Chromatin Accessibility and Transcriptomic Alterations in Murine Ovarian Granulosa Cells upon Deoxynivalenol Exposure
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals and Reagents
2.2. Isolation and Culture of Ovarian GCs
2.3. Treatment of GCs with DON and Assessment of Cell Viability
2.4. Ovarian GCs Morphology and Radioimmunoassay
2.5. Analyses of Apoptosis, Reactive Oxygen Species and Mitochondrial Membrane Potential by Flow Cytometry
2.6. ATAC-Seq Library Preparation and Data Analysis
2.7. RNA-Seq Library Preparation and Data Analysis
2.8. Integrative Analysis of ATAC-Seq and RNA-Seq Data
2.9. Gene Ontology, Pathway and PPI Network Analyses
2.10. RT-PCR, ChIP-qPCR and Western Blotting
2.11. Statistical Analysis
3. Results
3.1. Severe Toxicity Effect of DON on Murine Ovarian GCs
3.2. Numerous Genomic Loci Have Altered Chromatin Accessibility upon DON Exposure
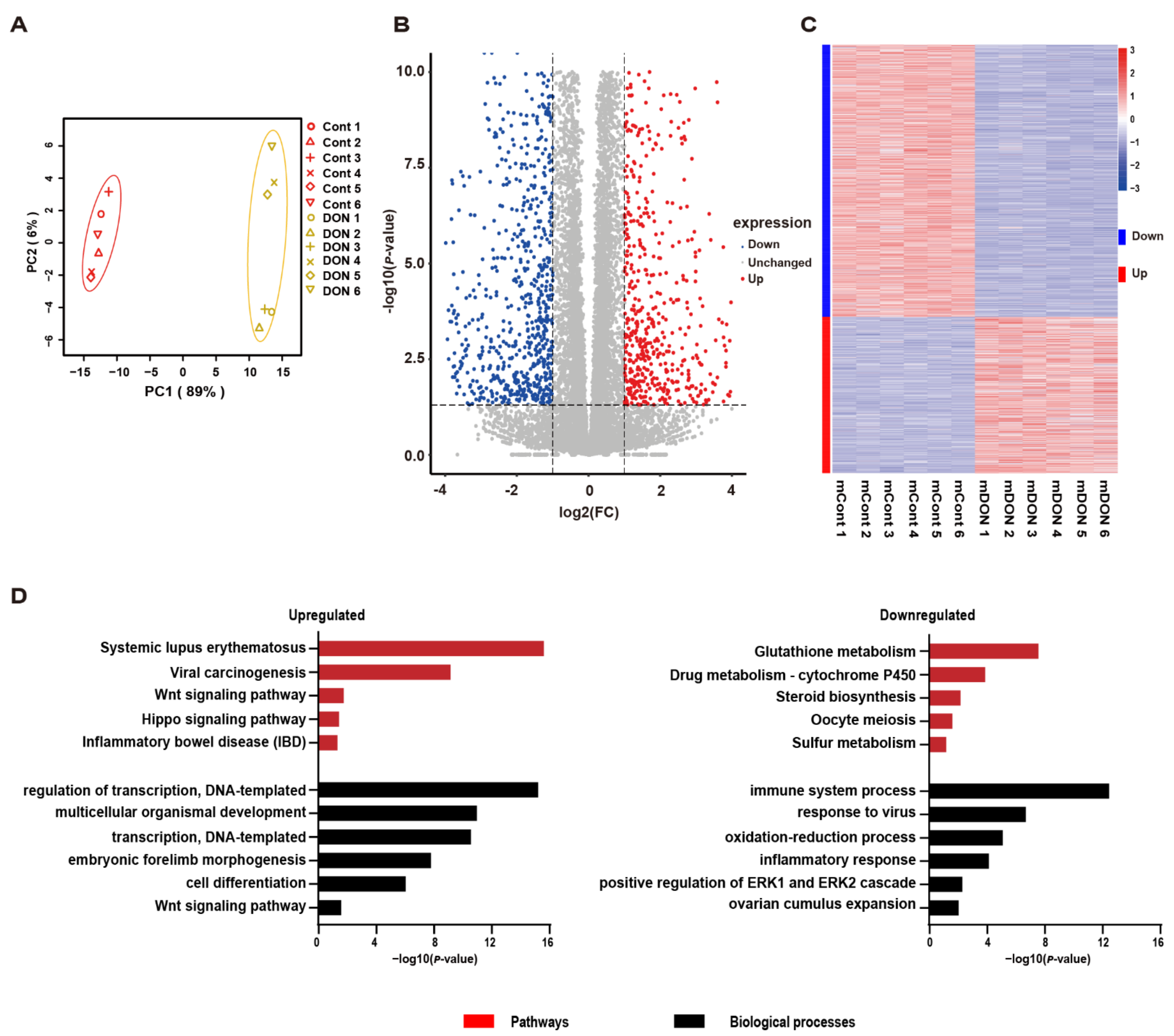
3.3. Thousands of Genes Have Altered Transcription in Response to DON Exposure
3.4. Integrative Analysis Identifies Putative Direct Targets in Response to DON Toxicity
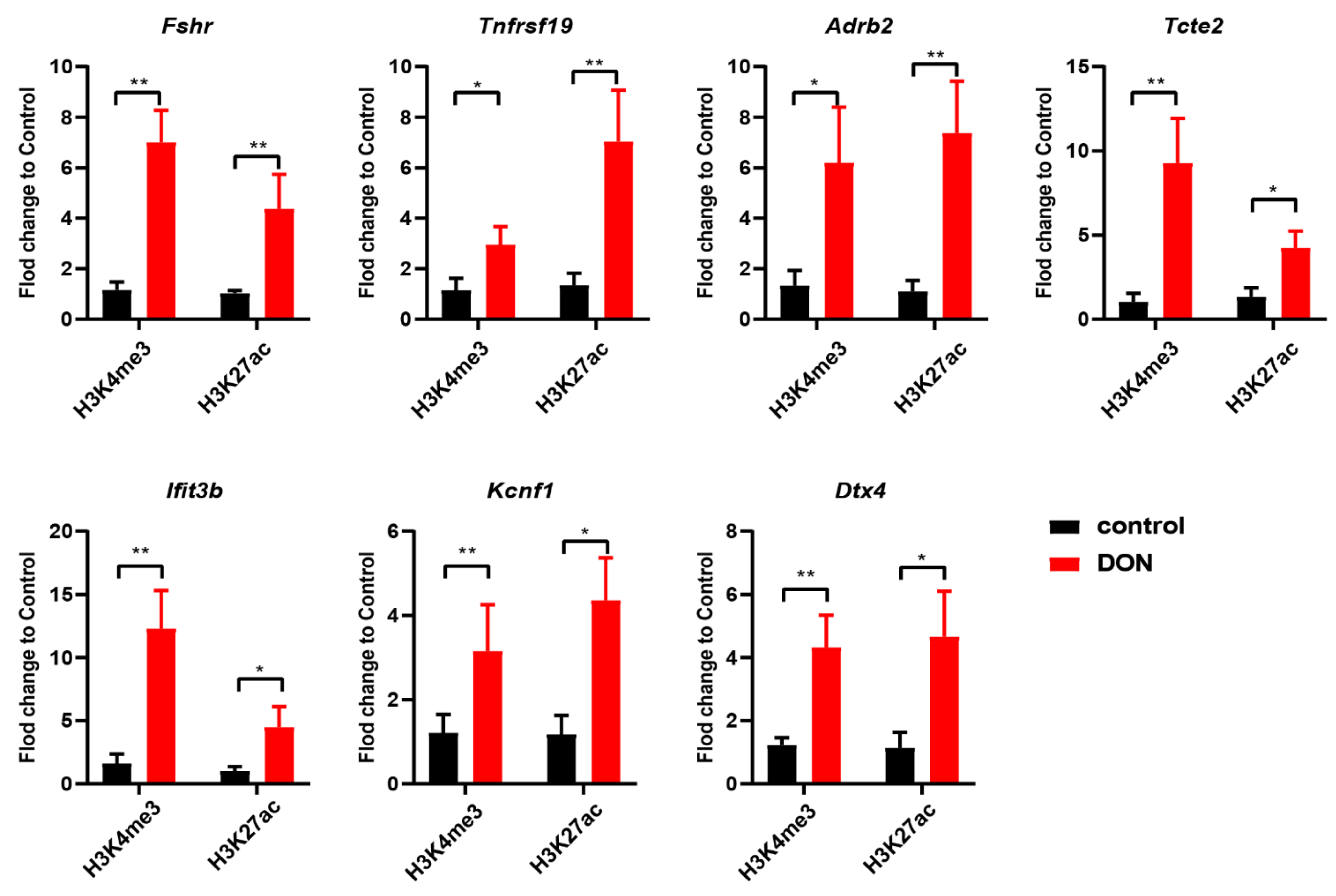
3.5. Active Histone Marks Are Enriched in Open Chromatin Regions in Murine Ovarian GCs upon DON Exposure
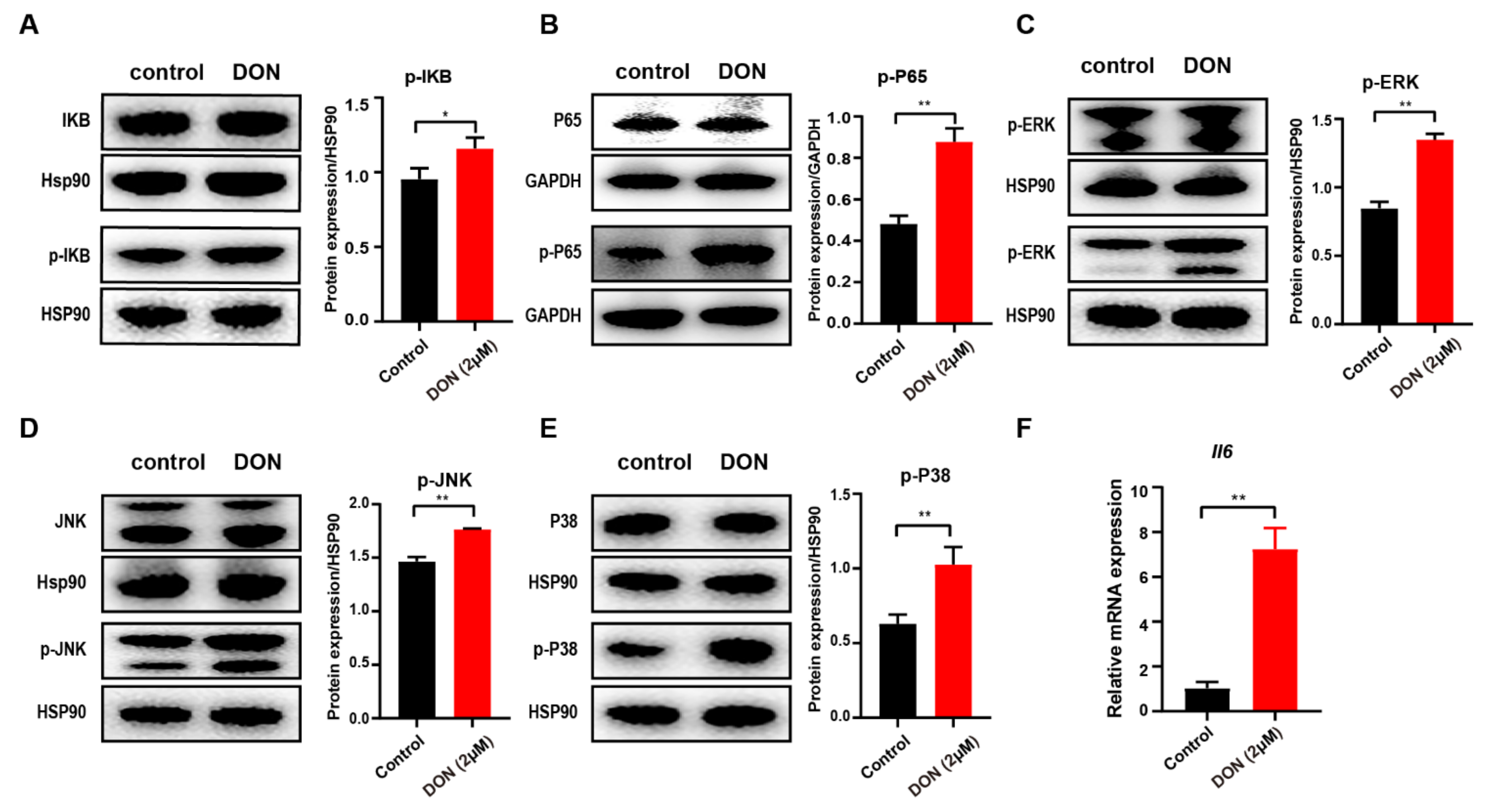
3.6. DON-Induced Inflammatory Response via Activation of NF-κB and MAPK Signaling Pathways
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Li, R.; Albertini, D.F. The road to maturation: Somatic cell interaction and self-organization of the mammalian oocyte. Nat. Rev. Mol. Cell. Biol. 2013, 14, 141–152. [Google Scholar] [CrossRef] [PubMed]
- Manabe, N.; Goto, Y.; Matsuda-Minehata, F.; Inoue, N.; Maeda, A.; Sakamaki, K.; Miyano, T. Regulation Mechanism of Selective Atresia in Porcine Follicles: Regulation of Granulosa Cell Apoptosis during Atresia. J. Reprod. Dev. 2004, 50, 493–514. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Robinson, J.W.; Zhang, M.; Shuhaibar, L.C.; Norris, R.P.; Geerts, A.; Wunder, F.; Eppig, J.J.; Potter, L.R.; Jaffe, L.A. Luteinizing hormone reduces the activity of the NPR2 guanylyl cyclase in mouse ovarian follicles, contributing to the cyclic GMP decrease that promotes resumption of meiosis in oocytes. Dev. Biol. 2012, 366, 308–316. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cai, L.; Sun, A.; Li, H.; Tsinkgou, A.; Yu, J.; Ying, S.; Chen, Z.; Shi, Z. Molecular mechanisms of enhancing porcine granulosa cell proliferation and function by treatment in vitro with anti-inhibin alpha subunit antibody. Reprod. Biol. Endocrinol. 2015, 13, 26. [Google Scholar] [CrossRef] [Green Version]
- Gore-Langton, R.E. Follicle-stimulating hormone and estradiol regulate antrum-like reorganization of granulosa cells in rat preantral follicle cultures. Biol. Reprod. 1990, 43, 65–72. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Urbanek, K.A.; Habrowska-Gorczynska, D.E.; Kowalska, K.; Stanczyk, A.; Dominska, K.; Piastowska-Ciesielska, A.W. Deoxynivalenol as potential modulator of human steroidogenesis. J. Appl. Toxicol. 2018, 38, 1450–1459. [Google Scholar] [CrossRef]
- Han, J.; Wang, Q.C.; Zhu, C.C.; Liu, J.; Zhang, Y.; Cui, X.S.; Kim, N.H.; Sun, S.C. Deoxynivalenol exposure induces autophagy/apoptosis and epigenetic modification changes during porcine oocyte maturation. Toxicol. Appl. Pharm. 2016, 300, 70–76. [Google Scholar] [CrossRef]
- Medvedova, M.; Kolesarova, A.; Capcarova, M.; Labuda, R.; Sirotkin, A.V.; Kovacik, J.; Bulla, J. The effect of deoxynivalenol on the secretion activity, proliferation and apoptosis of porcine ovarian granulosa cells in vitro. J. Env. Sci. Health B 2011, 46, 213–219. [Google Scholar] [CrossRef]
- Wang, H.; Zong, Q.; Wang, S.; Zhao, C.; Wu, S.; Bao, W. Genome-Wide DNA Methylome and Transcriptome Analysis of Porcine Intestinal Epithelial Cells upon Deoxynivalenol Exposure. J. Agric. Food Chem. 2019, 67, 6423–6431. [Google Scholar] [CrossRef]
- Wang, Z.; Wu, Q.; Kuca, K.; Dohnal, V.; Tian, Z. Deoxynivalenol: Signaling pathways and human exposure risk assessment–An update. Arch. Toxicol. 2014, 88, 1915–1928. [Google Scholar] [CrossRef]
- EFSA; Knutsen, H.K.; Alexander, J.; Barregard, L.; Bignami, M.; Bruschweiler, B.; Ceccatelli, S.; Cottrill, B.; Dinovi, M.; Grasl-Kraupp, B.; et al. Risks to Human and Animal Health Related to the Presence of Deoxynivalenol and Its Acetylated and Modified Forms in Food and Feed, 2017/09/11 ed.; John Wiley and Sons Ltd.: Hoboken, NJ, USA, 2017; Volume 15, p. e04718. [Google Scholar]
- Wang, X.; Liu, Q.; Ihsan, A.; Huang, L.; Dai, M.; Hao, H.; Cheng, G.; Liu, Z.; Wang, Y.; Yuan, Z. JAK/STAT pathway plays a critical role in the proinflammatory gene expression and apoptosis of RAW264.7 cells induced by trichothecenes as DON and T-2 toxin. Toxicol. Sci. 2012, 127, 412–424. [Google Scholar] [CrossRef] [PubMed]
- Pestka, J.J.; Amuzie, C.J. Tissue distribution and proinflammatory cytokine gene expression following acute oral exposure to deoxynivalenol: Comparison of weanling and adult mice. Food Chem. Toxicol. 2008, 46, 2826–2831. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, D.; Ma, H.; Ye, Y.; Ji, C.; Tang, X.; Ouyang, D.; Chen, J.; Li, Y.; Ma, Y. Deoxynivalenol induces apoptosis in mouse thymic epithelial cells through mitochondria-mediated pathway. Env. Toxicol. Pharm. 2014, 38, 163–171. [Google Scholar] [CrossRef]
- Li, D.; Ye, Y.; Lin, S.; Deng, L.; Fan, X.; Zhang, Y.; Deng, X.; Li, Y.; Yan, H.; Ma, Y. Evaluation of deoxynivalenol-induced toxic effects on DF-1 cells in vitro: Cell-cycle arrest, oxidative stress, and apoptosis. Env. Toxicol. Pharm. 2014, 37, 141–149. [Google Scholar] [CrossRef]
- Borutova, R.; Faix, S.; Placha, I.; Gresakova, L.; Cobanova, K.; Leng, L. Effects of deoxynivalenol and zearalenone on oxidative stress and blood phagocytic activity in broilers. Arch. Anim. Nutr. 2008, 62, 303–312. [Google Scholar] [CrossRef] [PubMed]
- Guerrero-Netro, H.M.; Chorfi, Y.; Price, C.A. Effects of the mycotoxin deoxynivalenol on steroidogenesis and apoptosis in granulosa cells. Reproduction 2015, 149, 555–561. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, M.; Wu, X.; Zhang, W.; Ye, P.; Wang, Y.; Zhu, W.; Tao, Q.; Xu, Y.; Shang, J.; Zhao, D.; et al. Transcriptional analysis of deoxynivalenol-induced apoptosis of sow ovarian granulosa cell. Reprod. Domest. Anim. 2020, 55, 217–228. [Google Scholar] [CrossRef]
- Dai, Y.; Xie, H.; Xu, Y. Evaluation of deoxynivalenol-induced toxic effects on mouse endometrial stromal cells: Cell apoptosis and cell cycle. Biochem. Biophys. Res. Commun. 2017, 483, 572–577. [Google Scholar] [CrossRef] [PubMed]
- Alm, H.; Greising, T.; Brussow, K.-P.; Torner, H.; Tiemann, U. The influence of the mycotoxins deoxynivalenol and zearalenol on in vitro maturation of pig oocytes and in vitro culture of pig zygotes. Toxicol. Vitr. 2002, 16, 643–648. [Google Scholar] [CrossRef]
- Minervinia, F.; Dell’Aquilab, M.E.; Maritatob, F.; Minoiab, P.; Viscontia, A. Toxic effects of the mycotoxin zearalenone and its derivatives on in vitro maturation of bovine oocytes and 17β-estradiol levels in mural granulosa cell cultures. Toxicol. Vitr. 2001, 15, 489–495. [Google Scholar] [CrossRef]
- Alm, H.; Brüssow, K.-P.; Torner, H.; Vanselow, J.; Tomek, W.; Dänicke, S.; Tiemann, U. Influence of Fusarium-toxin contaminated feed on initial quality and meiotic competence of gilt oocytes. Reprod. Toxicol. 2006, 22, 44–50. [Google Scholar] [CrossRef]
- Lan, M.; Han, J.; Pan, M.H.; Wan, X.; Pan, Z.N.; Sun, S.C. Melatonin protects against defects induced by deoxynivalenol during mouse oocyte maturation. J. Pineal Res. 2018, 65, e12477. [Google Scholar] [CrossRef]
- Malekinejad, H.; Schoevers, E.J.; Daemen, I.J.; Zijlstra, C.; Colenbrander, B.; Fink-Gremmels, J.; Roelen, B.A. Exposure of oocytes to the Fusarium toxins zearalenone and deoxynivalenol causes aneuploidy and abnormal embryo development in pigs. Biol. Reprod. 2007, 77, 840–847. [Google Scholar] [CrossRef] [Green Version]
- Li, B.; Carey, M.; Workman, J.L. The role of chromatin during transcription. Cell 2007, 128, 707–719. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Voss, T.C.; Hager, G.L. Dynamic regulation of transcriptional states by chromatin and transcription factors. Nat. Rev. Genet. 2014, 15, 69–81. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, X.; Song, Q.; Hou, Y.; Liu, J.; Sun, Y.; Wang, P. Characterization of the chromatin accessibility in an Alzheimer’s disease (AD) mouse model. Alzheimers Res. 2020, 12, 29. [Google Scholar] [CrossRef] [PubMed]
- Buenrostro, J.; Wu, B.; Chang, H.; Greenleaf, W. ATAC-seq: A Method for Assaying Chromatin Accessibility Genome-Wide. Curr. Protoc. Mol. Biol. 2015, 109, 21–29. [Google Scholar] [CrossRef] [PubMed]
- Ruiz, J.L.; Tena, J.J.; Bancells, C.; Cortes, A.; Gomez-Skarmeta, J.L.; Gomez-Diaz, E. Characterization of the accessible genome in the human malaria parasite Plasmodium falciparum. Nucleic Acids Res. 2018, 46, 9414–9431. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lowe, E.K.; Cuomo, C.; Voronov, D.; Arnone, M.I. Using ATAC-seq and RNA-seq to increase resolution in GRN connectivity. Methods Cell Biol. 2019, 151, 115–126. [Google Scholar] [PubMed]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Liu, T.; Meyer, C.A.; Eeckhoute, J.; Johnson, D.S.; Bernstein, B.E.; Nusbaum, C.; Myers, R.M.; Brown, M.; Li, W.; et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 2008, 9, R137. [Google Scholar] [CrossRef] [Green Version]
- Ramirez, F.; Dundar, F.; Diehl, S.; Gruning, B.A.; Manke, T. deepTools: A flexible platform for exploring deep-sequencing data. Nucleic Acids Res. 2014, 42, W187–W191. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Quinlan, A.R.; Hall, I.M. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 2010, 26, 841–842. [Google Scholar] [CrossRef] [Green Version]
- Heinz, S.; Benner, C.; Spann, N.; Bertolino, E.; Lin, Y.C.; Laslo, P.; Cheng, J.X.; Murre, C.; Singh, H.; Glass, C.K. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 2010, 38, 576–589. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR ultrafast universal RNA seq aligner. Bioinformatics 2013, 29, 15–29. [Google Scholar] [CrossRef] [PubMed]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq–A Python framework to work with high-throughput sequencing data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef] [PubMed]
- Anders, S.; Huber, W. Differential expression analysis for sequence count data. Genome Biol. 2010, 11, R106. [Google Scholar] [CrossRef] [Green Version]
- McLean, C.Y.; Bristor, D.; Hiller, M.; Clarke, S.L.; Schaar, B.T.; Lowe, C.B.; Wenger, A.M.; Bejerano, G. GREAT improves functional interpretation of cis-regulatory regions. Nat. Biotechnol. 2010, 28, 495–501. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Draghici, S.; Khatri, P.; Tarca, A.L.; Amin, K.; Done, A.; Voichita, C.; Georgescu, C.; Romero, R. A systems biology approach for pathway level analysis. Genome Res. 2007, 17, 1537–1545. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stark, C.; Breitkreutz, B.J.; Reguly, T.; Boucher, L.; Breitkreutz, A.; Tyers, M. BioGRID: A general repository for interaction datasets. Nucleic Acids Res. 2006, 34, D535–D539. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Turei, D.; Korcsmaros, T.; Saez-Rodriguez, J. OmniPath: Guidelines and gateway for literature-curated signaling pathway resources. Nat. Methods 2016, 13, 966–967. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, S.; Pohl, M.O.; Zhou, Y.; Rodriguez-Frandsen, A.; Wang, G.; Stein, D.A.; Moulton, H.M.; DeJesus, P.; Che, J.; Mulder, L.C.; et al. Meta- and Orthogonal Integration of Influenza “OMICs” Data Defines a Role for UBR4 in Virus Budding. Cell Host Microbe 2015, 18, 723–735. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Ba Der, G.D.; Hogue, C. An Automated Method for Finding Molecular Complexes in Large Protein Interaction Networks. BMC Bioinform. 2003, 4, 2. [Google Scholar]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2 (-Delta Delta C (T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Sahu, M.R.; Mondal, A.C. The emerging role of Hippo signaling in neurodegeneration. J. Neurosci. Res. 2020, 98, 796–814. [Google Scholar] [CrossRef]
- Wang, S.P.; Wang, L.H. Disease implication of hyper-Hippo signalling. Open Biol. 2016, 6, 160119. [Google Scholar] [CrossRef] [Green Version]
- Zhang, C.; Large, M.J.; Duggavathi, R.; DeMayo, F.J.; Lydon, J.P.; Schoonjans, K.; Kovanci, E.; Murphy, B.D. Liver receptor homolog-1 is essential for pregnancy. Nat. Med. 2013, 19, 1061–1066. [Google Scholar] [CrossRef] [Green Version]
- Duggavathi, R.; Volle, D.H.; Mataki, C.; Antal, M.C.; Messaddeq, N.; Auwerx, J.; Murphy, B.D.; Schoonjans, K. Liver receptor homolog 1 is essential for ovulation. Genes Dev. 2008, 22, 1871–1876. [Google Scholar] [CrossRef] [Green Version]
- Calo, E.; Wysocka, J. Modification of enhancer chromatin: What, how, and why? Mol. Cell 2013, 49, 825–837. [Google Scholar] [CrossRef] [Green Version]
- Dey, N.; Liu, T.; Garofalo, R.P.; Casola, A. TAK1 regulates NF-KappaB and AP-1 activation in airway epithelial cells following RSV infection. Virology 2011, 418, 93–101. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, R.; Zhang, M.; Zhou, W.; Ly, P.T.; Cai, F.; Song, W. NF-kappaB signaling inhibits ubiquitin carboxyl-terminal hydrolase L1 gene expression. J. Neurochem. 2011, 116, 1160–1170. [Google Scholar] [CrossRef] [PubMed]
- Mikami, O.; Yamamoto, S.; Yamanaka, N.; Nakajima, Y. Porcine hepatocyte apoptosis and reduction of albumin secretion induced by deoxynivalenol. Toxicology 2004, 204, 241–249. [Google Scholar] [CrossRef] [PubMed]
- Diesing, A.K.; Nossol, C.; Panther, P.; Walk, N.; Post, A.; Kluess, J.; Kreutzmann, P.; Danicke, S.; Rothkotter, H.J.; Kahlert, S. Mycotoxin deoxynivalenol (DON) mediates biphasic cellular response in intestinal porcine epithelial cell lines IPEC-1 and IPEC-J2. Toxicol. Lett. 2011, 200, 8–18. [Google Scholar] [CrossRef] [PubMed]
- Kang, R.; Li, R.; Dai, P.; Li, Z.; Li, Y.; Li, C. Deoxynivalenol induced apoptosis and inflammation of IPEC-J2 cells by promoting ROS production. Env. Pollut. 2019, 251, 689–698. [Google Scholar] [CrossRef] [PubMed]
- Fan., H.; Wang, S.; Wang, H.; Sun, M.; Wu, S.; Bao, W. Melatonin Ameliorates the Toxicity Induced by Deoxynivalenol in Murine Ovary Granulosa Cells by Antioxidative and Anti-Inflammatory Effects. Antioxididants 2021, 10, 1045. [Google Scholar]
- Dechassa, M.L.; Tryndyak, V.; de Conti, A.; Xiao, W.; Beland, F.A.; Pogribny, I.P. Identification of chromatin-accessible domains in non-alcoholic steatohepatitis-derived hepatocellular carcinoma. Mol. Carcinog. 2018, 57, 978–987. [Google Scholar] [CrossRef] [PubMed]
- Qu, Y.L.; Deng, C.H.; Luo, Q.; Shang, X.Y.; Wu, J.X.; Shi, Y.; Wang, L.; Han, Z.G. Arid1a regulates insulin sensitivity and lipid metabolism. EBioMedicine 2019, 42, 481–493. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Merz, C.; Saller, S.; Kunz, L.; Xu, J.; Yeoman, R.R.; Ting, A.Y.; Lawson, M.S.; Stouffer, R.L.; Hennebold, J.D.; Pau, F.; et al. Expression of the beta-2 adrenergic receptor (ADRB-2) in human and monkey ovarian follicles: A marker of growing follicles? J. Ovarian Res. 2015, 8, 8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Agac, D.; Estrada, L.D.; Maples, R.; Hooper, L.V.; Farrar, J.D. The β2-adrenergic receptor controls inflammation by driving rapid IL-10 secretion. Brain Behav. Immun. 2018, 74, 176–185. [Google Scholar] [CrossRef]
- Mendelson, C.R.; Kamat, A. Mechanisms in the regulation of aromatase in developing ovary and placenta. J. Steroid Biochem. Mol. Biol. 2007, 106, 62–70. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Labelle-Dumais, C.; Jacob-Wagner, M.; Pare, J.F.; Belanger, L.; Dufort, D. Nuclear receptor NR5A2 is required for proper primitive streak morphogenesis. Dev. Dyn. 2006, 235, 3359–3369. [Google Scholar] [CrossRef] [PubMed]
- Val, P.; Lefrançois-Martinez, A.M.; Veyssière, G.; Martinez, A. SF1 a key player in the development and differentiation of steroidogenic tissues. Nucl. Recept. 2003, 1, 8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oh, Y.C.; Cho, W.K.; Jeong, Y.H.; Im, G.Y.; Kim, A.; Hwang, Y.H.; Kim, T.; Song, K.H.; Ma, J.Y. A Novel Herbal Medicine KIOM-MA Exerts an Anti-Inflammatory Effect in LPS-Stimulated RAW 264.7 Macrophage Cells. Evid. Based Complement Altern. Med. 2012, 2012, 462383. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaminska, B. MAPK signalling pathways as molecular targets for anti-inflammatory therapy--from molecular mechanisms to therapeutic benefits. Biochim. Biophys. Acta 2005, 1754, 253–262. [Google Scholar] [CrossRef]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fan, H.; Ren, Z.; Xu, C.; Wang, H.; Wu, Z.; Rehman, Z.u.; Wu, S.; Sun, M.-a.; Bao, W. Chromatin Accessibility and Transcriptomic Alterations in Murine Ovarian Granulosa Cells upon Deoxynivalenol Exposure. Cells 2021, 10, 2818. https://doi.org/10.3390/cells10112818
Fan H, Ren Z, Xu C, Wang H, Wu Z, Rehman Zu, Wu S, Sun M-a, Bao W. Chromatin Accessibility and Transcriptomic Alterations in Murine Ovarian Granulosa Cells upon Deoxynivalenol Exposure. Cells. 2021; 10(11):2818. https://doi.org/10.3390/cells10112818
Chicago/Turabian StyleFan, Hairui, Zhanshi Ren, Chao Xu, Haifei Wang, Zhengchang Wu, Zia ur Rehman, Shenglong Wu, Ming-an Sun, and Wenbin Bao. 2021. "Chromatin Accessibility and Transcriptomic Alterations in Murine Ovarian Granulosa Cells upon Deoxynivalenol Exposure" Cells 10, no. 11: 2818. https://doi.org/10.3390/cells10112818







