from the aboral physa of the anthozoan Cnidarian Nematostella vectensis. Dev. Dyn. 2013, 242, 1320–1331. [Google Scholar] [CrossRef]
- Burton, P.M.; Finnerty, J.R. Conserved and novel gene expression between regeneration and asexual fission in Nematostella vectensis. Dev. Genes Evol. 2009, 219, 79–87. [Google Scholar] [CrossRef] [PubMed]
- Newmark, A.P.; Alvarado, A.A.S. Regeneration in Planaria; John Wiley & Sons: Hoboken, NJ, USA, 2001; pp. 1–7. [Google Scholar]
- Galliot, B.; Chera, S. The Hydra model: Disclosing an apoptosis-driven generator of Wnt-based regeneration. Trends Cell Biol. 2010, 20, 514–523. [Google Scholar] [CrossRef] [Green Version]
- Ricci, L.; Srivastava, M. Wound-induced cell proliferation during animal regeneration. Wiley Interdiscip. Rev. Dev. Biol. 2018, 7, e321. [Google Scholar] [CrossRef] [PubMed]
- Chera, S.; Ghila, L.; Dobretz, K.; Wenger, Y.; Bauer, C.; Buzgariu, W.; Martinou, J.C.; Brigitte Galliot, B. Apoptotic Cells Provide an Unexpected Source of Wnt3 Signaling to Drive Hydra Head Regeneration. Dev. Cell 2009, 17, 279–289. [Google Scholar] [CrossRef]
- E Fogarty, C.; Bergmann, A. Killers creating new life: Caspases drive apoptosis-induced proliferation in tissue repair and disease. Cell Death Differ. 2017, 24, 1390–1400. [Google Scholar] [CrossRef] [Green Version]
- Juliano, C.E.; Swartz, S.Z.; Wessel, G.M. A conserved germline multipotency program. Development 2010, 137, 4113–4126. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alié, A.; Leclère, L.; Jager, M.; Dayraud, C.; Chang, P.; Le Guyader, H.; Quéinnec, E.; Manuel, M. Somatic stem cells express Piwi and Vasa genes in an adult ctenophore: Ancient association of “germline genes” with stemness. Dev. Biol. 2011, 350, 183–197. [Google Scholar] [CrossRef] [Green Version]
- Alié, A.; Hayashi, T.; Sugimura, I.; Manuel, M.; Sugano, W.; Mano, A.; Satoh, N.; Agata, K.; Funayama, N. The ancestral gene repertoire of animal stem cells. Proc. Natl. Acad. Sci. USA 2015, 112, E7093–E7100. [Google Scholar] [CrossRef] [Green Version]
- Leclère, L.; Jager, M.; Barreau, C.; Chang, P.; Le Guyader, H.; Manuel, M.; Houliston, E. Maternally localized germ plasm mRNAs and germ cell/stem cell formation in the cnidarian Clytia. Dev. Biol. 2012, 364, 236–248. [Google Scholar] [CrossRef] [PubMed]
- Solana, J.; Kao, D.; Mihaylova, Y.; Jaber-Hijazi, F.; Malla, S.; Wilson, R.; Aboobaker, A. Defining the molecular profile of planarian pluripotent stem cells using a combinatorial RNA-seq, RNA interference and irradiation approach. Genome Biol. 2012, 13, R19. [Google Scholar] [CrossRef] [Green Version]
- Eisenhoffer, G.T.; Kang, H.; Alvarado, A.S. Molecular Analysis of Stem Cells and Their Descendants during Cell Turnover and Regeneration in the Planarian Schmidtea mediterranea. Cell Stem Cell 2008, 3, 327–339. [Google Scholar] [CrossRef] [Green Version]
- Gazave, E.; Béhague, J.; Laplane, L.; Guillou, A.; Préau, L.; Demilly, A.; Balavoine, G.; Vervoort, M. Posterior elongation in the annelid Platynereis dumerilii involves stem cells molecularly related to primordial germ cells. Dev. Biol. 2013, 382, 246–267. [Google Scholar] [CrossRef]
- Hemmrich, G.; Khalturin, K.; Boehm, A.M.; Puchert, M.; Anton-Erxleben, F.; Wittlieb, J.; C Klostermeier, U.C.; Rosenstiel, P.; Oberg, H.H.; Domazet-Loso, T.; et al. Molecular Signatures of the Three Stem Cell Lineages in Hydra and the Emergence of Stem Cell Function at the Base of Multicellularity. Mol. Biol. Evol. 2012, 29, 3267–3280. [Google Scholar] [CrossRef] [Green Version]
- Hartl, M.; Mitterstiller, A.-M.; Valovka, T.; Breuker, K.; Hobmayer, B.; Bister, K. Stem cell-specific activation of an ancestral myc protooncogene with conserved basic functions in the early metazoan Hydra. Proc. Natl. Acad. Sci. USA 2010, 107, 4051–4056. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chera, S.; Ghila, L.; Wenger, Y.; Galliot, B. Injury-induced activation of the MAPK/CREB pathway triggers apoptosis-induced compensatory proliferation in hydra head regeneration. Dev. Growth Differ. 2011, 53, 186–201. [Google Scholar] [CrossRef] [Green Version]
- Gao, J.; Fan, L.; Zhao, L.; Su, Y. The interaction of Notch and Wnt signaling pathways in vertebrate regeneration. Cell Regen. 2021, 10, 11. [Google Scholar] [CrossRef] [PubMed]
- Cazet, J.F.; Cho, A.; Juliano, E.C. Generic injuries are sufficient to induce ectopic Wnt organizers in Hydra. eLife 2021, 10, e60562. [Google Scholar] [CrossRef] [PubMed]
- Vogg, M.C.; Beccari, L.; Ollé, L.I.; Rampon, C.; Vriz, S.; Perruchoud, C.; Wenger, Y.; Galliot, B. An evolutionarily-conserved Wnt3/β-catenin/Sp5 feedback loop restricts head organizer activity in Hydra. Nat. Commun. 2019, 10, 312. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, S.Y.; Selck, C.; Friedrich, B.; Lutz, R.; Vila-Farré, M.; Dahl, A.; Brandl, H.; Lakshmanaperumal, N.; Henry, I.; Rink, J.C. Reactivating head regrowth in a regeneration-deficient planarian species. Nature 2013, 500, 81–84. [Google Scholar] [CrossRef] [PubMed]
- Sikes, J.M.; Newmark, P.A. Restoration of anterior regeneration in a planarian with limited regenerative ability. Nature 2013, 500, 77. [Google Scholar] [CrossRef]
- Umesono, Y.; Tasaki, J.; Nishimura, Y.; Hrouda, M.; Kawaguchi, E.; Yazawa, S.; Nishimura, O.; Hosoda, K.; Inoue, T.; Agata, K. The molecular logic for planarian regeneration along the anterior–posterior axis. Nature 2013, 500, 73–76. [Google Scholar] [CrossRef] [PubMed]
- Schaffer, A.A.; Bazarsky, M.; Levy, K.; Chalifa-Caspi, V.; Gat, U. A transcriptional time-course analysis of oral vs. aboral whole-body regeneration in the Sea anemone Nematostella vectensis. BMC Genom. 2016, 17, 718. [Google Scholar] [CrossRef] [PubMed] [Green Version]


| Culture and Experimentation | |
| Rearing and spawning | [28,31,32] |
| Inducing and assessing regeneration | [33,34,35] |
| Microinjection and micromanipulation | [36,37,38,39,40,41,42,43,44,45,46,47] |
| Genome, Resources, and Protocols for -Omics Analyses | |
| Annotated genome—V1.0 | [18] https://mycocosm.jgi.doe.gov/Nemve1/Nemve1.home.html (accessed on 5 October 2021) |
| Annotated genome—V2.0 | [48] https://genomes.stowers.org/starletseaanemone (accessed on 5 October 2021) |
| Microarrays | [49,50,51,52] |
| RNA-seq/transcriptomes | [20,53,54] http://nvertx.kahikai.org (accessed on 5 October 2021) |
| scRNA-seq | [55] |
| ATAC-seq | [55] |
| ChIP-seq | [56] |
| Spatio–Temporal Gene Expression Analysis | |
| mRNA in situ | [57,58,59,60,61,62,63,64] |
| Immunohistochemistry | [36,37,39,40,46,65,66,67] |
| Transgenic reporter | [68,69,70,71,72,73] |
| Tools to Study Gene Function | |
| mRNA over-expression | [40,47,49,51,65] |
| Morpholino | [36,39,41,46,47,49,51,63,65] |
| Short hairpin RNA | [74,75] |
| TALEN/Fok1, CRISPR/Cas9 | [69] |
| Heat-shock inducible promoters | [69] |
| Pharmaceutical treatments | [21,39,51,71,76,77,78,79,80] |
| Available Stable Reporter Lines | |
| NvMyHC::mCherry | [68] |
| NvElav::mOrange | [81] |
| NvElav::Cerulean | [82] |
| NvLWamide:mCherry | [73] |
| NvEf1a::mOrange-CAAX | [83] |
| NvFoxQ2d::mOrange-CAAX | [82] |
| NvNcol-3::memOrange2 | [84] |
| NvAnthox8::Gfp | [74] |
| NvPou4::mCherry | [70] |
| NvTBP::mCherry | [85] |
| Available Stable KO Lines | |
| NvAnthox1a−/− | [74] |
| NvAnthox6−/− | [74] |
| NvAnthox8a−/− | [74] |
| NvFgfrB+/− | [71] |
| NvPou4+/− | [70] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Röttinger, E. Nematostella vectensis, an Emerging Model for Deciphering the Molecular and Cellular Mechanisms Underlying Whole-Body Regeneration. Cells 2021, 10, 2692. https://doi.org/10.3390/cells10102692
Röttinger E. Nematostella vectensis, an Emerging Model for Deciphering the Molecular and Cellular Mechanisms Underlying Whole-Body Regeneration. Cells. 2021; 10(10):2692. https://doi.org/10.3390/cells10102692
Chicago/Turabian StyleRöttinger, Eric. 2021. "Nematostella vectensis, an Emerging Model for Deciphering the Molecular and Cellular Mechanisms Underlying Whole-Body Regeneration" Cells 10, no. 10: 2692. https://doi.org/10.3390/cells10102692
APA StyleRöttinger, E. (2021). Nematostella vectensis, an Emerging Model for Deciphering the Molecular and Cellular Mechanisms Underlying Whole-Body Regeneration. Cells, 10(10), 2692. https://doi.org/10.3390/cells10102692






