Involvement of Transforming Growth Factor Beta Family Genes in Gonadal Differentiation in Japanese Eel, Anguilla japonica, According to Sex-Related Gene Expressions
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animals
2.2. E2-Induced Feminized Eels
2.3. HCG-Treated Male Eels
2.4. Sampling Procedures
2.5. Cloning of the Partial-Length amh, amhr2 and Full-Length gsdf cDNAs from Japanese Eel
2.6. Sequence Alignment and Phylogenetic Analysis
2.7. Histology
2.8. Dual ISH and IHC
2.9. RNA Extraction and Reverse Transcription
2.10. Quantification of Gene Transcripts Using Real-Time Quantitative PCR
2.11. Data Analysis
3. Results
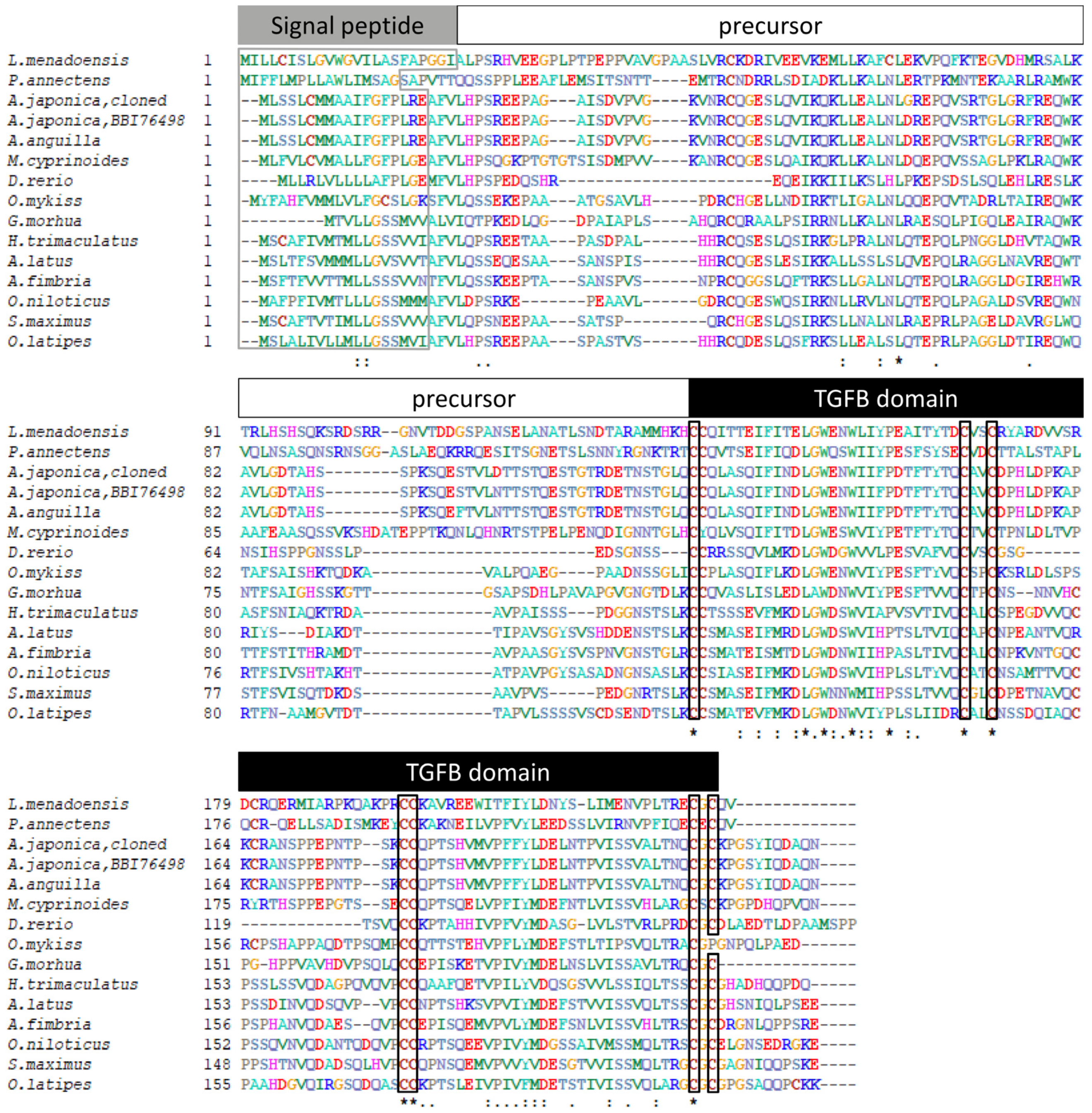
3.1. Sequence Analysis of Japanese Eel gsdf
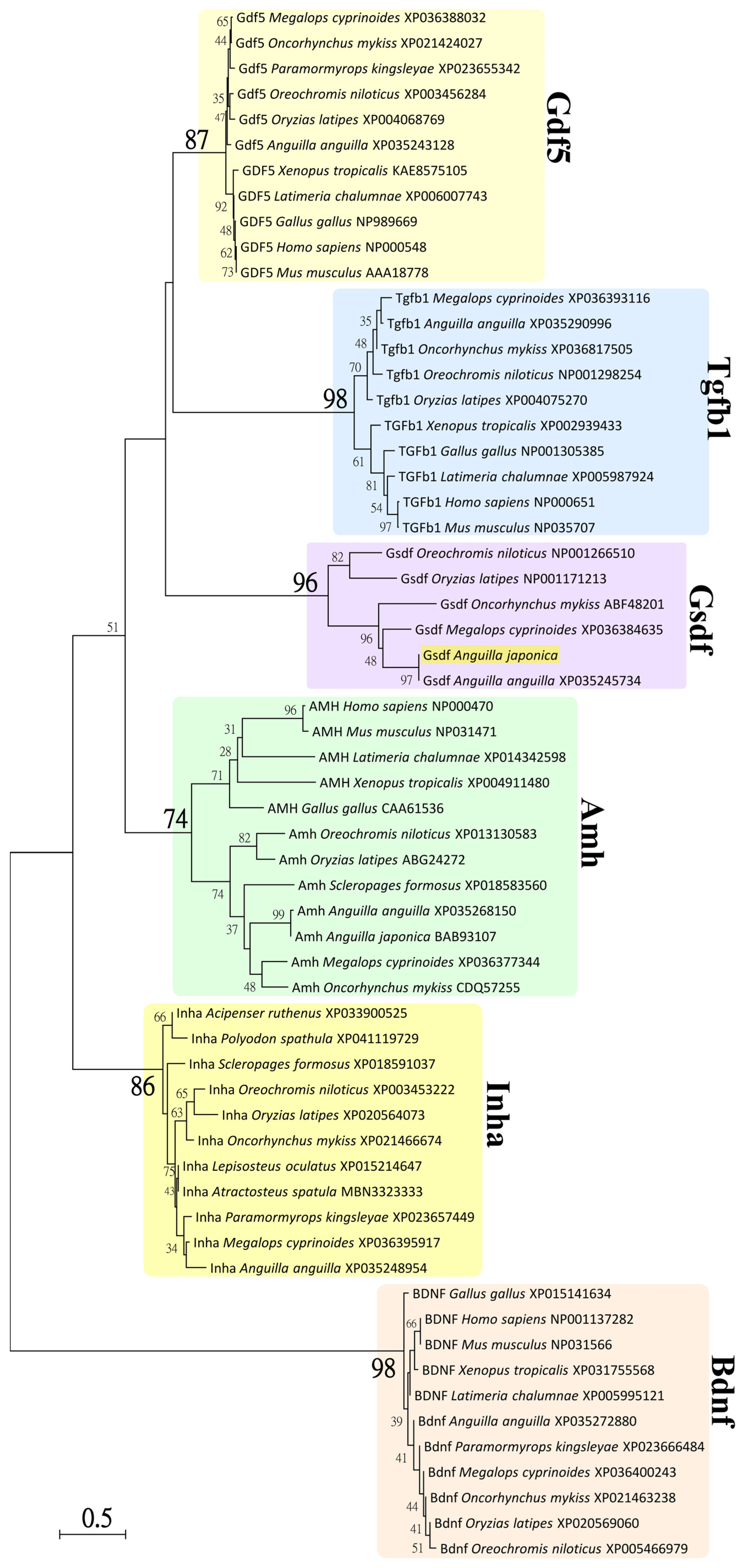
3.2. Phylogenetic Analysis of Japanese Eel Gsdf
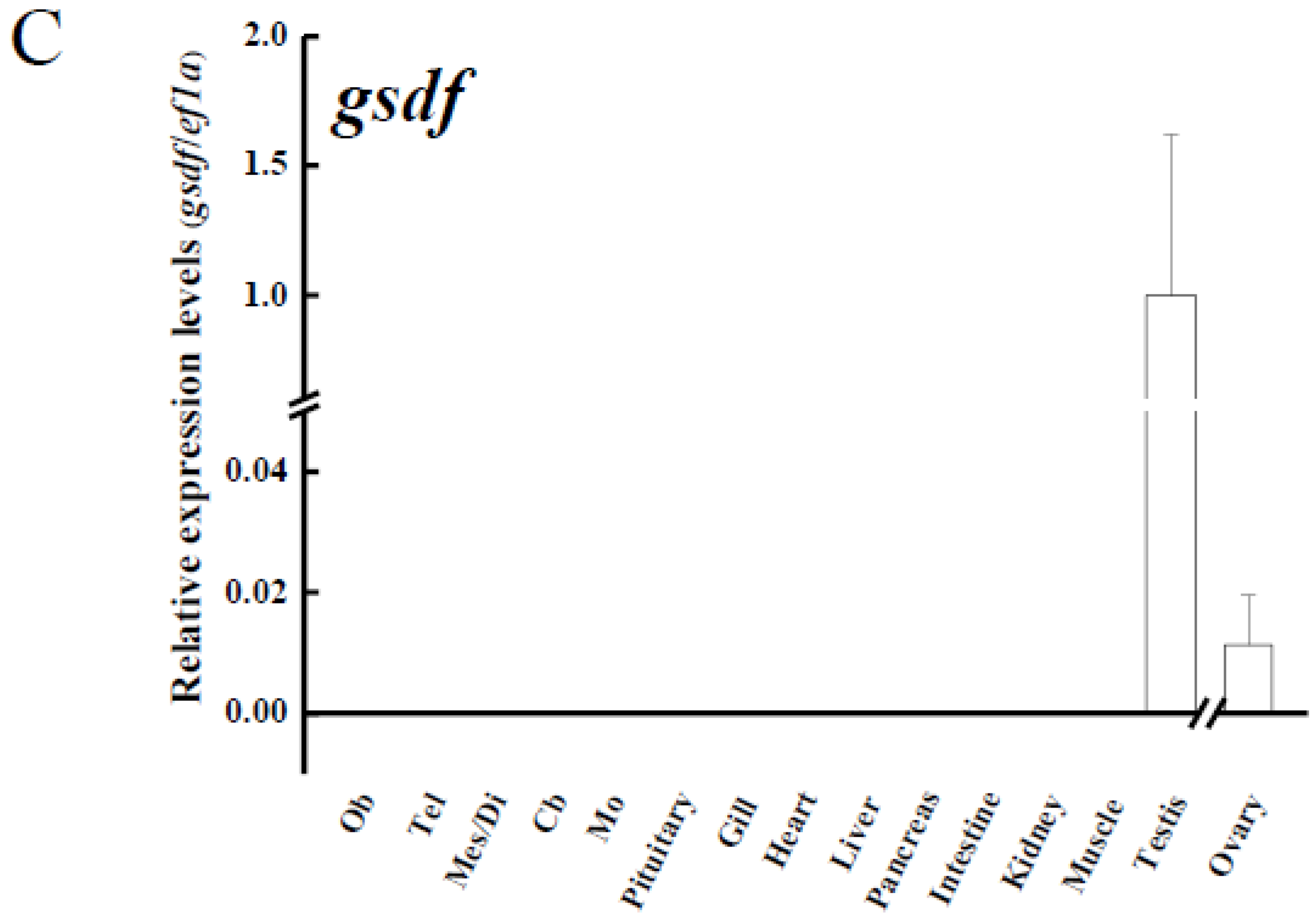
3.3. Tissue Distribution of amh, amhr2 and gsdf in Yellow Eels
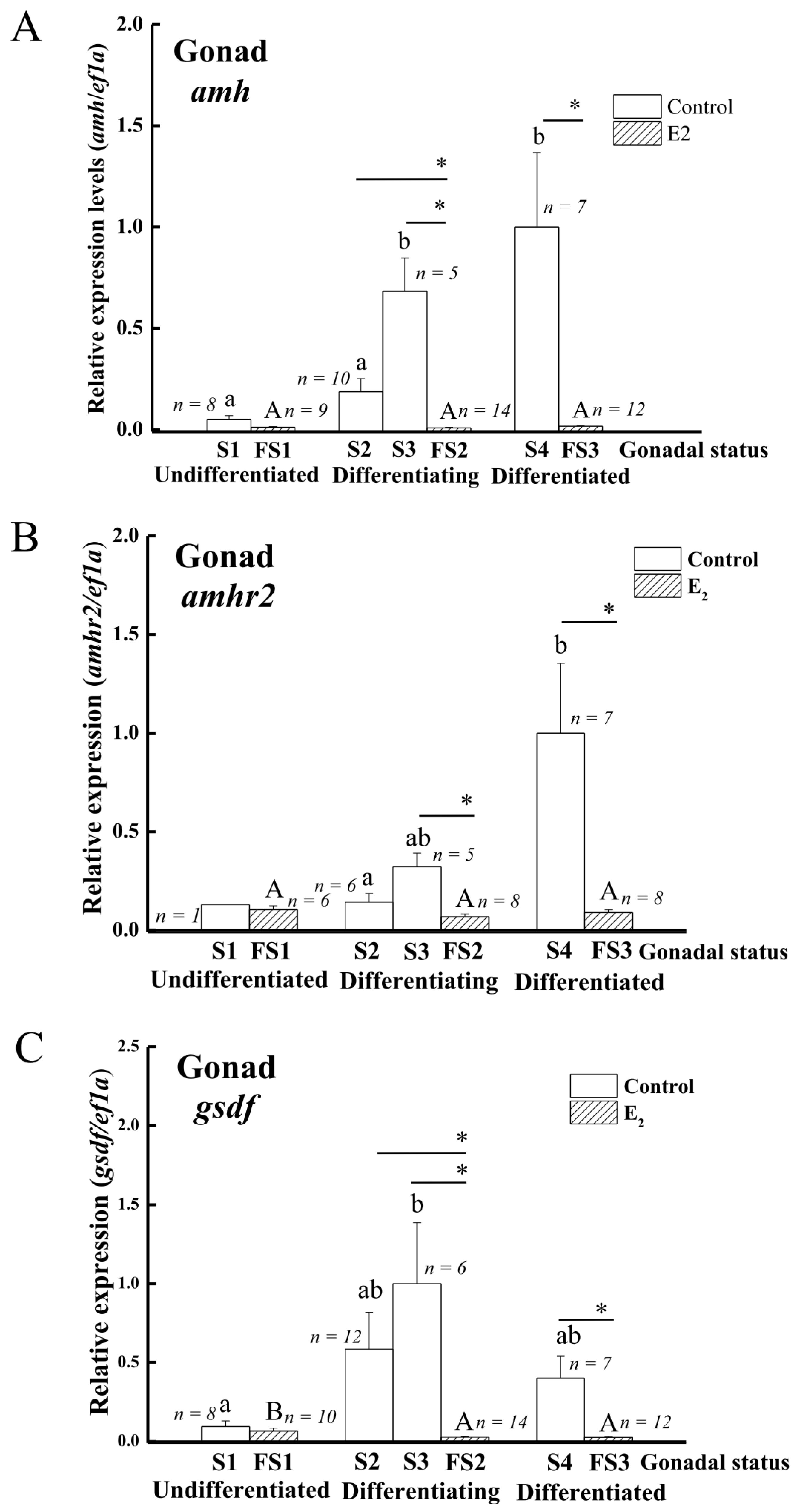
3.4. Expression of amh, amhr2, and gsdf in the Gonads during Sex Differentiation in Eels
3.5. Expression of amh, amhr2 and gsdf Genes in the Gonads during Eel Spermatogenesis
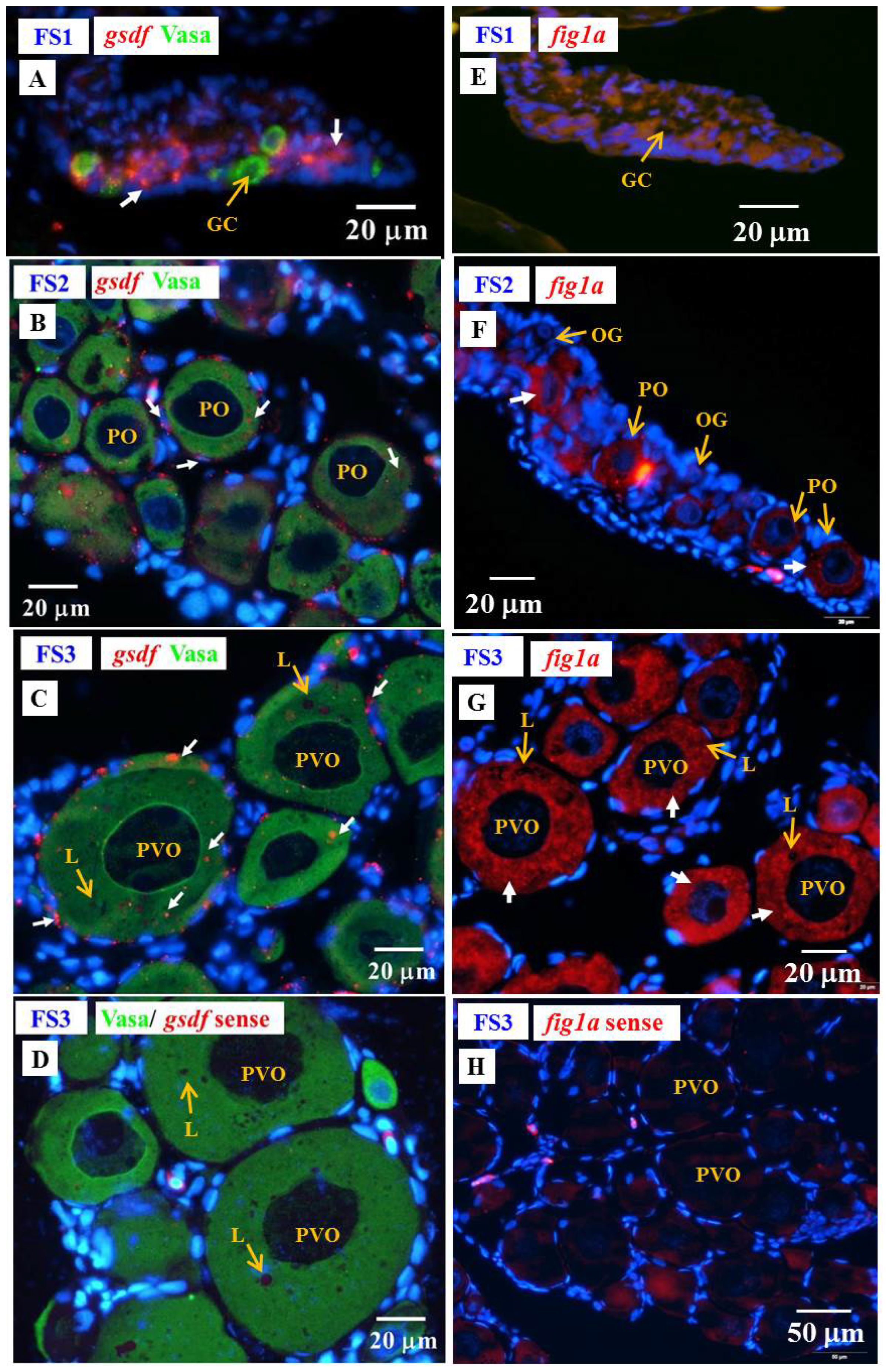
3.6. Cellular Localization of the amh and gsdf Transcripts in Eel Gonads
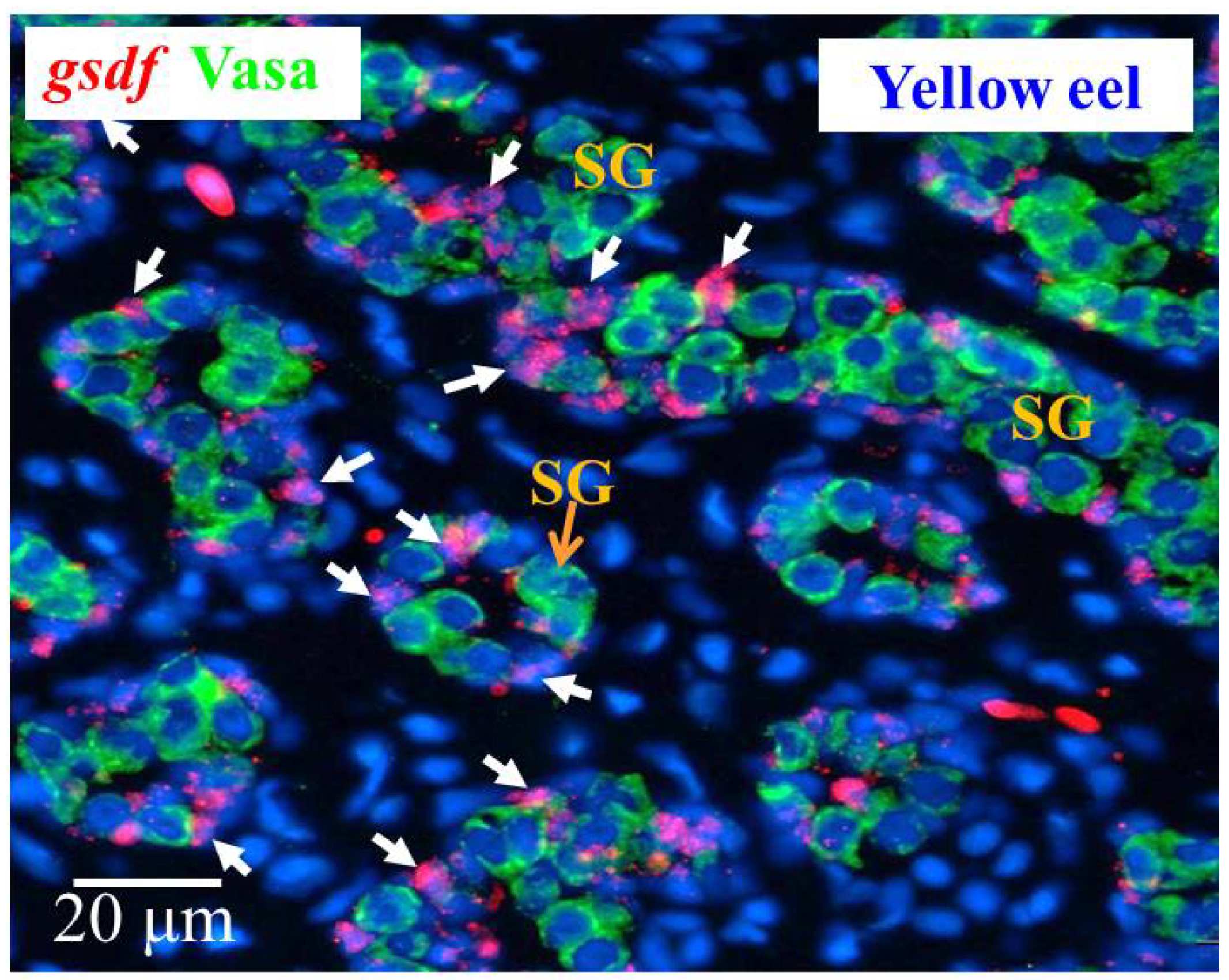
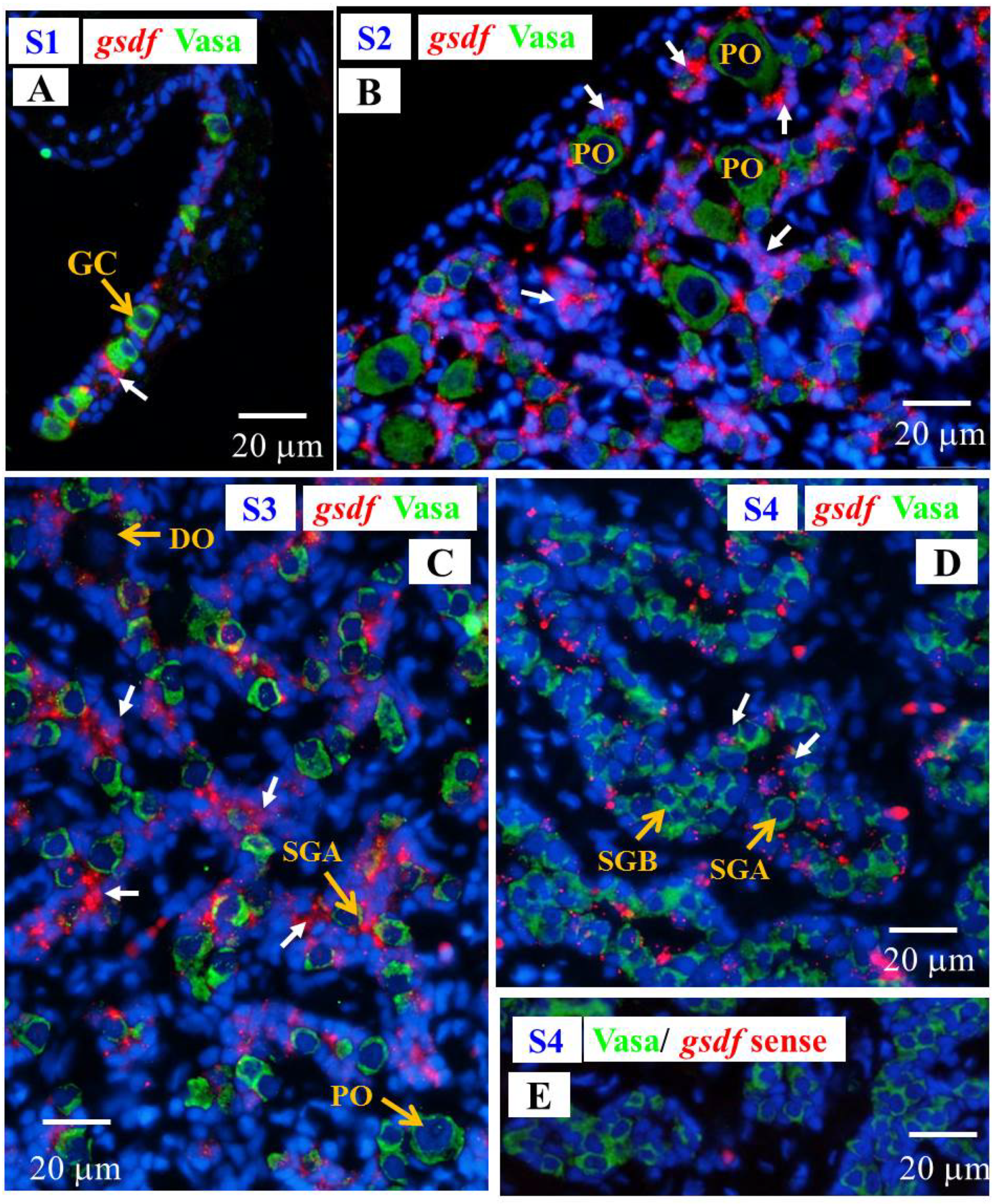
3.6.1. Cellular Localization of the gsdf Transcripts in the Testis of Yellow Eels
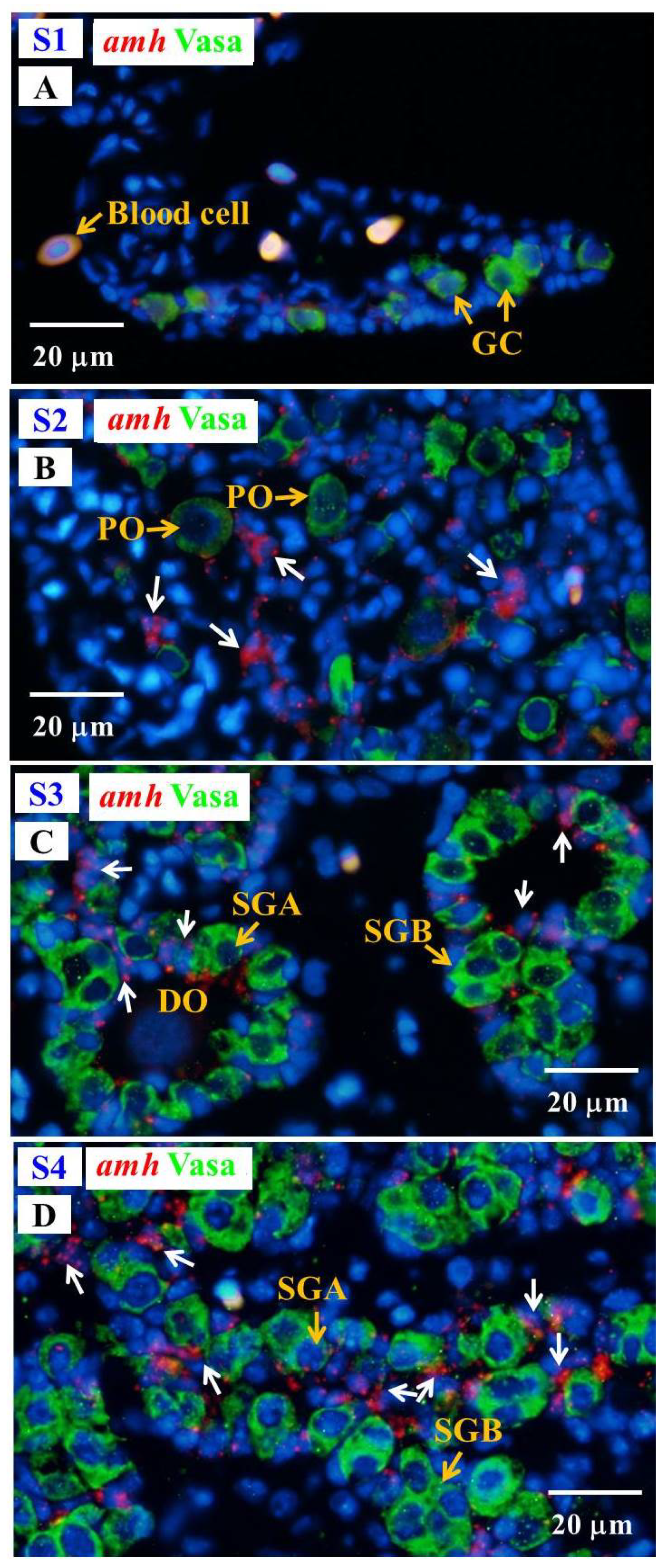
3.6.2. Cellular Localization of the amh and gsdf Transcripts during Testicular Differentiation in Eels
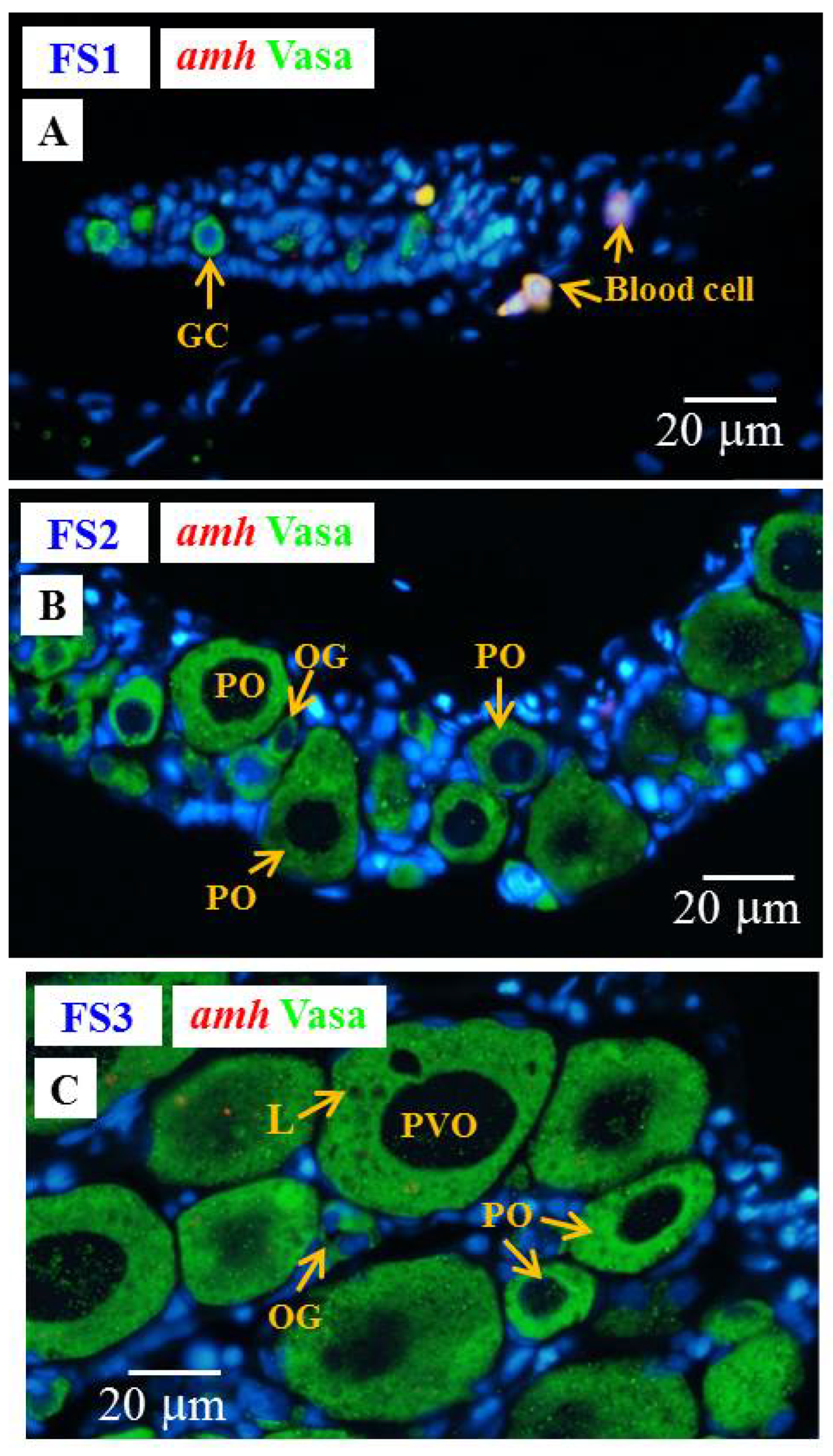
3.6.3. Cellular Localization of the amh, gsdf and fig1a Transcripts during Ovarian Development in E2-Feminized Japanese Eels
4. Discussion
4.1. Characterization of the gsdf cDNA and Phylogeny
4.2. amh, amhr2 and gsdf Are Dominantly Expressed in the Testis of Japanese Eel
4.3. amh/amhr2 Might Play Important Roles in Testicular Differentiation in Japanese Eel
4.4. gsdf Is Expressed in Sertoli Cells and Might Play an Indispensable Role in Testicular Differentiation in Japanese Eel
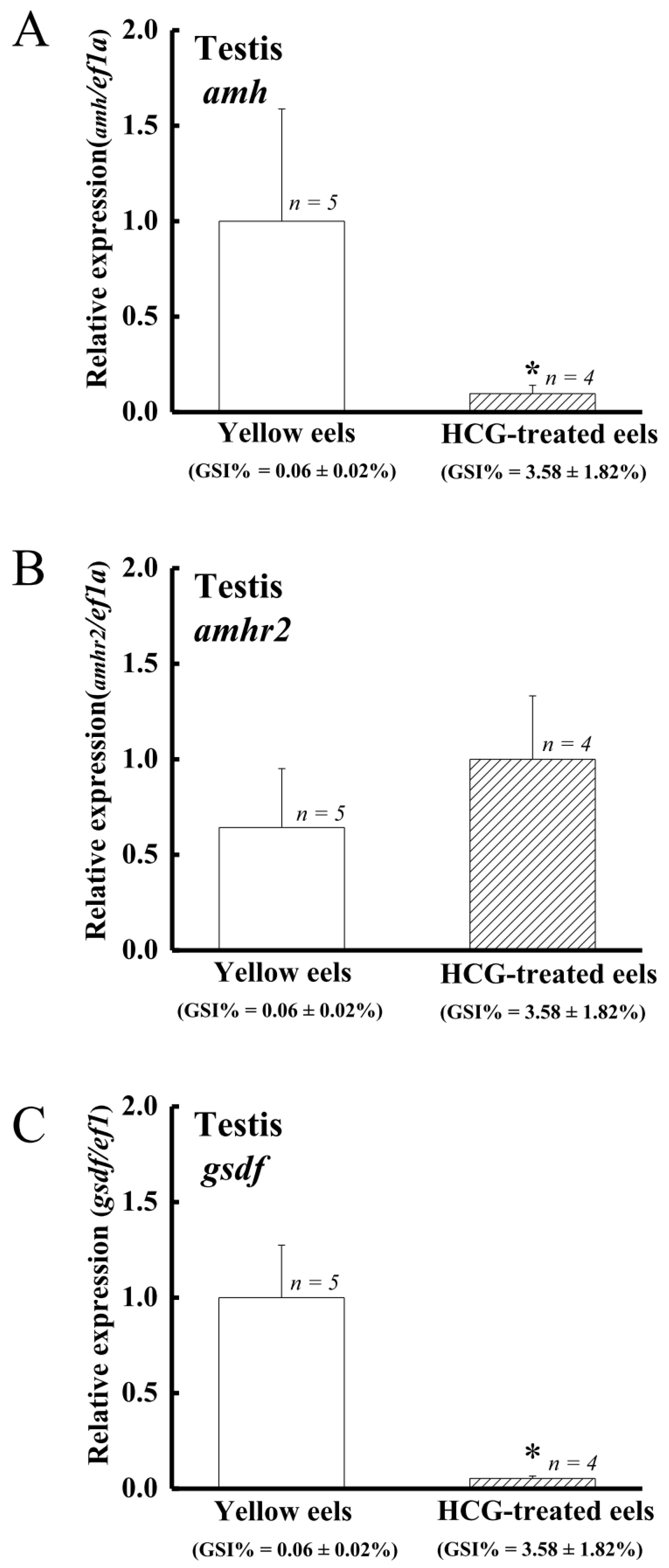
4.5. The Expression of amh and gsdf but Not amhr2 Is Suppressed by Gonadotropin
4.6. The Expressions of amh, amhr2, and gsdf in the Ovary Were Inhibited in E2-Induced Feminized Japanese Eels
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rousseau, K.; Lafont, A.G.; Pasquier, J.; Maugars, G.; Jolly, C.; Sébert, M.E.; Aroua, S.; Pasqualini, C.; Dufour, S. Advances in eel reproductive physiology and endocrinology. In Eel Physiology, 1st ed.; Trischitta, F., Takei, Y., Sébert, P., Eds.; CRC Press: Boca Raton, FL, USA, 2013; pp. 1–43. [Google Scholar]
- Colombo, G.; Grandidr, G. Histological study of the development and sex differentiation of the gonad in the European eel. J. Fish Biol. 1996, 48, 493–512. [Google Scholar] [CrossRef]
- Tesch, F.W. The Eel, 3rd ed.; Thorpe, J.E., Ed.; Blackwell Science Ltd.: Oxford, UK, 2003. [Google Scholar]
- Davey, A.J.; Jellyman, D. Sex determination in freshwater eels and management options for manipulation of Sex. Rev. Fish Biol. Fish. 2005, 15, 37–52. [Google Scholar] [CrossRef]
- Jellyman, D. The influence of growth rate on the size of migrating female eels in Lake Ellesmere, New Zealand. J. Fish Biol. 2001, 58, 725–736. [Google Scholar] [CrossRef]
- Jeng, S.R.; Wu, G.C.; Yueh, W.S.; Kuo, S.F.; Dufour, S.; Chang, C.F. Gonadal development and expression of sex-specific genes during sex differentiation in the Japanese eel. Gen. Comp. Endocrinol. 2018, 257, 74–85. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, K.; McCleave, J.D. Variation in population and life history traits of the American eel, Anguilla rostrata, in four rivers in Maine. Environ. Biol. Fishes 2000, 59, 141–151. [Google Scholar] [CrossRef]
- Nagahama, Y.; Chakraborty, T.; Paul-Prasanth, B.; Ohta, K.; Nakamura, M. Sex determination, gonadal sex differentiation, and plasticity in vertebrate species. Physiol. Rev. 2021, 101, 1237–1308. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.C.; Tomy, S.; Lee, M.F.; Lee, Y.H.; Yueh, W.S.; Lin, C.J.; Lau, E.L.; Chang, C.F. Sex differentiation and sex change in the protandrous black porgy, Acanthopagrus schlegeli. Gen. Comp. Endocrinol. 2010, 167, 417–421. [Google Scholar] [CrossRef]
- Wu, G.C.; Chang, C.F. The switch of secondary sex determination in protandrous black porgy, Acanthopagrus schlegeli. Fish Physiol. Biochem. 2013, 39, 33–38. [Google Scholar] [CrossRef]
- Wu, G.C.; Dufour, S.; Chang, C.F. Molecular and cellular regulation on sex change in hermaphroditic fish, with a special focus on protandrous black porgy, Acanthopagrus schlegelii. Mol. Cell. Endocrinol. 2021, 520, 111069. [Google Scholar] [CrossRef] [PubMed]
- Sinclair, A.H.; Berta, P.; Palmer, M.S.; Hawkins, J.R.; Griffiths, B.L.; Smith, M.J.; Foster, J.W.; Frischauf, A.M.; Lovell-Badge, R.; Goodfellow, P.N. A gene from the human sex-determining region encodes a protein with homology to a conserved DNA-binding motif. Nature 1990, 346, 240–244. [Google Scholar] [CrossRef] [Green Version]
- Smith, C.A.; Roeszler, K.N.; Ohnesorg, T.; Cummins, D.M.; Farlie, P.G.; Doran, T.J.; Sinclair, A.H. The avian Z-linked gene DMRT1 is required for male sex determination in the chicken. Nature 2009, 461, 267–271. [Google Scholar] [CrossRef] [PubMed]
- Matsuda, M.; Nagahama, Y.; Shinomiya, A.; Sato, T.; Matsuda, C.; Kobayashi, T.; Morrey, C.E.; Shibata, N.; Asakawa, S.; Shimizu, N. DMY is a Y-specific DM-domain gene required for male development in the medaka fish. Nature 2002, 417, 559–563. [Google Scholar] [CrossRef]
- Nanda, I.; Kondo, M.; Hornung, U.; Asakawa, S.; Winkler, C.; Shimizu, A.; Shan, Z.; Haaf, T.; Shimizu, N.; Shima, A. A duplicated copy of DMRT1 in the sex-determining region of the Y chromosome of the medaka, Oryzias latipes. Proc. Natl. Acad. Sci. USA 2002, 99, 11778–11783. [Google Scholar] [CrossRef] [Green Version]
- Yano, A.; Nicol, B.; Jouanno, E.; Quillet, E.; Fostier, A.; Guyomard, R.; Guiguen, Y. The sexually dimorphic on the Y-chromosome gene (sdY) is a conserved male-specific Y-chromosome sequence in many salmonids. Evol. Appl. 2013, 6, 486–496. [Google Scholar] [CrossRef] [PubMed]
- Takehana, Y.; Matsuda, M.; Myosho, T.; Suster, M.L.; Kawakami, K.; Shin, T.; Kohara, Y.; Kuroki, Y.; Toyoda, A.; Fujiyama, A. Co-option of Sox3 as the male-determining factor on the Y chromosome in the fish Oryzias dancena. Nat. Commun. 2014, 5, 4157. [Google Scholar] [CrossRef] [Green Version]
- Hattori, R.S.; Murai, Y.; Oura, M.; Masuda, S.; Majhi, S.K.; Sakamoto, T.; Fernandino, J.I.; Somoza, G.M.; Yokota, M.; Strüssmann, C.A. A Y-linked anti-Müllerian hormone duplication takes over a critical role in sex determination. Proc. Natl. Acad. Sci. USA 2012, 109, 2955–2959. [Google Scholar] [CrossRef] [Green Version]
- Kamiya, T.; Kai, W.; Tasumi, S.; Oka, A.; Matsunaga, T.; Mizuno, N.; Fujita, M.; Suetake, H.; Suzuki, S.; Hosoya, S. A trans-species missense SNP in Amhr2 is associated with sex determination in the tiger pufferfish, Takifugu rubripes (fugu). PLoS Genet. 2012, 8, e1002798. [Google Scholar] [CrossRef] [Green Version]
- Myosho, T.; Otake, H.; Masuyama, H.; Matsuda, M.; Kuroki, Y.; Fujiyama, A.; Naruse, K.; Hamaguchi, S.; Sakaizumi, M. Tracing the emergence of a novel sex-determining gene in medaka, Oryzias luzonensis. Genetics 2012, 191, 163–170. [Google Scholar] [CrossRef] [Green Version]
- Devlin, R.H.; Nagahama, Y. Sex determination and sex differentiation in fish: An overview of genetic, physiological, and environmental influences. Aquaculture 2002, 208, 191–364. [Google Scholar] [CrossRef]
- Bull, J.J.; Vogt, R.C. Temperature-dependent sex determination in turtles. Science 1979, 206, 1186–1188. [Google Scholar] [CrossRef] [PubMed]
- Eggert, C. Sex determination: The amphibian models. Reprod. Nutr. Dev. 2004, 44, 539–549. [Google Scholar] [CrossRef]
- Baroiller, J.F.; Chourrout, D.; Fostier, A.; Jalabert, B. Temperature and sex chromosomes govern sex ratios of the mouthbrooding Cichlid fish Oreochromis niloticus. J. Exp. Zool. 1995, 273, 216–223. [Google Scholar] [CrossRef]
- Struussmann, C.A.; Moriyama, S.; Hanke, E.F.; Cota, J.C.C.; Takashima, F. Evidence of thermolabile sex determination in pejerrey. J. Fish Biol. 1996, 48, 643–651. [Google Scholar] [CrossRef]
- Pavlidis, M.; Koumoundouros, G.; Sterioti, A.; Somarakis, S.; Divanach, P.; Kentouri, M. Evidence of temperature-dependent sex determination in the European sea bass (Dicentrarchus labrax L.). J. Exp. Zool. 2000, 287, 225–232. [Google Scholar] [CrossRef]
- Ospina-Alvarez, N.; Piferrer, F. Temperature-dependent sex determination in fish revisited: Prevalence, a single sex ratio response pattern, and possible effects of climate change. PLoS ONE 2008, 3, e2837. [Google Scholar] [CrossRef] [Green Version]
- Baroiller, J.F.; Guiguen, Y.; Fostier, A. Endocrine and environmental aspects of sex differentiation in fish. Cell. Mol. Life Sci. 1999, 55, 910–931. [Google Scholar] [CrossRef]
- Colombo, G.; Grandi, G. Sex differentiation in the European eel: Histological analysis of the effects of sex steroids on the gonad. J. Fish Biol. 1995, 47, 394–413. [Google Scholar] [CrossRef]
- Guiguen, Y.; Fostier, A.; Piferrer, F.; Chang, C.F. Ovarian aromatase and estrogens: A pivotal role for gonadal sex differentiation and sex change in fish. Gen. Comp. Endocrinol. 2010, 165, 352–366. [Google Scholar] [CrossRef]
- Chiba, H.; Iwatsuki, K.; Hayami, K.; Yamauchi, K. Effects of dietary estradiol-17β on feminization, growth and body composition in the Japanese eel (Anguilla japonica). Comp. Biochem. Physiol. A Physiol. 1993, 106, 367–371. [Google Scholar]
- Piferrer, F.; Baker, I.J.; Donaldson, E.M. Effects of natural, synthetic, aromatizable, and nonaromatizable androgens in inducing male sex differentiation in genotypic female chinook salmon (Oncorhynchus tshawytscha). Gen. Comp. Endocrinol. 1993, 91, 59–65. [Google Scholar] [CrossRef]
- Chang, C.F.; Lau, E.L.; Lin, B.Y. Estradiol-17beta suppresses testicular development and stimulates sex reversal in protandrous black porgy, Acanthopagrus schlegeli. Fish Physiol. Biochem. 1995, 14, 481–488. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.F.; Lin, B.Y. Estradiol-17β stimulates aromatase activity and reversible sex change in protandrous black porgy, Acanthopagrus schlegeli. J. Exp. Zool. 1998, 280, 165–173. [Google Scholar] [CrossRef]
- Lee, Y.H.; Du, J.L.; Yueh, W.S.; Lin, B.Y.; Huang, J.D.; Lee, C.Y.; Lee, M.F.; Lau, E.L.; Lee, F.Y.; Morrey, C.; et al. Sex change in the protandrous black porgy, Acanthopagrus schlegeli: A review in gonadal development, estradiol, estrogen receptor, aromatase activity and gonadotropin. J. Exp. Zool. 2001, 290, 715–726. [Google Scholar] [CrossRef]
- Guo, C.Y.; Tseng, P.W.; Hwang, J.S.; Wu, G.C.; Chang, C.F. Potential role of DNA methylation of cyp19a1a promoter during sex change in protogynous orange-spotted grouper, Epinephelus coioides. Gen. Comp. Endocrinol. 2021, 311, 113840. [Google Scholar] [CrossRef]
- Jørgensen, A.; Morthorst, J.E.; Andersen, O.; Rasmussen, L.J.; Bjerregaard, P. Expression profiles for six zebrafish genes during gonadal sex differentiation. Reprod. Biol. Endocrinol. 2008, 6, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Wang, D.S.; Zhou, L.Y.; Kobayashi, T.; Matsuda, M.; Shibata, Y.; Sakai, F.; Nagahama, Y. Doublesex-and Mab-3-related transcription factor-1 repression of aromatase transcription, a possible mechanism favoring the male pathway in tilapia. Endocrinology 2010, 151, 1331–1340. [Google Scholar] [CrossRef] [Green Version]
- Nakamoto, M.; Suzuki, A.; Matsuda, M.; Nagahama, Y.; Shibata, N. Testicular type Sox9 is not involved in sex determination but might be in the development of testicular structures in the medaka, Oryzias latipes. Biochem. Biophys. Res. Commun. 2005, 333, 729–736. [Google Scholar] [CrossRef] [PubMed]
- Chiang, E.F.; Pai, C.I.; Wyatt, M.; Yan, Y.L.; Postlethwait, J.; Chung, B. Two sox9 genes on duplicated zebrafish chromosomes: Expression of similar transcription activators in distinct sites. Dev. Biol. 2001, 231, 149–163. [Google Scholar] [CrossRef] [Green Version]
- Morais da Silva, S.; Hacker, A.; Harley, V.; Goodfellow, P.; Swain, A.; Lovell-Badge, R. Sox9 expression during gonadal development implies a conserved role for the gene in testis differentiation in mammals and birds. Nat. Genet. 1996, 14, 62–68. [Google Scholar] [CrossRef] [PubMed]
- Herpin, A.; Schartl, M. Dmrt1 genes at the crossroads: A widespread and central class of sexual development factors in fish. FEBS J. 2011, 278, 1010–1019. [Google Scholar] [CrossRef] [PubMed]
- Jeng, S.R.; Wu, G.C.; Yueh, W.S.; Kuo, S.F.; Dufour, S.; Chang, C.F. Dmrt1 (doublesex and mab-3-related transcription factor 1) expression during gonadal development and spermatogenesis in the Japanese eel. Gen. Comp. Endocrinol. 2019, 279, 154–163. [Google Scholar] [CrossRef] [PubMed]
- He, C.L.; Du, J.L.; Wu, G.C.; Lee, Y.H.; Sun, L.T.; Chang, C.F. Differential Dmrt1 transcripts in gonads of the protandrous black porgy, Acanthopagrus schlegeli. Cytogenet. Genome Res. 2003, 101, 309–313. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.C.; Chang, C.F. Primary males guide the femaleness through the regulation of testicular Dmrt1 and ovarian Cyp19a1a in protandrous black porgy. Gen. Comp. Endocrinol. 2018, 261, 198–202. [Google Scholar] [CrossRef]
- Wu, G.C.; Chiu, P.C.; Lin, C.J.; Lyu, Y.S.; Lan, D.S.; Chang, C.F. Testicular dmrt1 is involved in the sexual fate of the ovotestis in the protandrous black porgy. Biol. Reprod. 2012, 86, 41. [Google Scholar] [CrossRef]
- Webster, K.A.; Schach, U.; Ordaz, A.; Steinfeld, J.S.; Draper, B.W.; Siegfried, K.R. Dmrt1 is necessary for male sexual development in zebrafish. Dev. Biol. 2017, 422, 33–46. [Google Scholar] [CrossRef]
- Chang, C.F.; Lee, M.F.; Chen, G.R. Estradiol-17β associated with the sex reversal in protandrous black porgy, Acanthopagrus schlegeli. J. Exp. Zool. 1994, 268, 53–58. [Google Scholar] [CrossRef]
- Chang, C.F.; Lau, E.L.; Lin, B.Y. Stimulation of spermatogenesis or of sex reversal according to the dose of exogenous estradiol-17 beta in juvenile males of protandrous black porgy, Acanthopagrus schlegeli. Gen. Comp. Endocrinol. 1995, 100, 355–367. [Google Scholar] [CrossRef] [PubMed]
- Qiu, Y.; Sun, S.; Charkraborty, T.; Wu, L.; Sun, L.; Wei, J.; Nagahama, Y.; Wang, D.; Zhou, L. Figla favors ovarian differentiation by antagonizing spermatogenesis in a teleosts, Nile tilapia (Oreochromis niloticus). PLoS ONE 2015, 10, e0123900. [Google Scholar] [CrossRef] [Green Version]
- Soyal, S.M.; Amleh, A.; Dean, J. FIGalpha, a germ cell-specific transcription factor required for ovarian follicle formation. Development 2000, 127, 4645–4654. [Google Scholar] [CrossRef]
- Wu, G.C.; Tomy, S.; Nakamura, M.; Chang, C.F. Dual roles of cyp19a1a in gonadal sex differentiation and development in the protandrous black porgy, Acanthopagrus schlegeli. Biol. Reprod. 2008, 79, 1111–1120. [Google Scholar] [CrossRef] [Green Version]
- Johnson, P.; Kent, T.; Urick, M.; Giles, J. Expression and regulation of anti-Müllerian hormone in an oviparous species, the hen. Biol. Reprod. 2008, 78, 13–19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Josso, N.; di Clemente, N.; Gouédard, L. Anti-Müllerian hormone and its receptors. Mol. Cell. Endocrinol. 2001, 179, 25–32. [Google Scholar] [CrossRef]
- Miura, T.; Miura, C.; Konda, Y.; Yamauchi, K. Spermatogenesis-preventing substance in Japanese eel. Development 2002, 129, 2689–2697. [Google Scholar] [CrossRef]
- Yoshinaga, N.; Shiraishi, E.; Yamamoto, T.; Iguchi, T.; Abe, S.i.; Kitano, T. Sexually dimorphic expression of a teleost homologue of Müllerian inhibiting substance during gonadal sex differentiation in Japanese flounder, Paralichthys olivaceus. Biochem. Biophys. Res. Commun. 2004, 322, 508–513. [Google Scholar] [CrossRef]
- Rodríguez-Marí, A.; Yan, Y.L.; BreMiller, R.A.; Wilson, C.; Canestro, C.; Postlethwait, J.H. Characterization and expression pattern of zebrafish anti-Müllerian hormone (amh) relative to sox9a, sox9b, and cyp19a1a, during gonad development. Gene Expr. Patterns 2005, 5, 655–667. [Google Scholar] [CrossRef]
- Wu, G.C.; Chiu, P.C.; Lyu, Y.S.; Chang, C.F. The expression of amh and amhr2 is associated with the development of gonadal tissue and sex change in the protandrous black porgy, Acanthopagrus schlegeli. Biol. Reprod. 2010, 83, 443–453. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, G.C.; Li, H.W.; Tey, W.G.; Lin, C.J.; Chang, C.F. Expression profile of amh/Amh during bi-directional sex change in the protogynous orange-spotted grouper Epinephelus coioides. PLoS ONE 2017, 12, e0185864. [Google Scholar] [CrossRef]
- Maugars, G.; Schmitz, M. Gene expression profiling during spermatogenesis in early maturing male Atlantic salmon parr testes. Gen. Comp. Endocrinol. 2008, 159, 178–187. [Google Scholar] [CrossRef]
- Klüver, N.; Pfennig, F.; Pala, I.; Storch, K.; Schlieder, M.; Froschauer, A.; Gutzeit, H.O.; Schartl, M. Differential expression of anti-Müllerian hormone (amh) and anti-Müllerian hormone receptor type II (amhrII) in the teleost medaka. Dev. Dyn. 2007, 236, 271–281. [Google Scholar] [CrossRef]
- Sawatari, E.; Shikina, S.; Takeuchi, T.; Yoshizaki, G. A novel transforming growth factor-β superfamily member expressed in gonadal somatic cells enhances primordial germ cell and spermatogonial proliferation in rainbow trout (Oncorhynchus mykiss). Dev. Biol. 2007, 301, 266–275. [Google Scholar] [CrossRef] [Green Version]
- Kobayashi, T.; Chiba, A.; Sato, T.; Myosho, T.; Yamamoto, J.; Okamura, T.; Onishi, Y.; Sakaizumi, M.; Hamaguchi, S.; Iguchi, T.; et al. Estrogen alters gonadal soma-derived factor (Gsdf)/Foxl2 expression levels in the testes associated with testis-ova differentiation in adult medaka, Oryzias latipes. Aquat. Toxicol. 2017, 191, 209–218. [Google Scholar] [CrossRef]
- Shibata, Y.; Paul-Prasanth, B.; Suzuki, A.; Usami, T.; Nakamoto, M.; Matsuda, M.; Nagahama, Y. Expression of gonadal soma derived factor (GSDF) is spatially and temporally correlated with early testicular differentiation in medaka. Gene Expr. Patterns 2010, 10, 283–289. [Google Scholar] [CrossRef]
- Kaneko, H.; Ijiri, S.; Kobayashi, T.; Izumi, H.; Kuramochi, Y.; Wang, D.S.; Mizuno, S.; Nagahama, Y. Gonadal soma-derived factor (gsdf), a TGF-beta superfamily gene, induces testis differentiation in the teleost fish Oreochromis niloticus. Mol. Cell. Endocrinol. 2015, 415, 87–99. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Zhang, W.; Du, X.; Zhao, J.; Liu, X.; Li, X.; Zhang, Q.; Wang, X. Sexually dimorphic expression in developing and adult gonads shows an important role of gonadal soma-derived factor during sex differentiation in olive flounder (Paralichthys olivaceus). Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2017, 210, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Meng, L.; Xu, W.; Cui, Z.; Zhang, N.; Guo, H.; Wang, N.; Shao, C.; Chen, S. The autosomal Gsdf gene plays a role in male gonad development in Chinese tongue sole (Cynoglossus semilaevis). Sci. Rep. 2018, 8, 17716. [Google Scholar] [CrossRef]
- Jiang, D.N.; Mustapha, U.F.; Shi, H.J.; Huang, Y.Q.; Si-Tu, J.X.; Wang, M.; Deng, S.P.; Chen, H.P.; Tian, C.X.; Zhu, C.H. Expression and transcriptional regulation of gsdf in spotted scat (Scatophagus argus). Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2019, 233, 35–45. [Google Scholar] [CrossRef]
- Jiang, D.N.; Yang, H.H.; Li, M.H.; Shi, H.J.; Zhang, X.B.; Wang, D.S. gsdf is a downstream gene of dmrt1 that functions in the male sex determination pathway of the Nile tilapia. Mol. Reprod. Dev. 2016, 83, 497–508. [Google Scholar] [CrossRef] [Green Version]
- Wu, G.C.; Jeng, S.R.; Pan, Y.T.; Li, H.W.; Ku, W.L.; Lin, C.J.; Chang, C.F. The germline-specific expression of Foxl3a and its paralogous Foxl3b are associated with male gonadal differentiation in the Japanese eel, Anguilla japonica. Gen. Comp. Endocrinol. 2019, 277, 56–65. [Google Scholar] [CrossRef] [PubMed]
- Jeng, S.R.; Yueh, W.S.; Pen, Y.T.; Gueguen, M.M.; Pasquier, J.; Dufour, S.; Chang, C.F.; Kah, O. Expression of aromatase in radial glial cells in the brain of the Japanese eel provides insight into the evolution of the cyp19a1 gene in Actinopterygians. PLoS ONE 2012, 7, e44750. [Google Scholar] [CrossRef] [Green Version]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miura, T.; Yamauchi, K.; Takahashi, H.; Nagahama, Y. Human chorionic gonadotropin induces all stages of spermatogenesis in vitro in the male Japanese eel (Anguilla japonica). Dev. Biol. 1991, 146, 258–262. [Google Scholar] [CrossRef]
- Daopin, S.; Piez, K.A.; Ogawa, Y.; Davies, D.R. Crystal structure of transforming growth factor-beta 2: An unusual fold for the superfamily. Science 1992, 257, 369–373. [Google Scholar] [CrossRef] [Green Version]
- Kingsley, D.M. The TGF-beta superfamily: New members, new receptors, and new genetic tests of function in different organisms. Genes Dev. 1994, 8, 133–146. [Google Scholar] [CrossRef] [Green Version]
- Crespo, B.; Gómez, A.; Mazón, M.J.; Carrillo, M.; Zanuy, S. Isolation and characterization of Ff1 and Gsdf family genes in European sea bass and identification of early gonadal markers of precocious puberty in males. Gen. Comp. Endocrinol. 2013, 191, 155–167. [Google Scholar] [CrossRef] [Green Version]
- Suzuki, H.; Kazeto, Y.; Gen, K.; Ozaki, Y. Functional analysis of recombinant single-chain Japanese eel Fsh and Lh produced in FreeStyle 293-F cell lines: Binding specificities to their receptors and differential efficacy on testicular steroidogenesis. Gen. Comp. Endocrinol. 2020, 285, 113241. [Google Scholar] [CrossRef] [PubMed]
- Inaba, H.; Hara, S.; Horiuchi, M.; Ijiri, S.; Kitano, T. Gonadal expression profiles of sex-specific genes during early sexual differentiation in Japanese eel Anguilla japonica. Fish. Sci. 2021, 87, 203–209. [Google Scholar] [CrossRef]
- Curzon, A.Y.; Shirak, A.; Dor, L.; Zak, T.; Perelberg, A.; Seroussi, E.; Ron, M. A duplication of the Anti-Müllerian hormone gene is associated with genetic sex determination of different Oreochromis niloticus strains. Heredity 2020, 125, 317–327. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Sun, Y.; Zhao, J.; Shi, H.; Zeng, S.; Ye, K.; Jiang, D.; Zhou, L.; Sun, L.; Tao, W. A tandem duplicate of anti-Müllerian hormone with a missense SNP on the Y chromosome is essential for male sex determination in Nile tilapia, Oreochromis niloticus. PLoS Genet. 2015, 11, e1005678. [Google Scholar] [CrossRef] [Green Version]
- Pfennig, F.; Standke, A.; Gutzeit, H.O. The role of Amh signaling in teleost fish–multiple functions not restricted to the gonads. Gen. Comp. Endocrinol. 2015, 223, 87–107. [Google Scholar] [CrossRef]
- Geffroy, B.; Guilbaud, F.; Amilhat, E.; Beaulaton, L.; Vignon, M.; Huchet, E.; Rives, J.; Bobe, J.; Fostier, A.; Guiguen, Y.; et al. Sexually dimorphic gene expressions in eels: Useful markers for early sex assessment in a conservation context. Sci. Rep. 2016, 6, 34041. [Google Scholar] [CrossRef] [Green Version]
- Ijiri, S.; Kaneko, H.; Kobayashi, T.; Wang, D.S.; Sakai, F.; Paul-Prasanth, B.; Nakamura, M.; Nagahama, Y. Sexual dimorphic expression of genes in gonads during early differentiation of a teleost fish, the Nile tilapia Oreochromis niloticus. Biol. Reprod. 2008, 78, 333–341. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pfennig, F.; Kurth, T.; Meißner, S.; Standke, A.; Hoppe, M.; Zieschang, F.; Reitmayer, C.; Göbel, A.; Kretzschmar, G.; Gutzeit, H.O. The social status of the male Nile tilapia (Oreochromis niloticus) influences testis structure and gene expression. Reproduction 2012, 143, 71–84. [Google Scholar] [CrossRef]
- Poonlaphdecha, S.; Pepey, E.; Huang, S.H.; Canonne, M.; Soler, L.; Mortaji, S.; Morand, S.; Pfennig, F.; Melard, C.; Baroiller, J.F.; et al. Elevated amh gene expression in the brain of male tilapia (Oreochromis niloticus) during testis differentiation. Sex. Dev. 2011, 5, 33–47. [Google Scholar] [CrossRef] [Green Version]
- Yamamoto, Y.; Zhang, Y.; Sarida, M.; Hattori, R.S.; Strussmann, C.A. Coexistence of genotypic and temperature-dependent sex determination in pejerrey Odontesthes bonariensis. PLoS ONE 2014, 9, e102574. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Halm, S.; Rocha, A.; Miura, T.; Prat, F.; Zanuy, S. Anti-Müllerian hormone (AMH/AMH) in the European sea bass: Its gene structure, regulatory elements, and the expression of alternatively-spliced isoforms. Gene 2007, 388, 148–158. [Google Scholar] [CrossRef]
- Johnsen, H.; Tveiten, H.; Torgersen, J.S.; Andersen, Ø. Divergent and sex-dimorphic expression of the paralogs of the Sox9-Amh-Cyp19a1 regulatory cascade in developing and adult atlantic cod (Gadus morhua L.). Mol. Reprod. Dev. 2013, 80, 358–370. [Google Scholar] [CrossRef] [PubMed]
- Poonlaphdecha, S.; Pepey, E.; Canonne, M.; De Verdal, H.; Baroiller, J.F.; D’cotta, H. Temperature induced-masculinisation in the Nile tilapia causes rapid up-regulation of both dmrt1 and amh expressions. Gen. Comp. Endocrinol. 2013, 193, 234–242. [Google Scholar] [CrossRef]
- Fernandino, J.I.; Hattori, R.S.; Kimura, H.; Strüssmann, C.A.; Somoza, G.M. Expression profile and estrogenic regulation of anti-Müllerian hormone during gonadal development in pejerrey Odontesthes bonariensis, a teleost fish with strong temperature-dependent sex determination. Dev. Dyn. 2008, 237, 3192–3199. [Google Scholar] [CrossRef] [PubMed]
- Mankiewicz, J.L.; Godwin, J.; Holler, B.L.; Turner, P.M.; Murashige, R.; Shamey, R.; Daniels, H.V.; Borski, R.J. Masculinizing effect of background color and cortisol in a flatfish with environmental sex-determination. Integr. Comp. Biol. 2013, 53, 755–765. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shiraishi, E.; Yoshinaga, N.; Miura, T.; Yokoi, H.; Wakamatsu, Y.; Abe, S.I.; Kitano, T. Müllerian inhibiting substance is required for germ cell proliferation during early gonadal differentiation in medaka (Oryzias latipes). Endocrinology 2008, 149, 1813–1819. [Google Scholar] [CrossRef] [Green Version]
- Han, Y.; Zhao, M.; Wang, L.; Yu, Z.; Wang, J.; Yu, Q.; Xiao, L.; Lu, M.; Li, S.; Zhang, Y. Overexpression of anti-Müllerian hormone gene in vivo affects gonad sex differentiation in undifferentiated orange-spotted groupers (Epinephelus coioides). Front. Endocrinol. 2019, 10, 210. [Google Scholar] [CrossRef] [Green Version]
- di Clemente, N.; Wilson, C.; Faure, E.; Boussin, L.; Carmillo, P.; Tizard, R.; Picard, J.Y.; Vigier, B.; Josso, N.; Cate, R. Cloning, expression, and alternative splicing of the receptor for anti-Müllerian hormone. Mol. Endocrinol. 1994, 8, 1006–1020. [Google Scholar] [PubMed]
- Rocha, A.; Zanuy, S.; Gómez, A. Conserved anti-Müllerian hormone: Anti-Müllerian hormone type-2 receptor specific interaction and intracellular signaling in Teleosts. Biol. Reprod. 2016, 94, 141. [Google Scholar] [CrossRef] [Green Version]
- Morinaga, C.; Saito, D.; Nakamura, S.; Sasaki, T.; Asakawa, S.; Shimizu, N.; Mitani, H.; Furutani-Seiki, M.; Tanaka, M.; Kondoh, H. The hotei mutation of medaka in the anti-Müllerian hormone receptor causes the dysregulation of germ cell and sexual development. Proc. Natl. Acad. Sci. USA 2007, 104, 9691–9696. [Google Scholar] [CrossRef] [Green Version]
- Nakamura, S.; Watakabe, I.; Nishimura, T.; Picard, J.Y.; Toyoda, A.; Taniguchi, Y.; di Clemente, N.; Tanaka, M. Hyperproliferation of mitotically active germ cells due to defective anti-Müllerian hormone signaling mediates sex reversal in medaka. Development 2012, 139, 2283–2287. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kleppe, L.; Edvardsen, R.B.; Furmanek, T.; Andersson, E.; Skaftnesmo, K.O.; Segafredo, F.T.; Wargelius, A. Transcriptomic analysis of dead end knockout testis reveals germ cell and gonadal somatic factors in Atlantic salmon. BMC Genom. 2020, 21, 99. [Google Scholar] [CrossRef] [Green Version]
- Guo, C.Y.; Tseng, P.W.; Hwang, J.S.; Wu, G.C.; Chang, C.F. The characteristics and expression profile of gonadal soma-derived factor (gsdf/Gsdf) during androgen-induced reversible sex change in the protogynous orange-spotted grouper, Epinephelus coioides. Aquaculture 2021, 542, 736832. [Google Scholar] [CrossRef]
- Skaar, K.; Nobrega, R.; Magaraki, A.; Olsen, L.; Schulz, R.; Male, R. Proteolytically activated, recombinant anti-Müllerian hormone inhibits androgen secretion, proliferation, and differentiation of spermatogonia in adult zebrafish testis organ cultures. Endocrinology 2011, 152, 3527–3540. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, G.C.; Li, H.W.; Luo, J.W.; Chen, C.; Chang, C.F. The potential role of Amh to prevent ectopic female development in testicular tissue of the protandrous black porgy, Acanthopagrus schlegelii. Biol. Reprod. 2015, 92, 158. [Google Scholar] [CrossRef]
- Rolland, A.D.; Lareyre, J.J.; Goupil, A.S.; Montfort, J.; Ricordel, M.J.; Esquerré, D.; Hugot, K.; Houlgatte, R.; Chalmel, F.; Le Gac, F. Expression profiling of rainbow trout testis development identifies evolutionary conserved genes involved in spermatogenesis. BMC Genom. 2009, 10, 546. [Google Scholar] [CrossRef] [Green Version]
- Sambroni, E.; Rolland, A.D.; Lareyre, J.J.; Le Gac, F. Fsh and Lh have common and distinct effects on gene expression in rainbow trout testis. J. Mol. Endocrinol. 2013, 50, 1–18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chi, M.L.; Wen, H.S.; Ni, M.; He, F.; Li, J.F.; Qian, K.; Zhang, P.; Chai, S.H.; Ding, Y.X.; Yin, X.H. Molecular identification of genes involved in testicular steroid synthesis and characterization of the responses to hormones stimulation in testis of Japanese sea bass (Lateolabrax japonicas). Steroids 2014, 84, 92–102. [Google Scholar] [CrossRef]
- Chen, Y.; Hong, W.S.; Wang, Q.; Chen, S.X. Cloning and expression pattern of gsdf during the first maleness reproductive phase in the protandrous Acanthopagrus latus. Gen. Comp. Endocrinol. 2015, 217–218, 71–80. [Google Scholar] [CrossRef]
- Aris, A.Z.; Shamsuddin, A.S.; Praveena, S.M. Occurrence of 17alpha-ethynylestradiol (EE2) in the environment and effect on exposed biota: A review. Environ. Int. 2014, 69, 104–119. [Google Scholar] [CrossRef]
- Schulz, R.W.; Bogerd, J.; Male, R.; Ball, J.; Fenske, M.; Olsen, L.C.; Tyler, C.R. Estrogen-induced alterations in amh and dmrt1 expression signal for disruption in male sexual development in the zebrafish. Environ. Sci. Technol. 2007, 41, 6305–6310. [Google Scholar] [CrossRef]
- Gautier, A.; Le Gac, F.; Lareyre, J.J. The gsdf gene locus harbors evolutionary conserved and clustered genes preferentially expressed in fish previtellogenic oocytes. Gene 2011, 472, 7–17. [Google Scholar] [CrossRef]
- Yan, Y.L.; Desvignes, T.; Bremiller, R.; Wilson, C.; Dillon, D.; High, S.; Draper, B.; Buck, C.L.; Postlethwait, J. Gonadal soma controls ovarian follicle proliferation through Gsdf in zebrafish. Dev. Dyn. 2017, 246, 925–945. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Myosho, T.; Hattori, M.; Yamamoto, J.; Toda, M.; Okamura, T.; Onishi, Y.; Takehana, Y.; Kobayashi, T. Effects of synthetic sex steroid hormone exposures on gonadal sex differentiation and dynamics of a male-related gene, Gonadal soma-derived factor (Gsdf) and an estrogen up-regulated gene, Choriogenine-H (ChgH) gene expression in the euryhaline Javafish medaka, Oryzias javanicus, based on genetic sexes. Chemosphere 2021, 274, 129893. [Google Scholar] [PubMed]
- Schartl, M. A comparative view on sex determination in medaka. Mech. Dev. 2004, 121, 639–645. [Google Scholar] [CrossRef]



| Gene | Sequences | Amplicon Size |
|---|---|---|
| ef1a | Sense: 5′-TGTGGGAGTCAACAAGATGGA-3′ | 58 bp |
| Antisense: 5′-CTCAAAACGCTTCTGGCTGTA-3′ | ||
| amh | Sense: 5′-TCCTGGTCAGCACTGCGTATC-3′ | 60 bp |
| Antisense: 5′-TCCCGCACCGACAGACA-3′ | ||
| amhr2 | Sense: 5′-TCTGCATGGTGGTGGTTCCT-3′ | 66 bp |
| Antisense: 5′-TGGATCTGTGGCACATGAAGA-3′ | ||
| gsdf | Sense: 5′-GAGCCAAACACCCCTTCAAA-3′ | 82 bp |
| Antisense: 5′-GCGTGTTGAGCTCATCCAAGT-3 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lin, C.-J.; Jeng, S.-R.; Lei, Z.-Y.; Yueh, W.-S.; Dufour, S.; Wu, G.-C.; Chang, C.-F. Involvement of Transforming Growth Factor Beta Family Genes in Gonadal Differentiation in Japanese Eel, Anguilla japonica, According to Sex-Related Gene Expressions. Cells 2021, 10, 3007. https://doi.org/10.3390/cells10113007
Lin C-J, Jeng S-R, Lei Z-Y, Yueh W-S, Dufour S, Wu G-C, Chang C-F. Involvement of Transforming Growth Factor Beta Family Genes in Gonadal Differentiation in Japanese Eel, Anguilla japonica, According to Sex-Related Gene Expressions. Cells. 2021; 10(11):3007. https://doi.org/10.3390/cells10113007
Chicago/Turabian StyleLin, Chien-Ju, Shan-Ru Jeng, Zhen-Yuan Lei, Wen-Shiun Yueh, Sylvie Dufour, Guan-Chung Wu, and Ching-Fong Chang. 2021. "Involvement of Transforming Growth Factor Beta Family Genes in Gonadal Differentiation in Japanese Eel, Anguilla japonica, According to Sex-Related Gene Expressions" Cells 10, no. 11: 3007. https://doi.org/10.3390/cells10113007
APA StyleLin, C. -J., Jeng, S. -R., Lei, Z. -Y., Yueh, W. -S., Dufour, S., Wu, G. -C., & Chang, C. -F. (2021). Involvement of Transforming Growth Factor Beta Family Genes in Gonadal Differentiation in Japanese Eel, Anguilla japonica, According to Sex-Related Gene Expressions. Cells, 10(11), 3007. https://doi.org/10.3390/cells10113007






