Mitochondrial Phospholipid Homeostasis Is Regulated by the i-AAA Protease PaIAP and Affects Organismic Aging
Abstract
1. Introduction
2. Materials and Methods
2.1. P. anserina Strains and Cultivation
2.2. Lifespan Analysis
2.3. Cloning Procedure and Generation of P. anserina Mutants
2.4. Southern Blot Analysis
2.5. Mitochondria Isolation
2.6. Cycloheximide Assay
2.7. Total Protein Extraction
2.8. Lipidomic Analysis
2.9. Thin-Layer-Chromatography (TLC)
2.10. Western Blot Analysis
2.11. Blue-Native Polyacrylamide Gel Electrophoresis (BN-PAGE)
2.12. Quantitative Real-Time PCR (qPCR)
2.13. Stress Sensitivity Assays
2.14. Determination of ROS Release
2.15. Statistical Analysis
3. Results and Discussion
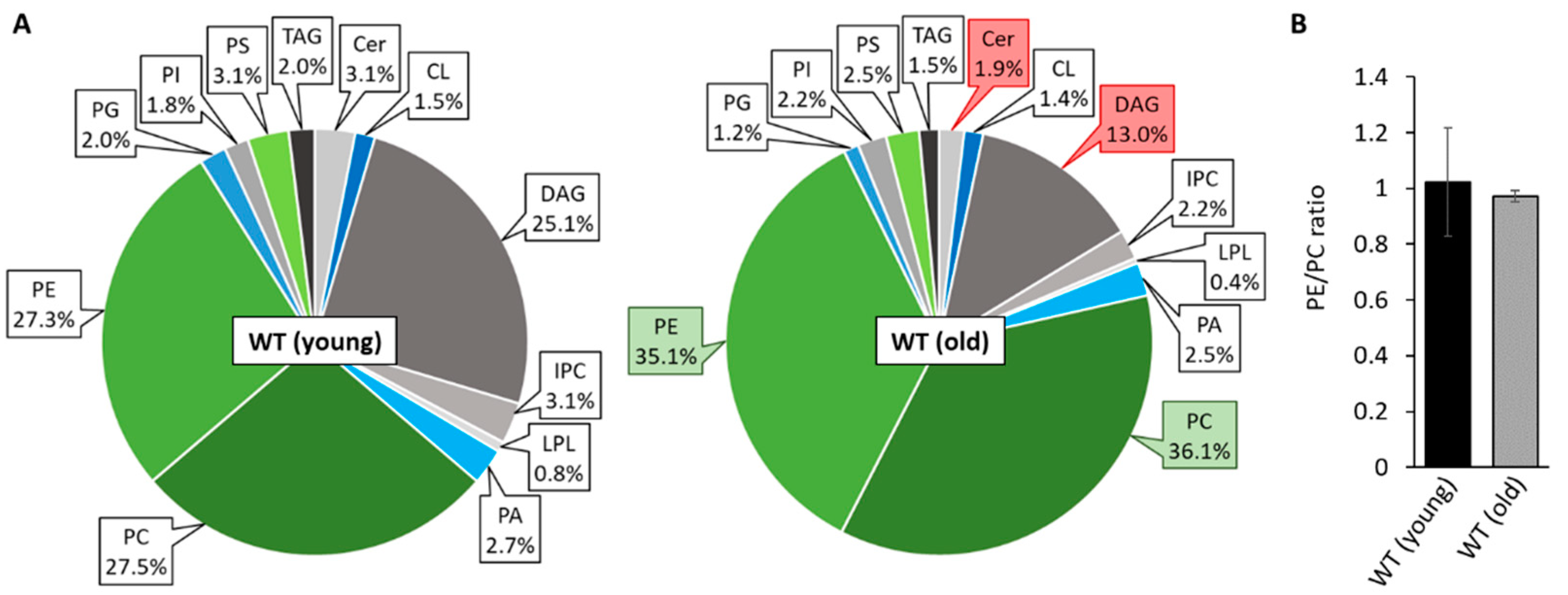
3.1. Unravelling Age-Dependent Alterations in the Mitochondrial Lipid Composition of P. anserina
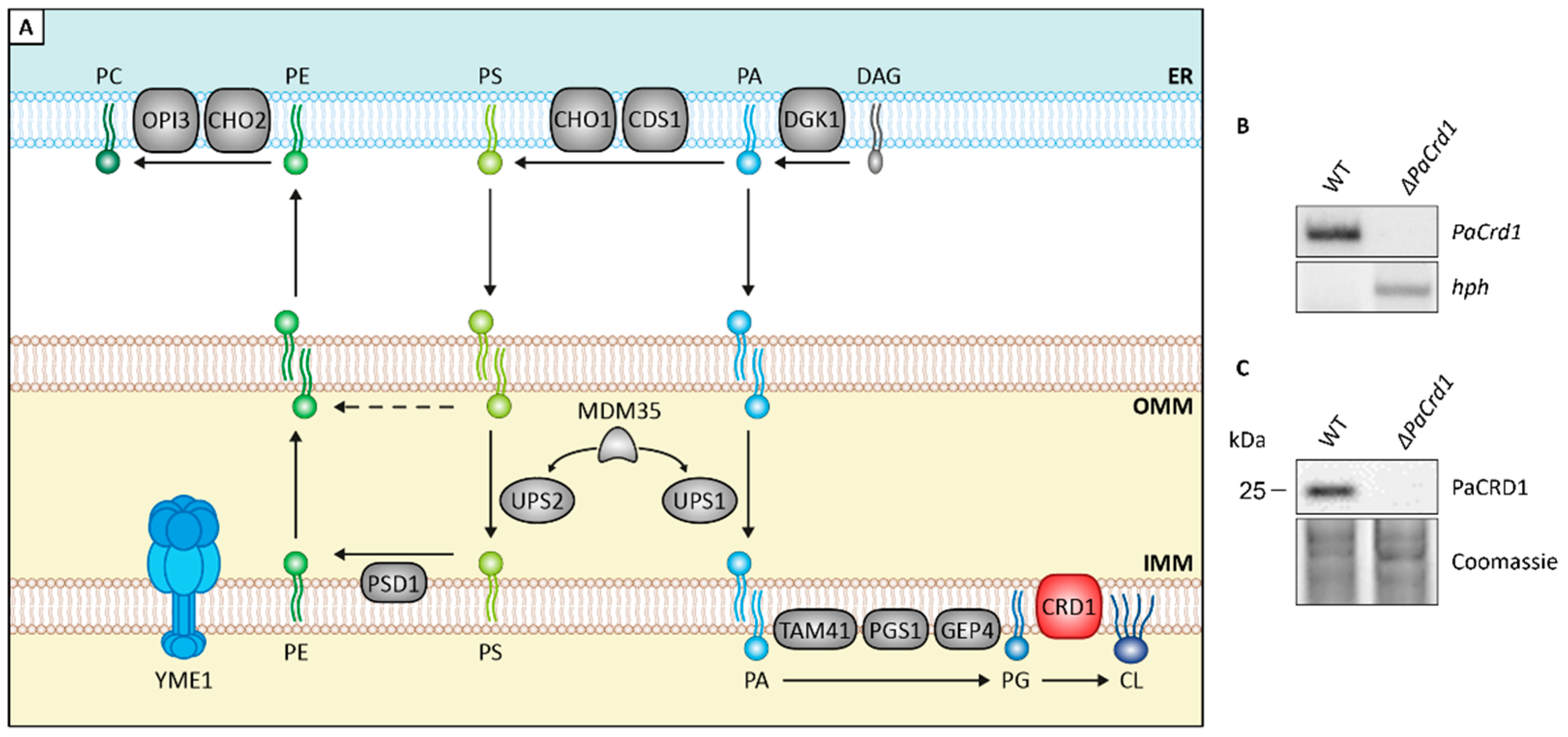
3.2. Manipulating Components of PL Biosynthesis Modulates Mitochondrial Lipid Composition and Affects Lifespan
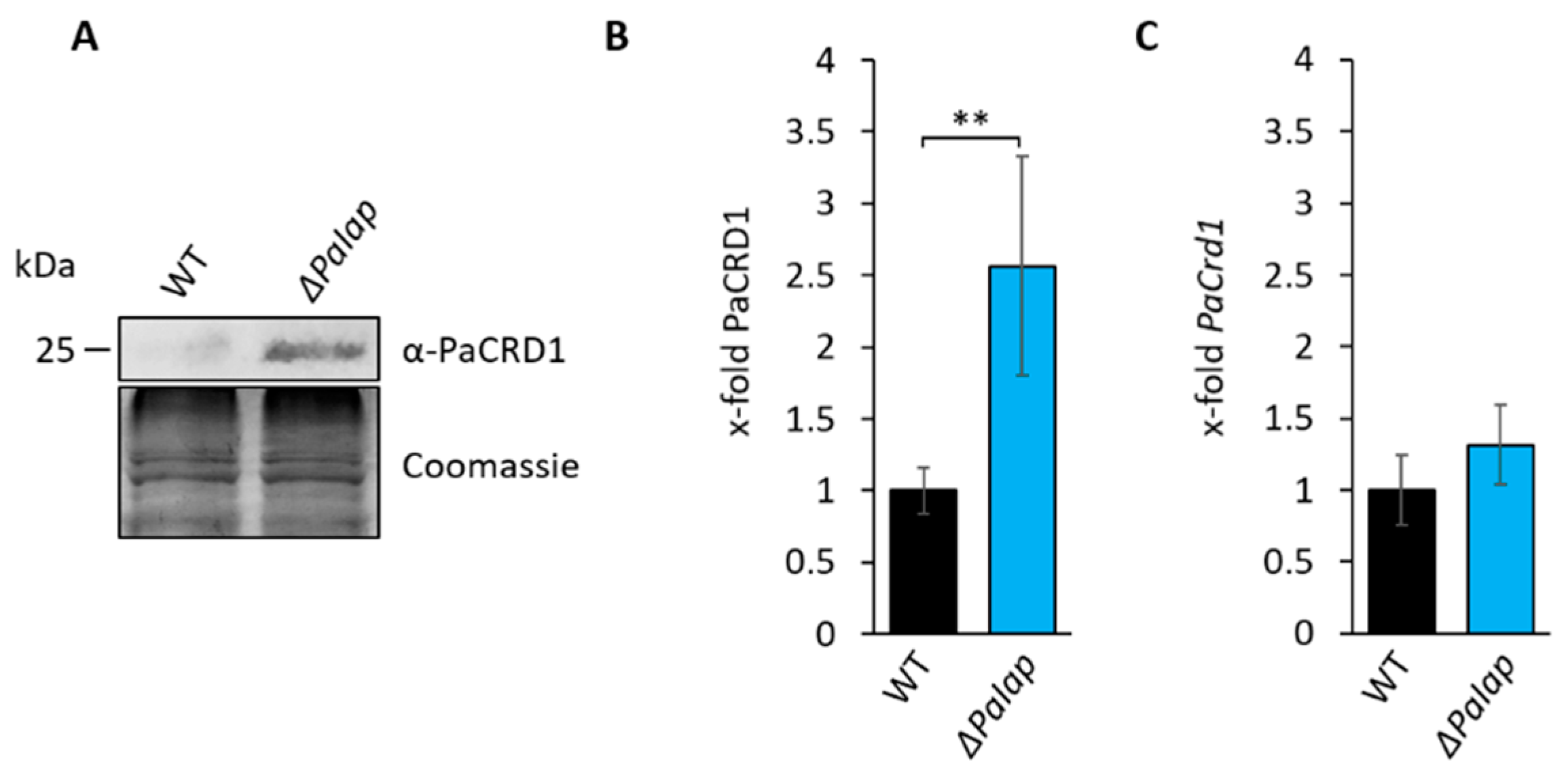
3.3. Absence of PaIAP Promotes Cardiolipin Synthase Activity by Loss of Proteolytic Turnover
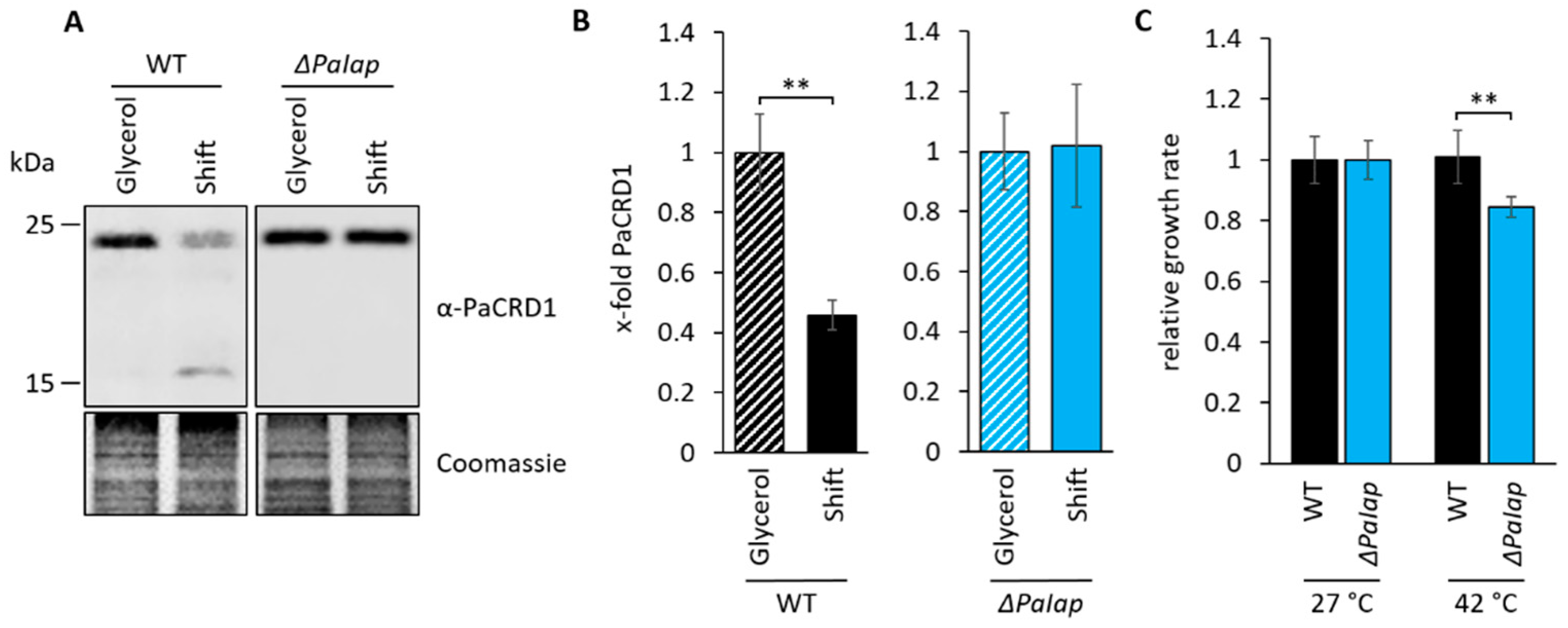
3.4. The Effect of ΔPaIap on Longevity Is Artificially Achieved in Wild-Type Strains Growing on Non-Fermentable Glycerol
3.5. PaIAP as an Adaptive Regulator Is Necessary for Short-Term Reaction on Changing Environmental Conditions
3.6. Elevated Mitochondrial Turnover Ensures Proper Quality Control despite Absence of PaIAP
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rizet, G. Impossibility of obtaining uninterrupted and unlimited multiplication of the ascomycete Podospora anserina. C. R. Hebd. Seances Acad. Sci. 1953, 237, 838–840. [Google Scholar]
- Bernhardt, D.; Hamann, A.; Osiewacz, H. The role of mitochondria in fungal aging. Curr. Opin. Microbiol. 2014, 22, 1–7. [Google Scholar] [CrossRef]
- Luce, K.; Weil, A.C.; Osiewacz, H.D. Mitochondrial protein quality control systems in aging and disease. Adv. Exp. Med. Biol 2010, 694, 108–125. [Google Scholar] [CrossRef]
- Fischer, F.; Hamann, A.; Osiewacz, H.D. Mitochondrial quality control: An integrated network of pathways. Trends Biochem. Sci. 2012, 37, 284–292. [Google Scholar] [CrossRef]
- Scheckhuber, C.Q.; Osiewacz, H.D. Podospora anserina: A model organism to study mechanisms of healthy ageing. Mol. Genet. Genomics 2008, 280, 365–374. [Google Scholar] [CrossRef]
- Osiewacz, H.D. Aging in fungi: Role of mitochondria in Podospora anserina. Mech. Ageing Dev. 2002, 123, 755–764. [Google Scholar] [CrossRef]
- Kück, U.; Osiewacz, H.D.; Schmidt, U.; Kappelhoff, B.; Schulte, E.; Stahl, U.; Esser, K. The onset of senescence is affected by DNA rearrangements of a discontinuous mitochondrial gene in Podospora anserina. Curr. Genet. 1985, 9, 373–382. [Google Scholar] [CrossRef] [PubMed]
- Fischer, F.; Weil, A.; Hamann, A.; Osiewacz, H.D. Human CLPP reverts the longevity phenotype of a fungal ClpP deletion strain. Nat. Commun 2013, 4, 1397. [Google Scholar] [CrossRef]
- Luce, K.; Osiewacz, H.D. Increasing organismal healthspan by enhancing mitochondrial protein quality control. Nat. Cell Biol. 2009, 11, 852–858. [Google Scholar] [CrossRef] [PubMed]
- Weil, A.; Luce, K.; Dröse, S.; Wittig, I.; Brandt, U.; Osiewacz, H.D. Unmasking a temperature-dependent effect of the P. anserina i-AAA protease on aging and development. Cell Cycle 2011, 10, 4280–4290. [Google Scholar] [CrossRef] [PubMed]
- Osman, C.; Voelker, D.R.; Langer, T. Making heads or tails of phospholipids in mitochondria. J. Cell Biol 2011, 192, 7–16. [Google Scholar] [CrossRef]
- Colina-Tenorio, L.; Horten, P.; Pfanner, N.; Rampelt, H. Shaping the mitochondrial inner membrane in health and disease. J. Intern. Med. 2020, 287, 645–664. [Google Scholar] [CrossRef]
- Van der Veen, J.N.; Kennelly, J.P.; Wan, S.; Vance, J.E.; Vance, D.E.; Jacobs, R.L. The critical role of phosphatidylcholine and phosphatidylethanolamine metabolism in health and disease. Biochim. Biophys. Acta Biomembr. 2017, 1859, 1558–1572. [Google Scholar] [CrossRef]
- Baker, C.D.; Basu Ball, W.; Pryce, E.N.; Gohil, V.M. Specific requirements of nonbilayer phospholipids in mitochondrial respiratory chain function and formation. Mol. Biol. Cell 2016, 27, 2161–2171. [Google Scholar] [CrossRef]
- Schuler, M.-H.; Di Bartolomeo, F.; Mårtensson, C.U.; Daum, G.; Becker, T. Phosphatidylcholine affects inner membrane protein translocases of mitochondria. J. Biol. Chem. 2016, 291, 18718–18729. [Google Scholar] [CrossRef]
- Ban, T.; Ishihara, T.; Kohno, H.; Saita, S.; Ichimura, A.; Maenaka, K.; Oka, T.; Mihara, K.; Ishihara, N. Molecular basis of selective mitochondrial fusion by heterotypic action between OPA1 and cardiolipin. Nat. Cell Biol. 2017, 19, 856–863. [Google Scholar] [CrossRef]
- Panov, A.V.; Dikalov, S.I. Cardiolipin, perhydroxyl radicals, and lipid peroxidation in mitochondrial dysfunctions and aging. Oxid. Med. Cell Longev. 2020, 2020, 1323028. [Google Scholar] [CrossRef] [PubMed]
- Ademowo, O.S.; Dias, H.K.I.; Burton, D.G.A.; Griffiths, H.R. Lipid (per) oxidation in mitochondria: An emerging target in the ageing process? Biogerontology 2017, 18, 859–879. [Google Scholar] [CrossRef] [PubMed]
- Calzada, E.; Onguka, O.; Claypool, S.M. Phosphatidylethanolamine metabolism in health and disease. Int. Rev. Cell Mol. Biol. 2016, 321, 29–88. [Google Scholar] [CrossRef] [PubMed]
- Esser, K. Podospora anserina. In Handbook of Genetics; King, R.C., Ed.; Springer: New York, NY, USA, 1997; Volume 1, pp. 531–551. [Google Scholar]
- Osiewacz, H.D.; Hamann, A.; Zintel, S. Assessing organismal aging in the filamentous fungus Podospora anserina. Methods Mol. Biol. 2013, 965, 439–462. [Google Scholar] [CrossRef]
- El-Khoury, R.; Sellem, C.H.; Coppin, E.; Boivin, A.; Maas, M.F.; Debuchy, R.; Sainsard-Chanet, A. Gene deletion and allelic replacement in the filamentous fungus Podospora anserina. Curr. Genet. 2008, 53, 249–258. [Google Scholar] [CrossRef]
- Kunstmann, B.; Osiewacz, H.D. The S-adenosylmethionine dependent O-methyltransferase PaMTH1: A longevity assurance factor protecting Podospora anserina against oxidative stress. Aging 2009, 1, 328–334. [Google Scholar] [CrossRef][Green Version]
- Osiewacz, H.D.; Skaletz, A.; Esser, K. Integrative transformation of the ascomycete Podospora anserina: Identification of the mating-type locus on chromosome VII of electrophoretically separated chromosomes. Appl. Microbiol. Biotechnol. 1991, 35, 38–45. [Google Scholar] [CrossRef]
- Averbeck, N.B.; Borghouts, C.; Hamann, A.; Specke, V.; Osiewacz, H.D. Molecular control of copper homeostasis in filamentous fungi: Increased expression of a metallothionein gene during aging of Podospora anserina. Mol. Gen. Genet. 2001, 264, 604–612. [Google Scholar] [CrossRef]
- Kunstmann, B.; Osiewacz, H.D. Over-expression of an S-adenosylmethionine-dependent methyltransferase leads to an extended lifespan of Podospora anserina without impairments in vital functions. Aging Cell 2008, 7, 651–662. [Google Scholar] [CrossRef]
- Zintel, S.; Schwitalla, D.; Luce, K.; Hamann, A.; Osiewacz, H.D. Increasing mitochondrial superoxide dismutase abundance leads to impairments in protein quality control and ROS scavenging systems and to lifespan shortening. Exp. Gerontol. 2010, 45, 525–532. [Google Scholar] [CrossRef] [PubMed]
- Knuppertz, L.; Warnsmann, V.; Hamann, A.; Grimm, C.; Osiewacz, H.D. Stress-dependent opposing roles for mitophagy in aging of the ascomycete Podospora anserina. Autophagy 2017, 13, 1037–1052. [Google Scholar] [CrossRef] [PubMed]
- Lecellier, G.; Silar, P. Rapid methods for nucleic acids extraction from petri dish-grown mycelia. Curr. Genet. 1994, 25, 122–123. [Google Scholar] [CrossRef]
- Sambrook, J.; Fritsch, E.F.; Maniatis, T. Molecular Cloning: A Laboratory Manual; Cold Spring Harbor Laboratory: Cold Spring Harbor, NY, USA, 1989. [Google Scholar]
- Warnsmann, V.; Marschall, L.M.; Osiewacz, H.D. Impaired F(1)F(o)-ATP-synthase dimerization leads to the induction of cyclophilin D-mediated autophagy-dependent cell death and accelerated aging. Cells 2021, 10, 757. [Google Scholar] [CrossRef] [PubMed]
- Ejsing, C.S.; Sampaio, J.L.; Surendranath, V.; Duchoslav, E.; Ekroos, K.; Klemm, R.W.; Simons, K.; Shevchenko, A. Global analysis of the yeast lipidome by quantitative shotgun mass spectrometry. Proc. Natl. Acad. Sci. USA 2009, 106, 2136–2141. [Google Scholar] [CrossRef] [PubMed]
- Klose, C.; Surma, M.A.; Gerl, M.J.; Meyenhofer, F.; Shevchenko, A.; Simons, K. Flexibility of a eukaryotic lipidome—Insights from yeast lipidomics. PLoS ONE 2012, 7, e35063. [Google Scholar] [CrossRef]
- Surma, M.A.; Herzog, R.; Vasilj, A.; Klose, C.; Christinat, N.; Morin-Rivron, D.; Simons, K.; Masoodi, M.; Sampaio, J.L. An automated shotgun lipidomics platform for high throughput, comprehensive, and quantitative analysis of blood plasma intact lipids. Eur. J. Lipid Sci. Technol. 2015, 117, 1540–1549. [Google Scholar] [CrossRef]
- Herzog, R.; Schwudke, D.; Schuhmann, K.; Sampaio, J.L.; Bornstein, S.R.; Schroeder, M.; Shevchenko, A. A novel informatics concept for high-throughput shotgun lipidomics based on the molecular fragmentation query language. Genome Biol. 2011, 12, R8. [Google Scholar] [CrossRef]
- Herzog, R.; Schuhmann, K.; Schwudke, D.; Sampaio, J.L.; Bornstein, S.R.; Schroeder, M.; Shevchenko, A. LipidXplorer: A software for consensual cross-platform lipidomics. PLoS ONE 2012, 7, e29851. [Google Scholar] [CrossRef]
- Bligh, E.G.; Dyer, W.J. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 1959, 37, 911–917. [Google Scholar] [CrossRef] [PubMed]
- Wittig, I.; Braun, H.-P.; Schägger, H. Blue native PAGE. Nat. Protoc. 2006, 1, 418–428. [Google Scholar] [CrossRef]
- Heinz, D.; Krotova, E.; Hamann, A.; Osiewacz, H.D. Simultaneous ablation of the catalytic AMPK α-subunit SNF1 and mitochondrial matrix protease CLPP results in pronounced lifespan extension. Front. Cell Dev. Biol. 2021, 9, 616520. [Google Scholar] [CrossRef] [PubMed]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef] [PubMed]
- Warnsmann, V.; Hainbuch, S.; Osiewacz, H.D. Quercetin-induced lifespan extension in Podospora anserina requires methylation of the flavonoid by the o-methyltransferase PaMTH1. Front. Genet. 2018, 9, 160. [Google Scholar] [CrossRef] [PubMed]
- Scheckhuber, C.Q.; Erjavec, N.; Tinazli, A.; Hamann, A.; Nyström, T.; Osiewacz, H.D. Reducing mitochondrial fission results in increased life span and fitness of two fungal ageing models. Nat. Cell Biol. 2007, 9, 99–105. [Google Scholar] [CrossRef] [PubMed]
- Daum, B.; Walter, A.; Horst, A.; Osiewacz, H.D.; Kühlbrandt, W. Age-dependent dissociation of ATP synthase dimers and loss of inner-membrane cristae in mitochondria. Proc. Natl. Acad. Sci. USA 2013, 110, 15301–15306. [Google Scholar] [CrossRef] [PubMed]
- Eiyama, A.; Aaltonen, M.J.; Nolte, H.; Tatsuta, T.; Langer, T. Disturbed intramitochondrial phosphatidic acid transport impairs cellular stress signaling. J. Biol. Chem. 2021, 296, 100335. [Google Scholar] [CrossRef] [PubMed]
- Hallermayer, G.; Neupert, W. Lipid composition of mitochondrial outer and inner membranes of Neurospora crassa. Hoppe Seylers Z Physiol. Chem. 1974, 355, 279–288. [Google Scholar] [CrossRef]
- Chen, E.; Kiebish, M.A.; McDaniel, J.; Niedzwiecka, K.; Kucharczyk, R.; Ravasz, D.; Gao, F.; Narain, N.R.; Sarangarajan, R.; Seyfried, T.N.; et al. Perturbation of the yeast mitochondrial lipidome and associated membrane proteins following heterologous expression of Artemia-ANT. Sci. Rep. 2018, 8, 5915. [Google Scholar] [CrossRef]
- Horvath, S.E.; Daum, G. Lipids of mitochondria. Prog. Lipid Res. 2013, 52, 590–614. [Google Scholar] [CrossRef]
- Lev, S. Nonvesicular lipid transfer from the endoplasmic reticulum. CSH Perspect. Biol. 2012, 4, a013300. [Google Scholar] [CrossRef]
- Connerth, M.; Tatsuta, T.; Haag, M.; Klecker, T.; Westermann, B.; Langer, T. Intramitochondrial transport of phosphatidic acid in yeast by a lipid transfer protein. Science 2012, 338, 815–818. [Google Scholar] [CrossRef]
- Miyata, N.; Watanabe, Y.; Tamura, Y.; Endo, T.; Kuge, O. Phosphatidylserine transport by Ups2-Mdm35 in respiration-active mitochondria. J. Cell Biol. 2016, 214, 77–88. [Google Scholar] [CrossRef]
- Potting, C.; Wilmes, C.; Engmann, T.; Osman, C.; Langer, T. Regulation of mitochondrial phospholipids by Ups1/PRELI-like proteins depends on proteolysis and Mdm35. EMBO J. 2010, 29, 2888–2898. [Google Scholar] [CrossRef]
- Schlame, M.; Haldar, D. Cardiolipin is synthesized on the matrix side of the inner membrane in rat liver mitochondria. J. Biol. Chem. 1993, 268, 74–79. [Google Scholar] [CrossRef]
- Horvath, S.E.; Böttinger, L.; Vögtle, F.N.; Wiedemann, N.; Meisinger, C.; Becker, T.; Daum, G. Processing and topology of the yeast mitochondrial phosphatidylserine decarboxylase 1. J. Biol. Chem. 2012, 287, 36744–36755. [Google Scholar] [CrossRef] [PubMed]
- Aaltonen, M.J.; Friedman, J.R.; Osman, C.; Salin, B.; di Rago, J.P.; Nunnari, J.; Langer, T.; Tatsuta, T. MICOS and phospholipid transfer by Ups2-Mdm35 organize membrane lipid synthesis in mitochondria. J. Cell Biol. 2016, 213, 525–534. [Google Scholar] [CrossRef]
- Birner, R.; Bürgermeister, M.; Schneiter, R.; Daum, G. Roles of phosphatidylethanolamine and of its several biosynthetic pathways in Saccharomyces cerevisiae. Mol. Biol. Cell 2001, 12, 997–1007. [Google Scholar] [CrossRef] [PubMed]
- Garlid, A.O.; Schaffer, C.T.; Kim, J.; Bhatt, H.; Guevara-Gonzalez, V.; Ping, P. TAZ encodes tafazzin, a transacylase essential for cardiolipin formation and central to the etiology of Barth syndrome. Gene 2020, 726, 144148. [Google Scholar] [CrossRef] [PubMed]
- Jiang, F.; Ryan, M.T.; Schlame, M.; Zhao, M.; Gu, Z.; Klingenberg, M.; Pfanner, N.; Greenberg, M.L. Absence of cardiolipin in the crd1 null mutant results in decreased mitochondrial membrane potential and reduced mitochondrial function. J. Biol. Chem. 2000, 275, 22387–22394. [Google Scholar] [CrossRef] [PubMed]
- Pokorná, L.; Čermáková, P.; Horváth, A.; Baile, M.G.; Claypool, S.M.; Griač, P.; Malínský, J.; Balážová, M. Specific degradation of phosphatidylglycerol is necessary for proper mitochondrial morphology and function. Biochim. Biophys. Acta 2016, 1857, 34–45. [Google Scholar] [CrossRef]
- Escribá, P.V.; Ozaita, A.; Ribas, C.; Miralles, A.; Fodor, E.; Farkas, T.; García-Sevilla, J.A. Role of lipid polymorphism in G protein-membrane interactions: Nonlamellar-prone phospholipids and peripheral protein binding to membranes. Proc. Natl. Acad. Sci. USA 1997, 94, 11375–11380. [Google Scholar] [CrossRef]
- Cullis, P.R.; de Kruijff, B. Lipid polymorphism and the functional roles of lipids in biological membranes. Biochim. Biophys. Acta 1979, 559, 399–420. [Google Scholar] [CrossRef]
- Tasseva, G.; Bai, H.D.; Davidescu, M.; Haromy, A.; Michelakis, E.; Vance, J.E. Phosphatidylethanolamine deficiency in mammalian mitochondria impairs oxidative phosphorylation and alters mitochondrial morphology. J. Biol. Chem. 2013, 288, 4158–4173. [Google Scholar] [CrossRef] [PubMed]
- Fu, S.; Yang, L.; Li, P.; Hofmann, O.; Dicker, L.; Hide, W.; Lin, X.; Watkins, S.M.; Ivanov, A.R.; Hotamisligil, G.S. Aberrant lipid metabolism disrupts calcium homeostasis causing liver endoplasmic reticulum stress in obesity. Nature 2011, 473, 528–531. [Google Scholar] [CrossRef]
- Gao, X.; van der Veen, J.N.; Vance, J.E.; Thiesen, A.; Vance, D.E.; Jacobs, R.L. Lack of phosphatidylethanolamine N-methyltransferase alters hepatic phospholipid composition and induces endoplasmic reticulum stress. Biochim. Biophys. Acta 2015, 1852, 2689–2699. [Google Scholar] [CrossRef]
- Thibault, G.; Shui, G.; Kim, W.; McAlister, G.C.; Ismail, N.; Gygi, S.P.; Wenk, M.R.; Ng, D.T. The membrane stress response buffers lethal effects of lipid disequilibrium by reprogramming the protein homeostasis network. Mol. Cell 2012, 48, 16–27. [Google Scholar] [CrossRef]
- Ogunbona, O.B.; Onguka, O.; Calzada, E.; Claypool, S.M. Multitiered and cooperative surveillance of mitochondrial phosphatidylserine decarboxylase 1. Mol. Cell Biol. 2017, 37, e00049-17. [Google Scholar] [CrossRef]
- Weil, A. Untersuchungen zur Bedeutung der mitochondrialen Proteinqualitätskontrolle für Alterungsprozesse bei dem Ascomyceten Podospora anserina. Ph.D. Thesis, Goethe-University, Frankfurt am Main, Germany, 2013. [Google Scholar]
- Maranzana, E.; Barbero, G.; Falasca, A.I.; Lenaz, G.; Genova, M.L. Mitochondrial respiratory supercomplex association limits production of reactive oxygen species from complex I. Antioxid. Redox Signal. 2013, 19, 1469–1480. [Google Scholar] [CrossRef] [PubMed]
- Genova, M.L.; Lenaz, G. The interplay between respiratory supercomplexes and ROS in aging. Antioxid. Redox Signal. 2015, 23, 208–238. [Google Scholar] [CrossRef]
- Zhang, M.; Mileykovskaya, E.; Dowhan, W. Gluing the respiratory chain together. Cardiolipin is required for supercomplex formation in the inner mitochondrial membrane. J. Biol. Chem. 2002, 277, 43553–43556. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Lou, W.; Grevel, A.; Böttinger, L.; Liang, Z.; Ji, J.; Patil, V.A.; Liu, J.; Ye, C.; Hüttemann, M.; et al. Cardiolipin-deficient cells have decreased levels of the iron-sulfur biogenesis protein frataxin. J. Biol. Chem. 2020, 295, 11928–11937. [Google Scholar] [CrossRef] [PubMed]
- Stefanyk, L.E.; Coverdale, N.; Roy, B.D.; Peters, S.J.; LeBlanc, P.J. Skeletal muscle type comparison of subsarcolemmal mitochondrial membrane phospholipid fatty acid composition in rat. J. Membr. Biol. 2010, 234, 207–215. [Google Scholar] [CrossRef] [PubMed]
- Chicco, A.J.; Sparagna, G.C.; McCune, S.A.; Johnson, C.A.; Murphy, R.C.; Bolden, D.A.; Rees, M.L.; Gardner, R.T.; Moore, R.L. Linoleate-rich high-fat diet decreases mortality in hypertensive heart failure rats compared with lard and low-fat diets. Hypertension 2008, 52, 549–555. [Google Scholar] [CrossRef]
- Mulligan, C.M.; Sparagna, G.C.; Le, C.H.; De Mooy, A.B.; Routh, M.A.; Holmes, M.G.; Hickson-Bick, D.L.; Zarini, S.; Murphy, R.C.; Xu, F.Y.; et al. Dietary linoleate preserves cardiolipin and attenuates mitochondrial dysfunction in the failing rat heart. Cardiovasc. Res. 2012, 94, 460–468. [Google Scholar] [CrossRef]
- Ernst, R.; Ejsing, C.S.; Antonny, B. Homeoviscous adaptation and the regulation of membrane lipids. J. Mol. Biol. 2016, 428, 4776–4791. [Google Scholar] [CrossRef]
- Opalińska, M.; Parys, K.; Jańska, H. Identification of physiological substrates and binding partners of the plant mitochondrial protease FTSH4 by the trapping approach. Int. J. Mol. Sci. 2017, 18, 2455. [Google Scholar] [CrossRef]
- Claypool, S.M.; Whited, K.; Srijumnong, S.; Han, X.; Koehler, C.M. Barth syndrome mutations that cause tafazzin complex lability. J. Cell Biol. 2011, 192, 447–462. [Google Scholar] [CrossRef] [PubMed]
- Stiburek, L.; Cesnekova, J.; Kostkova, O.; Fornuskova, D.; Vinsova, K.; Wenchich, L.; Houstek, J.; Zeman, J. YME1L controls the accumulation of respiratory chain subunits and is required for apoptotic resistance, cristae morphogenesis, and cell proliferation. Mol. Biol. Cell 2012, 23, 1010–1023. [Google Scholar] [CrossRef] [PubMed]
- Aguer, C.; Gambarotta, D.; Mailloux, R.; Moffat, C.; Dent, R.; McPherson, R.; Harper, M.-E. Galactose enhances oxidative metabolism and reveals mitochondrial dysfunction in human primary muscle cells. PLoS ONE 2011, 6, e28536. [Google Scholar] [CrossRef] [PubMed]
- Tudzynski, P.; Esser, K. Inhibitors of mitochondrial function prevent senescence in the ascomycete Podospora anserina. Mol. Gen. Genet. 1977, 153, 111–113. [Google Scholar] [CrossRef] [PubMed]
- Tuller, G.; Hrastnik, C.; Achleitner, G.; Schiefthaler, U.; Klein, F.; Daum, G. YDL142c encodes cardiolipin synthase (Cls1p) and is non-essential for aerobic growth of Saccharomyces cerevisiae. FEBS Lett. 1998, 421, 15–18. [Google Scholar] [CrossRef]
- Tuller, G.; Nemec, T.; Hrastnik, C.; Daum, G. Lipid composition of subcellular membranes of an FY1679-derived haploid yeast wild-type strain grown on different carbon sources. Yeast 1999, 15, 1555–1564. [Google Scholar] [CrossRef]
- Bazán, S.; Mileykovskaya, E.; Mallampalli, V.K.; Heacock, P.; Sparagna, G.C.; Dowhan, W. Cardiolipin-dependent reconstitution of respiratory supercomplexes from purified Saccharomyces cerevisiae complexes III and IV. J. Biol. Chem. 2013, 288, 401–411. [Google Scholar] [CrossRef]
- Leonhard, K.; Herrmann, J.M.; Stuart, R.A.; Mannhaupt, G.; Neupert, W.; Langer, T. AAA proteases with catalytic sites on opposite membrane surfaces comprise a proteolytic system for the ATP-dependent degradation of inner membrane proteins in mitochondria. EMBO J. 1996, 15, 4218–4229. [Google Scholar] [CrossRef] [PubMed]
- Weber, E.R.; Hanekamp, T.; Thorsness, P.E. Biochemical and functional analysis of the YME1 gene product, an ATP and zinc-dependent mitochondrial protease from S. cerevisiae. Mol. Biol. Cell 1996, 7, 307–317. [Google Scholar] [CrossRef] [PubMed]
- Böttinger, L.; Horvath, S.E.; Kleinschroth, T.; Hunte, C.; Daum, G.; Pfanner, N.; Becker, T. Phosphatidylethanolamine and cardiolipin differentially affect the stability of mitochondrial respiratory chain supercomplexes. J. Mol. Biol. 2012, 423, 677–686. [Google Scholar] [CrossRef] [PubMed]
- Arnarez, C.; Marrink, S.J.; Periole, X. Molecular mechanism of cardiolipin-mediated assembly of respiratory chain supercomplexes. Chem. Sci. 2016, 7, 4435–4443. [Google Scholar] [CrossRef] [PubMed]
- Renner, L.D.; Weibel, D.B. Cardiolipin microdomains localize to negatively curved regions of Escherichia coli membranes. Proc. Natl. Acad. Sci. USA 2011, 108, 6264–6269. [Google Scholar] [CrossRef] [PubMed]
- Teague, W.E., Jr.; Soubias, O.; Petrache, H.; Fuller, N.; Hines, K.G.; Rand, R.P.; Gawrisch, K. Elastic properties of polyunsaturated phosphatidylethanolamines influence rhodopsin function. Faraday Discuss. 2013, 161, 383–395. [Google Scholar] [CrossRef] [PubMed]
- Ikon, N.; Ryan, R.O. Cardiolipin and mitochondrial cristae organization. Biochim. Biophys. Acta Biomembr. 2017, 1859, 1156–1163. [Google Scholar] [CrossRef] [PubMed]
- Chan, E.Y.; McQuibban, G.A. Phosphatidylserine decarboxylase 1 (Psd1) promotes mitochondrial fusion by regulating the biophysical properties of the mitochondrial membrane and alternative topogenesis of mitochondrial genome maintenance protein 1 (Mgm1). J. Biol. Chem. 2012, 287, 40131–40139. [Google Scholar] [CrossRef]
- Macdonald, P.J.; Stepanyants, N.; Mehrotra, N.; Mears, J.A.; Qi, X.; Sesaki, H.; Ramachandran, R. A dimeric equilibrium intermediate nucleates Drp1 reassembly on mitochondrial membranes for fission. Mol. Biol. Cell 2014, 25, 1905–1915. [Google Scholar] [CrossRef]
- Li, Z.; Agellon, L.B.; Allen, T.M.; Umeda, M.; Jewell, L.; Mason, A.; Vance, D.E. The ratio of phosphatidylcholine to phosphatidylethanolamine influences membrane integrity and steatohepatitis. Cell Metab. 2006, 3, 321–331. [Google Scholar] [CrossRef] [PubMed]
- Fajardo, V.A.; McMeekin, L.; LeBlanc, P.J. Influence of phospholipid species on membrane fluidity: A meta-analysis for a novel phospholipid fluidity index. J. Membr. Biol. 2011, 244, 97–103. [Google Scholar] [CrossRef]
- Dawaliby, R.; Trubbia, C.; Delporte, C.; Noyon, C.; Ruysschaert, J.M.; Van Antwerpen, P.; Govaerts, C. Phosphatidylethanolamine is a key regulator of membrane fluidity in eukaryotic cells. J. Biol. Chem. 2016, 291, 3658–3667. [Google Scholar] [CrossRef] [PubMed]
- MacVicar, T.; Ohba, Y.; Nolte, H.; Mayer, F.C.; Tatsuta, T.; Sprenger, H.G.; Lindner, B.; Zhao, Y.; Li, J.; Bruns, C.; et al. Lipid signalling drives proteolytic rewiring of mitochondria by YME1L. Nature 2019, 575, 361–365. [Google Scholar] [CrossRef]
- Francis, B.R.; White, K.H.; Thorsness, P.E. Mutations in the Atp1p and Atp3p subunits of yeast ATP synthase differentially affect respiration and fermentation in Saccharomyces cerevisiae. J. Bioenerg. Biomembr. 2007, 39, 127–144. [Google Scholar] [CrossRef] [PubMed]
- Yu, R.; Cao, X.; Sun, L.; Zhu, J.Y.; Wasko, B.M.; Liu, W.; Crutcher, E.; Liu, H.; Jo, M.C.; Qin, L.; et al. Inactivating histone deacetylase HDA promotes longevity by mobilizing trehalose metabolism. Nat. Commun. 2021, 12, 1981. [Google Scholar] [CrossRef] [PubMed]
- Thorsness, P.E.; Fox, T.D. Nuclear mutations in Saccharomyces cerevisiae that affect the escape of DNA from mitochondria to the nucleus. Genetics 1993, 134, 21–28. [Google Scholar] [CrossRef]
- Nebauer, R.; Schuiki, I.; Kulterer, B.; Trajanoski, Z.; Daum, G. The phosphatidylethanolamine level of yeast mitochondria is affected by the mitochondrial components Oxa1p and Yme1p. FEBS J. 2007, 274, 6180–6190. [Google Scholar] [CrossRef]
- Wang, K.; Jin, M.; Liu, X.; Klionsky, D.J. Proteolytic processing of Atg32 by the mitochondrial i-AAA protease Yme1 regulates mitophagy. Autophagy 2013, 9, 1828–1836. [Google Scholar] [CrossRef]
- Knuppertz, L.; Osiewacz, H.D. Autophagy compensates impaired energy metabolism in CLPXP-deficient Podospora anserina strains and extends healthspan. Aging Cell 2017, 16, 704–715. [Google Scholar] [CrossRef]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Löser, T.; Joppe, A.; Hamann, A.; Osiewacz, H.D. Mitochondrial Phospholipid Homeostasis Is Regulated by the i-AAA Protease PaIAP and Affects Organismic Aging. Cells 2021, 10, 2775. https://doi.org/10.3390/cells10102775
Löser T, Joppe A, Hamann A, Osiewacz HD. Mitochondrial Phospholipid Homeostasis Is Regulated by the i-AAA Protease PaIAP and Affects Organismic Aging. Cells. 2021; 10(10):2775. https://doi.org/10.3390/cells10102775
Chicago/Turabian StyleLöser, Timo, Aljoscha Joppe, Andrea Hamann, and Heinz D. Osiewacz. 2021. "Mitochondrial Phospholipid Homeostasis Is Regulated by the i-AAA Protease PaIAP and Affects Organismic Aging" Cells 10, no. 10: 2775. https://doi.org/10.3390/cells10102775
APA StyleLöser, T., Joppe, A., Hamann, A., & Osiewacz, H. D. (2021). Mitochondrial Phospholipid Homeostasis Is Regulated by the i-AAA Protease PaIAP and Affects Organismic Aging. Cells, 10(10), 2775. https://doi.org/10.3390/cells10102775





