Transcriptional and Metabolic Investigation in 5′-Nucleotidase Deficient Cancer Cell Lines
Abstract
:1. Introduction
2. Material and Methods
2.1. Cell Models and Culture
2.2. RNA Sequencing
2.3. Quantitative RT-PCR
2.4. Targeted Metabolomics
3. Results
3.1. Gene Expression in 5′-Nucleotidase-Deficient Cancer Cells
3.2. Pathway-Directed Analysis of Gene Expression Profiles
3.3. Transcriptional Regulation by Adenosine and AMP
3.4. Metabolic Modifications in Cell Models
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Flinn, A.M.; Gennery, A.R. Adenosine deaminase deficiency: A review. Orphanet J. Rare Dis. 2018, 13, 65. [Google Scholar] [CrossRef]
- Garone, C.; Taylor, R.W.; Nascimento, A.; Poulton, J.; Fratter, C.; Dominguez-Gonzalez, C.; Evans, J.C.; Loos, M.; Isohanni, P.; Suomalainen, A.; et al. Retrospective natural history of thymidine kinase 2 deficiency. J. Med. Genet. 2018, 55, 515–521. [Google Scholar] [CrossRef]
- Koker, S.A.; Oymak, Y.; Bianchi, P.; Fermo, E.; Karapinar, T.H.; Gozmen, S.; Ay, Y.; Vergin, R.C. A New Homozygous Mutation (c.393-394del TA/c.393-394del TA) in the NT5C3 Gene Associated With Pyrimidine-5′-Nucleotidase Deficiency: A Case Report. J. Pediatr. Hematol. Oncol. 2019, 41, e484–e486. [Google Scholar] [CrossRef] [PubMed]
- Perez-Torras, S.; Mata-Ventosa, A.; Drogemoller, B.; Tarailo-Graovac, M.; Meijer, J.; Meinsma, R.; van Cruchten, A.G.; Kulik, W.; Viel-Oliva, A.; Bidon-Chanal, A.; et al. Deficiency of perforin and hCNT1, a novel inborn error of pyrimidine metabolism, associated with a rapidly developing lethal phenotype due to multi-organ failure. Biochim. Biophys. Acta (BBA)-Mol. Basis Dis. 2019, 1865, 1182–1191. [Google Scholar] [CrossRef] [PubMed]
- St Hilaire, C.; Ziegler, S.G.; Markello, T.C.; Brusco, A.; Groden, C.; Gill, F.; Carlson-Donohoe, H.; Lederman, R.J.; Chen, M.Y.; Yang, D.; et al. NT5E mutations and arterial calcifications. N. Engl. J. Med. 2011, 364, 432–442. [Google Scholar] [CrossRef]
- Novarino, G.; Fenstermaker, A.G.; Zaki, M.S.; Hofree, M.; Silhavy, J.L.; Heiberg, A.D.; Abdellateef, M.; Rosti, B.; Scott, E.; Mansour, L.; et al. Exome sequencing links corticospinal motor neuron disease to common neurodegenerative disorders. Science 2014, 343, 506–511. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Minor, M.; Alcedo, K.P.; Battaglia, R.A.; Snider, N.T. Cell type- and tissue-specific functions of ecto-5′-nucleotidase (CD73). Am. J. Physiol.-Cell Physiol. 2019, 317, C1079–C1092. [Google Scholar] [CrossRef] [PubMed]
- Thompson, L.F.; Eltzschig, H.K.; Ibla, J.C.; Van De Wiele, C.J.; Resta, R.; Morote-Garcia, J.C.; Colgan, S.P. Crucial role for ecto-5′-nucleotidase (CD73) in vascular leakage during hypoxia. J. Exp. Med. 2004, 200, 1395–1405. [Google Scholar] [CrossRef]
- Johanns, M.; Kviklyte, S.; Chuang, S.J.; Corbeels, K.; Jacobs, R.; Herinckx, G.; Vertommen, D.; Schakman, O.; Duparc, T.; Cani, P.D.; et al. Genetic deletion of soluble 5′-nucleotidase II reduces body weight gain and insulin resistance induced by a high-fat diet. Mol. Genet. Metab. 2019, 126, 377–387. [Google Scholar] [CrossRef]
- Kviklyte, S.; Vertommen, D.; Yerna, X.; Andersen, H.; Xu, X.; Gailly, P.; Bohlooly, Y.M.; Oscarsson, J.; Rider, M.H. Effects of genetic deletion of soluble 5′-nucleotidases NT5C1A and NT5C2 on AMPK activation and nucleotide levels in contracting mouse skeletal muscles. Am. J. Physiol.-Endocrinol. Metab. 2017, 313, E48–E62. [Google Scholar] [CrossRef]
- Dumontet, C.; Peyrottes, S.; Rabeson, C.; Cros-Perrial, E.; Geant, P.Y.; Chaloin, L.; Jordheim, L.P. CD73 inhibition by purine cytotoxic nucleoside analogue-based diphosphonates. Eur. J. Med. Chem. 2018, 157, 1051–1055. [Google Scholar] [CrossRef] [PubMed]
- Ghoteimi, R.; Braka, A.; Rodriguez, C.; Cros-Perrial, E.; Tai Nguyen, V.; Uttaro, J.P.; Mathe, C.; Chaloin, L.; Menetrier-Caux, C.; Jordheim, L.P.; et al. 4-Substituted-1,2,3-triazolo nucleotide analogues as CD73 inhibitors, their synthesis, in vitro screening, kinetic and in silico studies. Bioorg. Chem. 2021, 107, 104577. [Google Scholar] [CrossRef]
- Ghoteimi, R.; Nguyen, V.T.; Rahimova, R.; Grosjean, F.; Cros-Perrial, E.; Uttaro, J.P.; Mathe, C.; Chaloin, L.; Jordheim, L.P.; Peyrottes, S. Synthesis of Substituted 5′-Aminoadenosine Derivatives and Evaluation of Their Inhibitory Potential toward CD73. ChemMedChem 2019, 14, 1431–1443. [Google Scholar] [CrossRef] [Green Version]
- Rahimova, R.; Fontanel, S.; Lionne, C.; Jordheim, L.P.; Peyrottes, S.; Chaloin, L. Identification of allosteric inhibitors of the ecto-5′-nucleotidase (CD73) targeting the dimer interface. PLoS Comput. Biol. 2018, 14, e1005943. [Google Scholar] [CrossRef] [PubMed]
- Guillon, R.; Rahimova, R.; Preeti; Egron, D.; Rouanet, S.; Dumontet, C.; Aghajari, N.; Jordheim, L.P.; Chaloin, L.; Peyrottes, S. Lead optimization and biological evaluation of fragment-based cN-II inhibitors. Eur. J. Med. Chem. 2019, 168, 28–44. [Google Scholar] [CrossRef] [Green Version]
- Jordheim, L.P.; Marton, Z.; Rhimi, M.; Cros-Perrial, E.; Lionne, C.; Peyrottes, S.; Dumontet, C.; Aghajari, N.; Chaloin, L. Identification and characterization of inhibitors of cytoplasmic 5′-nucleotidase cN-II issued from virtual screening. Biochem. Pharmacol. 2013, 85, 497–506. [Google Scholar] [CrossRef] [PubMed]
- Marton, Z.; Guillon, R.; Krimm, I.; Preeti; Rahimova, R.; Egron, D.; Jordheim, L.P.; Aghajari, N.; Dumontet, C.; Perigaud, C.; et al. Identification of Noncompetitive Inhibitors of Cytosolic 5′-Nucleotidase II Using a Fragment-Based Approach. J. Med. Chem. 2015, 58, 9680–9696. [Google Scholar] [CrossRef]
- Jordheim, L.P.; Puy, J.Y.; Cros-Perrial, E.; Peyrottes, S.; Lefebvre, I.; Perigaud, C.; Dumontet, C. Determination of the enzymatic activity of cytosolic 5′-nucleotidase cN-II in cancer cells: Development of a simple analytical method and related cell line models. Anal. Bioanal. Chem. 2015, 407, 5747–5758. [Google Scholar] [CrossRef]
- Bricard, G.; Cadassou, O.; Cassagnes, L.E.; Cros-Perrial, E.; Payen-Gay, L.; Puy, J.Y.; Lefebvre-Tournier, I.; Tozzi, M.G.; Dumontet, C.; Jordheim, L.P. The cytosolic 5′-nucleotidase cN-II lowers the adaptability to glucose deprivation in human breast cancer cells. Oncotarget 2017, 8, 67380–67393. [Google Scholar] [CrossRef] [Green Version]
- Cadassou, O.; Raza, M.Z.; Machon, C.; Gudefin, L.; Armanet, C.; Chettab, K.; Guitton, J.; Tozzi, M.G.; Dumontet, C.; Cros-Perrial, E.; et al. Enhanced migration of breast and lung cancer cells deficient for cN-II and CD73 via COX-2/PGE2/AKT axis regulation. Cell. Oncol. 2021, 44, 151–165. [Google Scholar] [CrossRef]
- Raza, M.Z.; Cadassou, O.; Dumontet, C.; Cros-Perrial, E.; Jordheim, L.P. CD73 and cN-II regulate the cellular response to chemotherapeutic and hypoxic stress in lung adenocarcinoma cells. Biochim. Biophys. Acta (BBA)-Gen. Subj. 2021, 1865, 129842. [Google Scholar] [CrossRef] [PubMed]
- Duarte, R.R.R.; Bachtel, N.D.; Cotel, M.C.; Lee, S.H.; Selvackadunco, S.; Watson, I.A.; Hovsepian, G.A.; Troakes, C.; Breen, G.D.; Nixon, D.F.; et al. The Psychiatric Risk Gene NT5C2 Regulates Adenosine Monophosphate-Activated Protein Kinase Signaling and Protein Translation in Human Neural Progenitor Cells. Biol. Psychiatry 2019, 86, 120–130. [Google Scholar] [CrossRef] [Green Version]
- Moriyama, T.; Liu, S.; Li, J.; Meyer, J.; Zhao, X.; Yang, W.; Shao, Y.; Heath, R.; Hnizda, A.; Carroll, W.L.; et al. Mechanisms of NT5C2-Mediated Thiopurine Resistance in Acute Lymphoblastic Leukemia. Mol. Cancer Ther. 2019, 18, 1887–1895. [Google Scholar] [CrossRef] [Green Version]
- Ueno, K.; Iwagawa, T.; Kuribayashi, H.; Baba, Y.; Nakauchi, H.; Murakami, A.; Nagasaki, M.; Suzuki, Y.; Watanabe, S. Transition of differential histone H3 methylation in photoreceptors and other retinal cells during retinal differentiation. Sci. Rep. 2016, 6, 29264. [Google Scholar] [CrossRef]
- Trapnell, C.; Williams, B.A.; Pertea, G.; Mortazavi, A.; Kwan, G.; van Baren, M.J.; Salzberg, S.L.; Wold, B.J.; Pachter, L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010, 28, 511–515. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garavaglia, S.; Bruzzone, S.; Cassani, C.; Canella, L.; Allegrone, G.; Sturla, L.; Mannino, E.; Millo, E.; De Flora, A.; Rizzi, M. The high-resolution crystal structure of periplasmic Haemophilus influenzae NAD nucleotidase reveals a novel enzymatic function of human CD73 related to NAD metabolism. Biochem. J. 2012, 441, 131–141. [Google Scholar] [CrossRef]
- Grozio, A.; Sociali, G.; Sturla, L.; Caffa, I.; Soncini, D.; Salis, A.; Raffaelli, N.; De Flora, A.; Nencioni, A.; Bruzzone, S. CD73 protein as a source of extracellular precursors for sustained NAD+ biosynthesis in FK866-treated tumor cells. J. Biol. Chem. 2013, 288, 25938–25949. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sociali, G.; Raffaghello, L.; Magnone, M.; Zamporlini, F.; Emionite, L.; Sturla, L.; Bianchi, G.; Vigliarolo, T.; Nahimana, A.; Nencioni, A.; et al. Antitumor effect of combined NAMPT and CD73 inhibition in an ovarian cancer model. Oncotarget 2016, 7, 2968–2984. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kulkarni, S.S.; Karlsson, H.K.; Szekeres, F.; Chibalin, A.V.; Krook, A.; Zierath, J.R. Suppression of 5′-nucleotidase enzymes promotes AMP-activated protein kinase (AMPK) phosphorylation and metabolism in human and mouse skeletal muscle. J. Biol. Chem. 2011, 286, 34567–34574. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pesi, R.; Allegrini, S.; Balestri, F.; Garcia-Gil, M.; Cividini, F.; Colombaioni, L.; Jordheim, L.P.; Camici, M.; Tozzi, M.G. Cytosolic 5′-Nucleotidase II Is a Sensor of Energy Charge and Oxidative Stress: A Possible Function as Metabolic Regulator. Cells 2021, 10, 182. [Google Scholar] [CrossRef] [PubMed]
- Yegutkin, G.G. Enzymes involved in metabolism of extracellular nucleotides and nucleosides: Functional implications and measurement of activities. Crit. Rev. Biochem. Mol. Biol. 2014, 49, 473–497. [Google Scholar] [CrossRef] [PubMed]
- Ipata, P.L.; Pesi, R. Nucleoside recycling in the brain and the nucleosidome: A complex metabolic and molecular cross-talk between the extracellular nucleotide cascade system and the intracellular nucleoside salvage. Metabolomics 2016, 12, 22. [Google Scholar] [CrossRef]
- Cividini, F.; Tozzi, M.G.; Galli, A.; Pesi, R.; Camici, M.; Dumontet, C.; Jordheim, L.P.; Allegrini, S. Cytosolic 5′-Nucleotidase II Interacts with the Leucin Rich Repeat of NLR Family Member Ipaf. PLoS ONE 2015, 10, e0121525. [Google Scholar] [CrossRef] [PubMed] [Green Version]
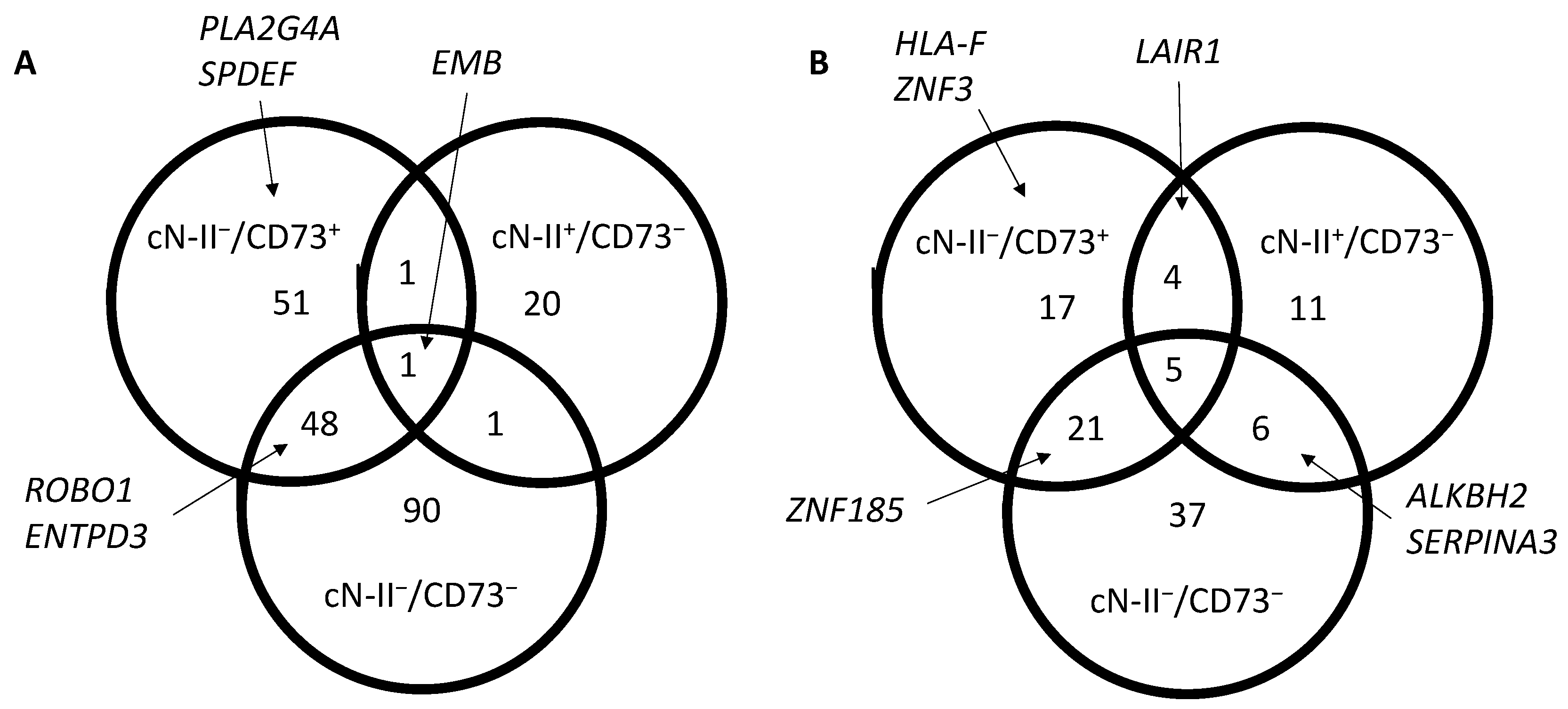



| RNA Sequencing | qRT-PCR | |||||||
|---|---|---|---|---|---|---|---|---|
| Gene | cN-II+/CD73+ | cN-II−/ CD73+ | cN-II+/ CD73− | cN-II−/ CD73− | cN-II+/ CD73+ | cN-II−/ CD73+ | cN-II+/ CD73− | cN-II−/ CD73− |
| ALKBH2 | 1.1 | 0.2 | 0.0003 a | 0.001 a | 0.8 ± 0.2 | 1.09 ± 0.05 a | 0.66 ± 0.04 | 1.4 ± 0.2 a |
| ALOX5AP | 3.2 | 0.7 a | 0.7 | 0.2 | 0.6 ± 0.3 | 0.7 ± 0.2 | 0.23 ± 0.01 | 0.8 ± 0.5 |
| EMB | 0.4 | 10.8 a | 2.5 a | 14.6 a | 0.7 ± 0.2 | 16.1 ± 3.9 a | 2.9 ± 0.7 | 22.2 ± 8.6 a |
| ENTPD3 | 0.4 | 2.1 a | 0.4 | 4.4 a | 0.8 ± 0.2 | 3.8 ± 1.1 a | 1.2 ± 0.4 | 13.9 ± 2.3 a |
| HLA-F | 1.3 | 0.1 a | 0.8 | 0.4 | 0.7 ± 0.2 | 0.6 ± 0.2 | 0.55 ± 0.08 | 0.6 ± 0.3 |
| LAIR1 | 1.4 | 0.0003 a | 0.08 a | 0.2 | 0.8 ± 0.2 | 1.1 ± 0.2 a | 0.30 ± 0.09 a | 0.83 ± 0.08 |
| LOX | 5.6 | 9.5 | 9.5 | 12.6 a | 0.8 ± 0.1 | 2.3 ± 0.7 a | 2.0 ± 0.3 a | 4.1 ± 0.7 a |
| PLA2G4A | 0.02 | 1.5 a | 0.02 | 1.6 a | 1.1 ± 0.4 | 79 ± 10 a | 3.1 ± 1.2 | 111 ± 10 a |
| ROBO1 | n.d. | 1.6 a | n.d. | 2.0 a | 1.0 ± 0.2 | 90 ± 16 a | 5.2 ± 2.2 | 225 ± 17 a |
| SERPINA3 | 2.5 | 0.8 a | 0.2 a | 0.1 a | 0.6 ± 0.3 | 0.6 ± 0.2 | 0.17 ± 0.04 a | 0.3 ± 0.2 |
| SPDEF | 0.1 | 15.0 a | 0.6 | 18.2 | 0.9 ± 0.3 | 14.5 ± 0.7 a | 2.0 ± 0.6 | 17.7 ± 2.2 a |
| ZNF185 | 1.6 | 0.1 a | 0.9 | n.d.a | 0.9 ± 0.1 | 1.3 ± 0.2 a | 1.30 ± 0.03 a | 0.82 ± 0.07 |
| ZNF3 | 1.4 | 0.1 a | 0.009 | 0.5 | 0.8 ± 0.1 | 0.8 ± 0.1 | 0.9 ± 0.1 | 0.8 ± 0.1 |
| NCI-H292 | RL | |||||
|---|---|---|---|---|---|---|
| Gene | cN-II+/ CD73+ | cN-II−/ CD73+ | cN-II+/ CD73− | cN-II−/ CD73− | cN-II+ | cN-II− |
| ALOX5AP | 0.6 ± 0.4 | 0.4 ± 0.2 | 0.6 ± 0.1 | 0.5 ± 0.3 | 0.9 ± 0.2 | 1.2 ± 0.3 |
| EMB | 1.4 ± 0.5 | 2.5 ± 0.7 a | 2.4 ± 0.1 a | 2.5 ± 0.5 a | 1.0 ± 0.2 | 1.4 ± 0.1 |
| ENTPD3 | 0.8 ± 0.2 | 1.5 ± 0.5 a | 1.2 ± 0.3 | 1.3 ± 0.5 | n.d. | n.d. |
| LAIR1 | n.d. | n.d. | n.d. | n.d. | 1.5 ± 0.7 | 0.8 ± 0.9 |
| LOX | 1.6 ± 0.8 | 3.17 ± 0.06 a | 2.6 ± 0.3 | 2.7 ± 1.2 | 0.9 ± 0.4 | 0.8 ± 0.4 |
| PLA2G4 | 1.4 ± 0.3 | 1.3 ± 0.2 | 1.4 ± 1.0 | 1.4 ± 0.5 | 0.7 ± 0.6 | 8.0 ± 8.2 |
| ROBO1 | 1.7 ± 1.2 | 2.9 ± 0.7 | 0.7 ± 0.2 a | 0.7 ± 0.2 a | 1.2 ± 0.2 | 0.8 ± 0.1 |
| SERPINA3 | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. |
| SPDEF | 0.8 ± 0.4 | 1.4 ± 0.3 | 1.7 ± 0.7 | 3.3 ± 2.3 a | n.d. | n.d. |
| Model | Pathway Term | Genes | p-Value |
|---|---|---|---|
| cN-II+/CD73− | Pantothenate and CoA biosynthesis | 4 | 0.00053 |
| HTLV-I infection | 6 | 0.0083 | |
| Herpes simplex infection | 8 | 0.025 | |
| Nicotinate and nicotinamide metabolism | 3 | 0.028 | |
| Intestinal immune network for IgA production | 3 | 0.068 | |
| Tuberculosis | 5 | 0.077 | |
| cN-II−/CD73+ | ECM-receptor interaction | 8 | 0.0065 |
| Hippo signaling pathway | 10 | 0.015 | |
| Protein digestion and absorption | 7 | 0.024 | |
| Complement and coagulation cascades | 6 | 0.031 | |
| Pathways in cancer | 17 | 0.042 | |
| Mucin type O-Glycan biosynthesis | 4 | 0.043 | |
| Ras signaling pathway | 11 | 0.062 | |
| Focal adhesion | 10 | 0.080 | |
| Rap1 signaling pathway | 10 | 0.087 | |
| cN-II−/CD73− | Complement and coagulation cascades | 8 | 0.0065 |
| Cell adhesion molecules | 11 | 0.016 | |
| Oxytocin signaling pathway | 11 | 0.023 | |
| Proteoglycans in cancer | 13 | 0.028 | |
| Staphylococcus aureus infection | 6 | 0.029 | |
| Viral myocarditis | 6 | 0.036 | |
| Rap1 signaling pathway | 13 | 0.039 | |
| Glucagon signaling pathway | 8 | 0.040 | |
| Protein digestion and absorption | 7 | 0.064 | |
| Malaria | 5 | 0.072 | |
| Inflammatory mediator regulation of TRP channels | 7 | 0.096 | |
| Epstein–Barr virus infection | 8 | 0.099 |
| MDA-MB-231 | NCI-H292 | RL | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Gene | cN-II+/ CD73+ | cN-II−/ CD73+ | cN-II+/ CD73− | cN-II−/ CD73− | cN-II+/ CD73+ | cN-II−/ CD73+ | cN-II+/ CD73− | cN-II−/ CD73− | cN-II+ | cN-II− |
| ACO1 | 1.1 ± 0.7 | 0.9 ± 0.3 | 0.9 ± 0.6 | 1.13 ± 0.09 | 1.2 ± 1.3 | 1.7 ± 0.8 | 0.7 ± 0.2 | 0.8 ± 0.2 | 2.5 ± 1.5 | 5.3 ± 0.3 |
| ASS1 | 1.5 ± 0.6 | 2.0 ± 1.8 | 2.5 ± 1.0 | 2.0 ± 0.2 | 1.0 ± 0.4 | 1.0 ± 0.6 | 0.8 ± 0.2 | 0.8 ± 0.2 | 0.7 ± 0.3 | 3.0 ± 1.0 |
| BCAT1 | 0.8 ± 0.2 | 1.1 ± 0.5 | 0.667 ± 0.002 | 0.83 ± 0.03 | 1.5 ± 0.7 | 1.5 ± 0.3 | 0.9 ± 0.4 | 0.8 ± 0.4 | 1.1 ± 0.2 | 1.3 ± 0.3 |
| CPS1 | 0.8 ± 0.4 | 1.7 ± 0.5 a | 0.7 ± 0.2 | 1.82 ± 0.08 a,b | 0.8 ± 0.4 | 0.7 ± 0.6 | 0.3 ± 0.1 | 0.4 ± 0.1 | 1.1 ± 0.2 | 1.4 ± 0.4 |
| CTH | 0.8 ± 0.2 | 0.5 ± 0.2 | 0.47 ± 0.01 | 0.5 ± 0.1 | 2.0 ± 1.2 | 1.7 ± 0.6 | 1.6 ± 0.5 | 1.5 ± 0.5 | 1.5 ± 0.7 | 1.8 ± 0.4 |
| ECHS1 | 0.7 ± 0.4 | 0.9 ± 0.5 | 0.66 ± 0.04 | 1.2 ± 0.3 | 1.1 ± 0.3 | 1.2 ± 0.5 | 1.0 ± 0.5 | 0.9 ± 0.4 | 1.0 ± 0.2 | 1.3 ± 0.3 |
| FH | 0.6 ± 0.5 | 0.6 ± 0.3 | 0.567 ± 0.004 | 0.7 ± 0.2 | 1.5 ± 0.7 | 1.6 ± 0.6 | 1.3 ± 0.5 | 1.1 ± 0.6 | 0.8 ± 0.2 | 1.0 ± 0.2 |
| GLUD1 | 0.9 ± 0.3 | 1.2 ± 0.4 | 0.1 ± 0.2 | 1.13 ± 0.08 | 1.2 ± 0.6 | 1.5 ± 1.3 | 0.9 ± 0.2 | 0.9 ± 0.3 | 1.1 ± 0.3 | 1.5 ± 0.3 |
| GNPNAT1 | 0.8 ± 0.2 | 0.8 ± 0.4 | 0.9 ± 0.4 | 0.9 ± 0.2 | 0.9 ± 0.3 | 2.0 ± 2.0 | 1.8 ± 0.5 | 1.2 ± 0.2 | 1.0 ± 0.3 | 1.7 ± 0.3 |
| IVD | 0.7 ± 0.4 | 0.7 ± 0.3 | 0.70 ± 0.06 | 0.98 ± 0.06 | 1.1 ± 0.4 | 1.1 ± 0.5 | 0.7 ± 0.3 | 0.8 ± 0.3 | 1.4 ± 0.2 | 2.5 ± 0.3 |
| PDHA1 | 0.94 ± 0.09 | 1.0 ± 0.4 | 1.7 ± 0.3 a | 1.15 ± 0.09 | 1.4 ± 0.6 | 1.3 ± 0.4 | 1.0 ± 0.3 | 1.0 ± 0.4 | 1.0 ± 0.2 | 1.2 ± 0.2 |
| TPI1 | 2.7 ± 2.4 | 2.1 ± 1.1 | 1.1 ± 0.2 | 1.4 ± 0.2 | 1.9 ± 1.0 | 2.0 ± 0.6 | 1.3 ± 0.3 | 1.5 ± 0.7 | 1.2 ± 0.2 | 1.5 ± 0.4 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cadassou, O.; Forey, P.; Machon, C.; Petrotto, E.; Chettab, K.; Tozzi, M.G.; Guitton, J.; Dumontet, C.; Cros-Perrial, E.; Jordheim, L.P. Transcriptional and Metabolic Investigation in 5′-Nucleotidase Deficient Cancer Cell Lines. Cells 2021, 10, 2918. https://doi.org/10.3390/cells10112918
Cadassou O, Forey P, Machon C, Petrotto E, Chettab K, Tozzi MG, Guitton J, Dumontet C, Cros-Perrial E, Jordheim LP. Transcriptional and Metabolic Investigation in 5′-Nucleotidase Deficient Cancer Cell Lines. Cells. 2021; 10(11):2918. https://doi.org/10.3390/cells10112918
Chicago/Turabian StyleCadassou, Octavia, Prescillia Forey, Christelle Machon, Edoardo Petrotto, Kamel Chettab, Maria Grazia Tozzi, Jérôme Guitton, Charles Dumontet, Emeline Cros-Perrial, and Lars Petter Jordheim. 2021. "Transcriptional and Metabolic Investigation in 5′-Nucleotidase Deficient Cancer Cell Lines" Cells 10, no. 11: 2918. https://doi.org/10.3390/cells10112918
APA StyleCadassou, O., Forey, P., Machon, C., Petrotto, E., Chettab, K., Tozzi, M. G., Guitton, J., Dumontet, C., Cros-Perrial, E., & Jordheim, L. P. (2021). Transcriptional and Metabolic Investigation in 5′-Nucleotidase Deficient Cancer Cell Lines. Cells, 10(11), 2918. https://doi.org/10.3390/cells10112918