Generation of Primordial Germ Cell-like Cells from iPSCs Derived from Turner Syndrome Patients
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Statements
2.2. PBMCs Expansion
2.3. Feeder Cells
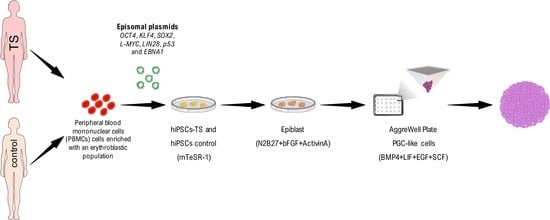
2.4. Reprograming and Culture of hiPSCs
2.5. Differentiation of hiPSCs into Embryoid Bodies
2.6. Differentiation of hiPSCs into PGCLCs
2.7. Screening for Vector Spontaneous Integration
2.8. Cytogenetic Analyses
2.9. Alkaline Phosphatase and Immunophenotypic Characterization
2.10. Analysis of Gene Transcription Levels by RT-qPCR
2.11. Statistical Analysis
3. Results
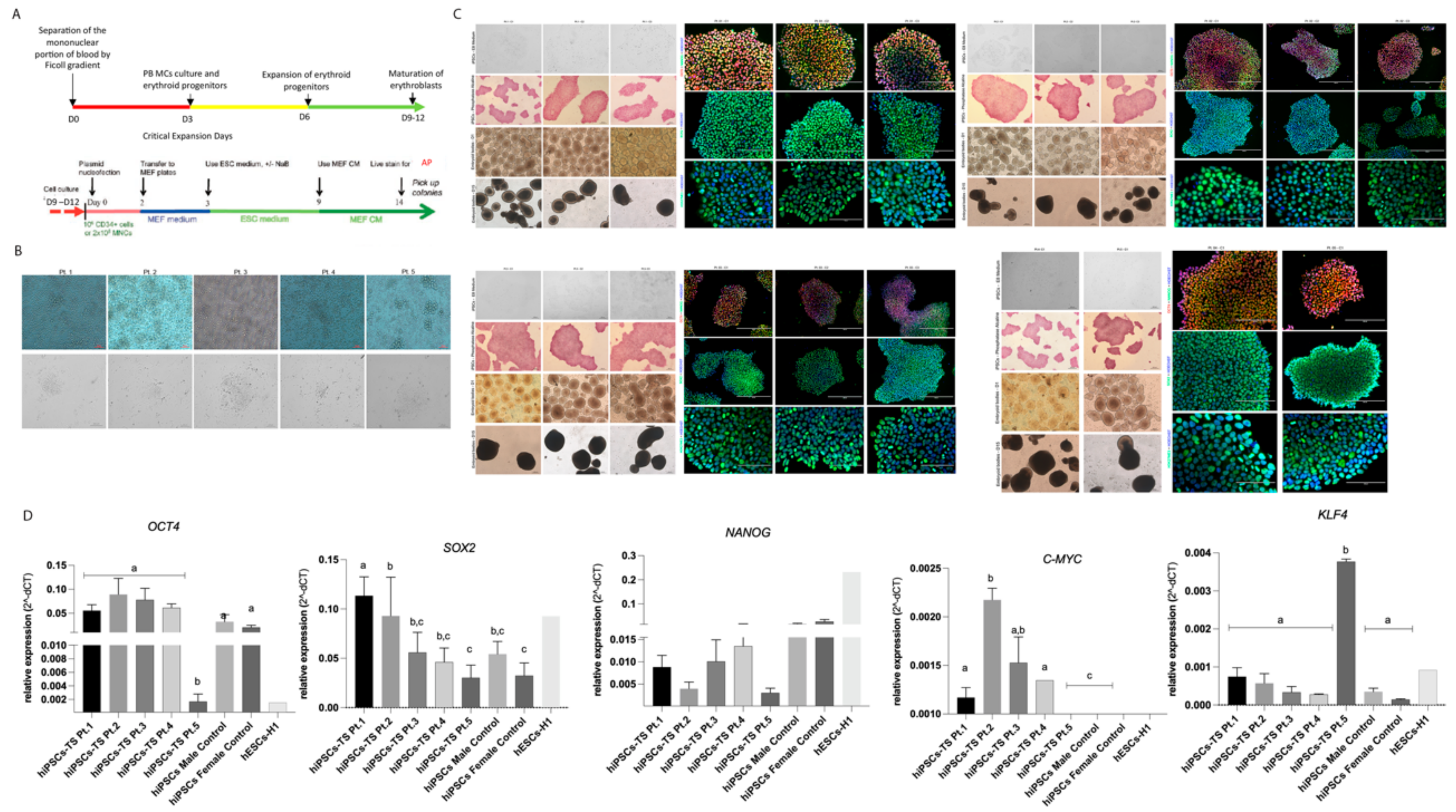
3.1. Generation and Characterization of hiPSCs-TS
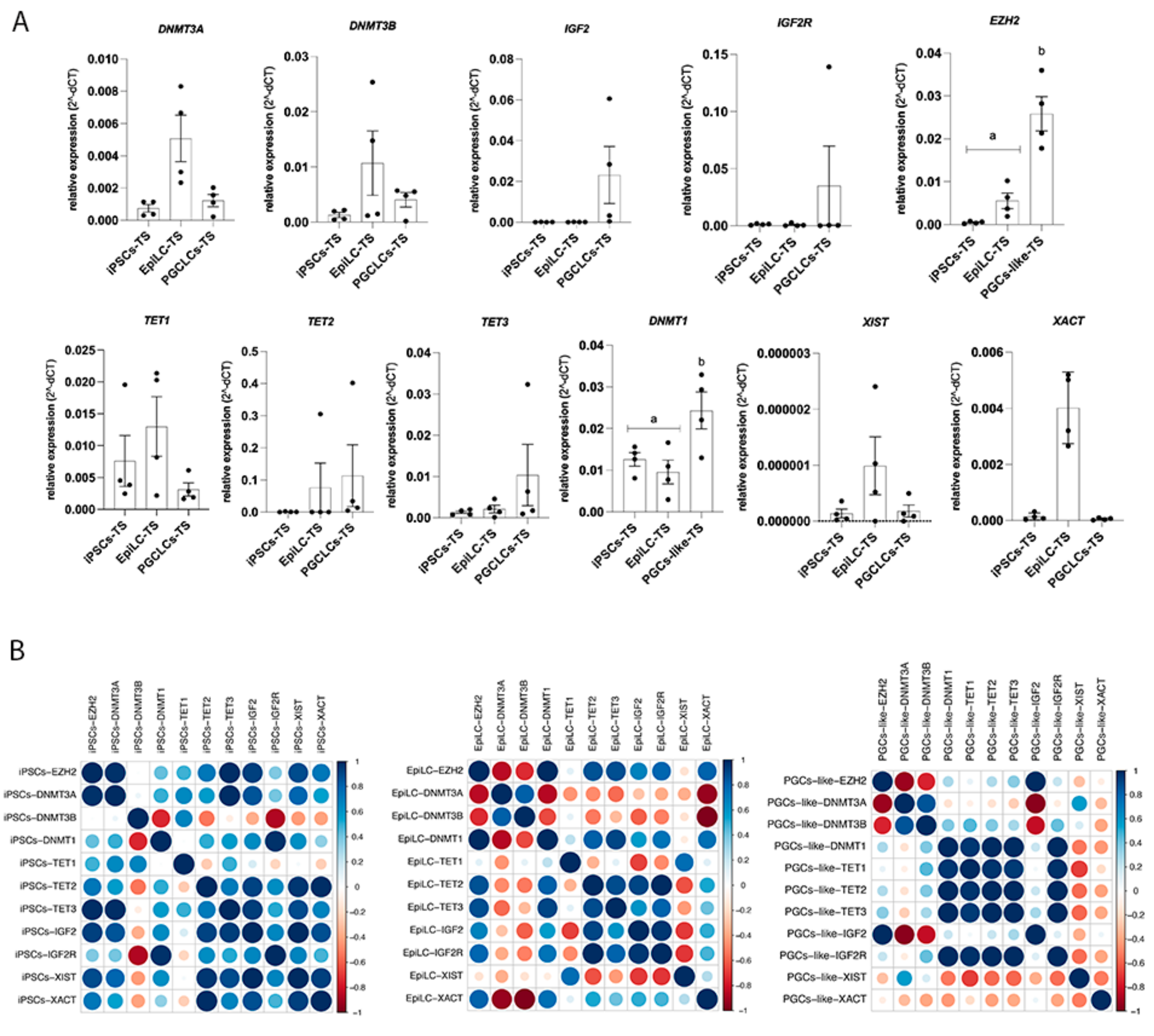
3.2. Epigenetic Signature of hiPSCs-TS
3.3. Induced and Characterization of hPGCLCs-TS
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Muntaj, S.; Ganaie, F.A.; Purva, S.V.; Radhika, S.; Tilak, P. Karyotypic Variables in Turner Syndrome: A Case Series. Int. J. Sci. Study 2015, 3, 171–175. [Google Scholar] [CrossRef]
- Nielsen, J.; Wohlert, M. Chromosome abnormalities found among 34910 newborn children: Results from a 13-year incidence study in Århus, Denmark. Hum. Genet. 1991, 87, 81–83. [Google Scholar] [CrossRef] [PubMed]
- Shankar, R.K.; Backeljauw, P.F. Current best practice in the management of Turner syndrome. Ther. Adv. Endocrinol. Metab. 2018, 9, 33–40. [Google Scholar] [CrossRef]
- Zinn, A.R.; Page, D.C.; Fisher, E.M.C. Turner syndrome: The case of the missing sex chromosome. Trends Genet. 1993, 9, 90–93. [Google Scholar] [CrossRef]
- Ross, J.; Roeltgen, D.; Zinn, A. Cognition and the sex chromosomes: Studies in Turner syndrome. Horm. Res. 2006, 65, 47–56. [Google Scholar] [CrossRef] [PubMed]
- Oktay, K.; Bedoschi, G.; Berkowitz, K.; Bronson, R.; Kashani, B.; McGovern, P.; Pal, L.; Quinn, G.; Rubin, K. Fertility Preservation in Women with Turner Syndrome: A Comprehensive Review and Practical Guidelines. J. Pediatr. Adolesc. Gynecol. 2016, 29, 409–416. [Google Scholar] [CrossRef] [PubMed]
- Álvarez-Nava, F.; Lanes, R. Epigenetics in Turner syndrome. Clin. Epigenetics 2018, 10, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Viuff, M.; Skakkebæk, A.; Nielsen, M.M.; Chang, S.; Gravholt, C.H. Epigenetics and genomics in Turner syndrome. Am. J. Med. Genet. Part C Semin. Med. Genet. 2019, 181, 68–75. [Google Scholar] [CrossRef]
- Trolle, C.; Nielsen, M.M.; Skakkebæk, A.; Lamy, P.; Vang, S.; Hedegaard, J.; Nordentoft, I.; Ørntoft, T.F.; Pedersen, J.S.; Gravholt, C.H. Widespread DNA hypomethylation and differential gene expression in Turner syndrome. Sci. Rep. 2016, 6, 1–14. [Google Scholar] [CrossRef]
- LeMaire-Adkins, R.; Hunt, P.A. Nonrandom segregation of the mouse univalent X chromosome: Evidence of spindle-mediated meiotic drive. Genet. Soc. Am. 2000, 156, 775–783. [Google Scholar] [CrossRef]
- Takahashi, K.; Yamanaka, S. Induction of Pluripotent Stem Cells from Mouse Embryonic and Adult Fibroblast Cultures by Defined Factors. Cell 2006, 2, 663–676. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, K.; Tanabe, K.; Ohnuki, M.; Narita, M.; Ichisaka, T.; Tomoda, K.; Yamanaka, S. Induction of Pluripotent Stem Cells from Adult Human Fibroblasts by Defined Factors. Cell 2007, 131, 861–872. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; David, B.T.; Trawczynski, M.; Fessler, R.G. Advances in Pluripotent Stem Cells: History, Mechanisms, Technologies, and Applications. Stem Cell Rev. Rep. 2020, 16, 3–32. [Google Scholar] [CrossRef]
- Aoi, T.; Yae, K.; Nakagawa, M.; Ichisaka, T.; Okita, K.; Takahashi, K.; Chiba, T.; Yamanaka, S. Generation of Pluripotent Stem Cells from Adult Mouse Liver and Stomach Cells. Science 2008, 321, 699–702. [Google Scholar] [CrossRef]
- Feng, B.; Jiang, J.; Kraus, P.; Ng, J.H.; Heng, J.C.D.; Chan, Y.S.; Yaw, L.P.; Zhang, W.; Loh, Y.H.; Han, J.; et al. Reprogramming of fibroblasts into induced pluripotent stem cells with orphan nuclear receptor Esrrb. Nat. Cell Biol. 2009, 11, 197–203. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.B.; Sebastiano, V.; Wu, G.; Araúzo-Bravo, M.J.; Sasse, P.; Gentile, L.; Ko, K.; Ruau, D.; Ehrich, M.; van den Boom, D.; et al. Oct4-Induced Pluripotency in Adult Neural Stem Cells. Cell 2009, 136, 411–419. [Google Scholar] [CrossRef]
- Yu, J.; Hu, K.; Smuga-Otto, K.; Tian, S.; Stewart, R.; Slukvin, I.I.; Thomson, J.A. Human Induced Pluripotent Stemm Cells Free of Vector and Trangene Sequence. Science 2009, 324, 797–802. [Google Scholar] [CrossRef]
- Nishimura, K.; Ohtaka, M.; Takada, H.; Kurisaki, A.; Tran, N.V.K.; Tran, Y.T.H.; Hisatake, K.; Sano, M.; Nakanishi, M. Simple and effective generation of transgene-free induced pluripotent stem cells using an auto-erasable Sendai virus vector responding to microRNA-302. Stem Cell Res. 2017, 23, 13–19. [Google Scholar] [CrossRef]
- Yusa, K.; Rad, R.; Takeda, J.; Bradley, A. Generation of transgene-free induced pluripotent mouse stem cells by the piggyBac transposon. Nat. Methods 2009, 6, 363–369. [Google Scholar] [CrossRef]
- Lindner, S.E.; Sugden, B. The plasmid replicon of Epstein-Barr virus: Mechanistic insights into efficient, licensed, extrachromosomal replication in human cells. Plasmid 2007, 58, 1–12. [Google Scholar] [CrossRef]
- Chou, B.-K.; Mali, P.; Huang, X.; Ye, Z.; Dowey, S.N.; Resar, L.M.; Zou, C.; Zhang, Y.A.; Tong, J.; Cheng, L. Efficient human iPS cell derivation by a non-integrating plasmid from blood cells with unique epigenetic and gene expression signatures. Cell Res. 2011, 21, 518–529. [Google Scholar] [CrossRef] [PubMed]
- Okita, K.; Yamakawa, T.; Matsumura, Y.; Sato, Y.; Amano, N.; Watanabe, A.; Goshima, N.; Yamanaka, S. An efficient nonviral method to generate integration-free human-induced pluripotent stem cells from cord blood and peripheral blood cells. Stem Cells 2013, 31, 458–466. [Google Scholar] [CrossRef] [PubMed]
- Tangprasittipap, A.; Jittorntrum, B.; Wongkummool, W.; Kitiyanant, N.; Tubsuwan, A. Generation of induced pluripotent stem cells from peripheral blood CD34 + hematopoietic progenitors of a 31 year old healthy woman. Stem Cell Res. 2017, 20, 91–93. [Google Scholar] [CrossRef] [PubMed]
- Molina, S.G.; Beltran, A.A.; Beltran, A.S. Generation of an integration-free induced pluripotent stem cell line (UNC001-A) from blood of a healthy individual. Stem Cell Res. 2020, 49, 102015. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhang, J.; Hu, J.; Li, G.; Lei, Y.; Zhao, J. Establishment of TUSMi008-A, an induced pluripotent stem cell (iPSC) line from a 76-year old Alzheimer’s disease (AD) patient with PAXIP1 gene mutation. Stem Cell Res. 2019, 36, 101391. [Google Scholar] [CrossRef]
- Ross, S.B.; Fraser, S.T.; Bagnall, R.D.; Semsarian, C. Peripheral blood derived induced pluripotent stem cells (iPSCs) from a female with familial hypertrophic cardiomyopathy. Stem Cell Res. 2017, 20, 76–79. [Google Scholar] [CrossRef]
- Luo, Y.; Zhu, D.; Xu, X.; Ge, L.; Sun, X.; Chen, G.; Chen, Y. Generation of an induced pluripotent stem cell line from an adult male with 45,X/46,XY mosaicism. Stem Cell Res. 2018, 27, 42–45. [Google Scholar] [CrossRef]
- Luo, Y.; Zhu, D.; Du, R.; Gong, Y.; Xie, C.; Xu, X.; Fan, Y.; Yu, B.; Sun, X.; Chen, Y. Uniparental disomy of the entire X chromosome in Turner syndrome patient-speci fi c induced pluripotent stem cells. Cell Discov. 2015, 1, 15022. [Google Scholar] [CrossRef]
- Li, W.; Wang, X.; Fan, W.; Zhao, P.; Chan, Y.C.; Chen, S.; Zhang, S.; Guo, X.; Zhang, Y.; Li, Y.; et al. Modeling abnormal early development with induced pluripotent stem cells from aneuploid syndromes. Hum. Mol. Genet. 2012, 21, 32–45. [Google Scholar] [CrossRef]
- Hayashi, K.; Ohta, H.; Kurimoto, K.; Aramaki, S.; Saitou, M. Reconstitution of the mouse germ cell specification pathway in culture by pluripotent stem cells. Cell 2011, 146, 519–532. [Google Scholar] [CrossRef]
- Hayashi, Y.; Saitou, M.; Yamanaka, S. Germline development from human pluripotent stem cells toward disease modeling of infertility. Fertil. Steril. 2012, 97, 1250–1259. [Google Scholar] [CrossRef]
- Yamashiro, C.; Sasaki, K.; Yabuta, Y.; Kojima, Y.; Nakamura, T.; Okamoto, I.; Yokobayashi, S.; Murase, Y.; Ishikura, Y.; Sasaki, H.; et al. Generation of human oogonia from induced pluripotent stem cells in vitro. Science 2018, 362, 356–360. [Google Scholar] [CrossRef]
- Yoshino, T.; Suzuki, T.; Nagamatsu, G.; Yabukami, H.; Ikegaya, M.; Kishima, M.; Kita, H.; Imamura, T.; Nakashima, K.; Nishinakamura, R.; et al. Generation of ovarian follicles from mouse pluripotent stem cells. Science 2021, 373, eabe0237. [Google Scholar] [CrossRef] [PubMed]
- Hwang, Y.S.; Suzuki, S.; Seita, Y.; Ito, J.; Sakata, Y.; Aso, H.; Sato, K.; Hermann, B.P.; Sasaki, K. Reconstitution of prospermatogonial specification in vitro from human induced pluripotent stem cells. Nat. Commun. 2020, 11, 5656. [Google Scholar] [CrossRef] [PubMed]
- Dominguez, A.A.; Chiang, H.R.; Sukhwani, M.; Orwig, K.E.; Pera, R.A.R. Human germ cell formation in xenotransplants of induced pluripotent stem cells carrying X chromosome aneuploidies. Sci. Rep. 2014, 4, 6432. [Google Scholar] [CrossRef]
- Yango, P.; Altman, E.; Nguyen, H.; Zamah, M.; Cedars, M.I.; Tran, N.D. Primordial germ cells (PGCs) derived from patients with turner syndrome and Klinefelter syndrome offer insight to the mechanism of premature gonadal failure. Fertil. Steril. 2013, 100, S40. [Google Scholar] [CrossRef]
- Thomson, J.A.; Itskovitz-Eldor, J.; Shapiro, S.S.; Waknitz, M.A.; Swiergiel, J.J.; Marshall, V.S.; Jones, J.M. Embryonic stem cell lines derived from human blastocysts. Science 1998, 282, 1145–1147. [Google Scholar] [CrossRef]
- Junqueira Reis, L.C.; Picanço-castro, V.; Paes, B.C.M.F.; Pereira, O.A.; Gerdes Gyuricza, I.; De Araújo, F.T.; Morato-Marques, M.; Moreira, L.F.; De Brito, E.; Costa, E.D.B.O.; et al. Induced Pluripotent Stem Cell for the Study and Treatment of Sickle Cell Anemia. Stem Cells Int. 2017, 2017, 1–30. [Google Scholar] [CrossRef]
- Chou, B.-K.; Gu, H.; Gao, Y.; Dowey, S.N.; Wang, Y.; Shi, J.; Li, Y.; Ye, Z.; Cheng, T.; Cheng, L. A Facile Method to Establish Human Induced Pluripotent Stem Cells From Adult Blood Cells Under Feeder-Free and Xeno-Free Culture Conditions: A Clinically Compliant Approach. Stem Cells Transl. Med. 2015, 4, 320–332. [Google Scholar] [CrossRef]
- Hayashi, K.; Ogushi, S.; Kurimoto, K.S.S.; Ohta, H.M.S. Offspring from Oocytes Derived from in Vitro Primordial Germ Cell–like Cells in Mice. Science 2012, 338, 971–976. [Google Scholar] [CrossRef] [PubMed]
- Irie, N.; Weinberger, L.; Tang, W.W.C.; Kobayashi, T.; Viukov, S.; Manor, Y.S.; Dietmann, S.; Hanna, J.H.; Surani, M.A. Article SOX17 Is a Critical Specifier of Human Primordial Germ Cell Fate. Cell 2015, 160, 253–268. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.Y.; Hysolli, E.; Tanaka, Y.; Wang, B.; Jung, Y.W.; Pan, X.; Weissman, S.M.; Park, I.H. X chromosome of female cells shows dynamic changes in status during human somatic cell reprogramming. Stem Cell Rep. 2014, 2, 896–909. [Google Scholar] [CrossRef][Green Version]
- Wang, X.; Liao, T.; Wan, C.; Yang, X.; Zhao, J.; Fu, R.; Yao, Z.; Huang, Y.; Shi, Y.; Chang, G.; et al. Efficient generation of human primordial germ cell-like cells from pluripotent stem cells in a methylcellulose-based 3D system at large scale. PeerJ 2019, 6, e6143. [Google Scholar] [CrossRef]
- Nettersheim, D.; Heukamp, L.C.; Fronhoffs, F.; Grewe, M.J.; Haas, N.; Waha, A.; Honecker, F.; Waha, A.; Kristiansen, G.; Schorle, H. Analysis of TET Expression/Activity and 5mC Oxidation during Normal and Malignant Germ Cell Development. PLoS ONE 2013, 8, e82881. [Google Scholar] [CrossRef]
- R Core Team. A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2020; Available online: https://www.R-project.org/ (accessed on 4 January 2021).
- Jung, M.; Peterson, H.; Chavez, L.; Kahlem, P.; Lehrach, H.; Vilo, J.; Adjaye, J. A data integration approach to mapping OCT4 gene regulatory networks operative in embryonic stem cells and embryonal carcinoma cells. PLoS ONE 2010, 5, e10709. [Google Scholar] [CrossRef]
- Ding, X.; Wang, X.; Sontag, S.; Qin, J.; Wanek, P.; Lin, Q.; Zenke, M. The polycomb protein Ezh2 impacts on induced pluripotent stem cell generation. Stem Cells Dev. 2014, 23, 931–940. [Google Scholar] [CrossRef] [PubMed]
- Harel, S.; Tu, E.Y.; Weisberg, S.; Esquilin, M.; Chambers, S.M.; Liu, B.; Carson, C.T.; Studer, L.; Reizis, B.; Tomishima, M.J. ZFX controls the self-renewal of human embryonic stem cells. PLoS ONE 2012, 7, 1–10. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Amps, K.; Andrews, P.; Anyfantis, G.; Armstrong, L.; Avery, S.; Baharvand, H.; Baker, J.; Baker, D.; Munoz, M.; Beil, S.; et al. Screening a large, ethnically diverse population of human embryonic stem cells identifies a chromosome 20 minimal amplicon that confers a growth advantage. Nat. Biotechnol. 2012, 29, 1132–1144. [Google Scholar] [CrossRef]
- Mayshar, Y.; Ben-David, U.; Lavon, N.; Biancotti, J.C.; Yakir, B.; Clark, A.T.; Plath, K.; Lowry, W.E.; Benvenisty, N. Identification and classification of chromosomal aberrations in human induced pluripotent stem cells. Cell Stem Cell 2010, 7, 521–531. [Google Scholar] [CrossRef]
- Sun, X.; Long, X.; Yin, Y.; Jiang, Y.; Chen, X.; Liu, W.; Zhang, W.; Du, H.; Li, S.; Zheng, Y.; et al. Similar biological characteristics of human embryonic stem cell lines with normal and abnormal karyotypes. Hum. Reprod. 2008, 23, 2185–2193. [Google Scholar] [CrossRef] [PubMed]
- Mitalipova, M.M.; Rao, R.R.; Hoyer, D.M.; Johnson, J.A.; Meisner, L.F.; Jones, K.L.; Dalton, S.; Stice, S.L. Preserving the genetic integrity of human embryonic stem cells. Nat. Biotechnol. 2005, 23, 19–20. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, K.; Yokobayashi, S.; Nakamura, T.; Okamoto, I.; Yabuta, Y.; Kurimoto, K.; Ohta, H.; Moritoki, Y.; Iwatani, C.; Tsuchiya, H.; et al. Robust In Vitro Induction of Human Germ Cell Fate from Pluripotent Stem Cells. Stem Cell 2015, 17, 178–194. [Google Scholar] [CrossRef]
- Chen, D.; Sun, N.; Hou, L.; Kim, R.; Faith, J.; Aslanyan, M.; Tao, Y.; Zheng, Y.; Fu, J.; Liu, W.; et al. Human Primordial Germ Cells Are Specified from Lineage-Primed Progenitors. Cell Rep. 2019, 29, 4568–4582. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.; Wei, Y.; Sun, H.-X.; Gu, Y.; Ross, P.J.; Correspondence, J.W.; Mahdi, A.K.; Arteaga, C.A.P.; Sakurai, M.; Schmitz, D.A.; et al. Derivation of Intermediate Pluripotent Stem Cells Amenable to Primordial Germ Cell Specification ll Derivation of Intermediate Pluripotent Stem Cells Amenable to Primordial Germ Cell Specification. Cell Stem Cell 2020, 28, 1–18. [Google Scholar] [CrossRef]
- Chen, D.; Liu, W.; Zimmerman, J.; Gell, J.J.; Jacobsen, S.E.; Clark, A.T.; Chen, D.; Liu, W.; Zimmerman, J.; Pastor, W.A.; et al. The TFAP2C-Regulated OCT4 Naive Enhancer Is Involved in Human Germline Formation. Cell Rep. 2018, 25, 3591–3602.e5. [Google Scholar] [CrossRef] [PubMed]
- Kojima, Y.; Sasaki, K.; Yokobayashi, S.; Sakai, Y.; Nakamura, T.; Yabuta, Y.; Nakaki, F.; Nagaoka, S.; Woltjen, K.; Hotta, A.; et al. Evolutionarily Distinctive Transcriptional and Signaling Programs Drive Human Germ Cell Lineage Specification from Pluripotent Stem Cells. Cell Stem Cell 2017, 21, 517–532.e5. [Google Scholar] [CrossRef]
- Chan, J.C.; Maze, I. Nothing Is yet Set in (Hi)stone: Novel Post-Translational Modifications Regulating Chromatin Function. Trends Biochem. Sci. 2020, 45, 829–844. [Google Scholar] [CrossRef]
- Kurimoto, K.; Saitou, M. Epigenome regulation during germ cell specification and development from pluripotent stem cells. Curr. Opin. Genet. Dev. 2018, 52, 57–64. [Google Scholar] [CrossRef]
- Eguizabal, C.; Herrera, L.; De Oñate, L.; Montserrat, N.; Hajkova, P.; Izpisua Belmonte, J.C. Characterization of the Epigenetic Changes During Human Gonadal Primordial Germ Cells Reprogramming. Stem Cells 2016, 34, 2418–2428. [Google Scholar] [CrossRef]
- Agger, K.; Cloos, P.A.C.; Christensen, J.; Pasini, D.; Rose, S.; Rappsilber, J.; Issaeva, I.; Canaani, E.; Salcini, A.E.; Helin, K. UTX and JMJD3 are histone H3K27 demethylases involved in HOX gene regulation and development. Nature 2007, 449, 731–734. [Google Scholar] [CrossRef]
- Shen, H.; Xu, W.; Lan, F. Histone lysine demethylases in mammalian embryonic development. Exp. Mol. Med. 2017, 49, e325. [Google Scholar] [CrossRef] [PubMed]
- Mansour, A.A.; Gafni, O.; Weinberger, L.; Zviran, A.; Ayyash, M.; Rais, Y.; Krupalnik, V.; Zerbib, M.; Amann-Zalcenstein, D.; Maza, I.; et al. The H3K27 demethylase Utx regulates somatic and germ cell epigenetic reprogramming. Nature 2012, 488, 409–413. [Google Scholar] [CrossRef] [PubMed]
- Nagaoka, S.; Saitou, M. Reconstitution of Female Germ Cell Fate Determination and Meiotic Initiation in Mammals. Cold Spring Harb. Symp. Quant. Biol. 2017, 82, 213–222. [Google Scholar] [CrossRef] [PubMed]
- Von Meyenn, F.; Berrens, R.V.; Andrews, S.; Santos, F.; Collier, A.J.; Krueger, F.; Osorno, R.; Dean, W.; Rugg-Gunn, P.J.; Reik, W. Comparative Principles of DNA Methylation Reprogramming during Human and Mouse In Vitro Primordial Germ Cell Specification. Dev. Cell 2016, 39, 104–115. [Google Scholar] [CrossRef]





| Primer | Forward Primer Sequence | Reverse Primer Sequence |
|---|---|---|
| EBNA-1 | ACGATGCTTTCCAAACCACC | CATCATCATCCGGGTCTCCA |
| Gene Name | Forward Primer Sequence | Reverse Primer Sequence | Annealing (°C) |
|---|---|---|---|
| 18S | GCGAGTACTCAACACCAACATCG | TCAAGTCTCCCCAGCCTTGC | 55 |
| Β-ACTIN | TGAAGTGTGACGTGGACATC | GGAGGAGCAATGATCTTGAT | 60 |
| OCT4 (POU5F1) | CCTCACTTCACTGCACTGTA | CAGGTTTTCTTTCCCTAGCT | 55 |
| SOX2 | CCCAGCAGACTTCACATGT | CCTCCCATTTCCCTCGTTTT | 55 |
| KLF4 | GATGAACTGACCAGGCACTA | GTGGGTCATATCCACTGTCT | 55 |
| MYC | TGCCTCAAATTGGACTTTGG | GATTGAAATTCTGTGTAACTGC | 55 |
| NANOG | TGAACCTCAGCTACAAACAG | TGGTGGTAGGAAGAGTAAAG | 55 |
| RUNX1 | CCCTAGGGGATGTTCCAGAT | TGAAGCTTTTCCCTCTTCCA | 60 |
| CD34 | TGAAGCCTAGCCTGTCACCT | CGCACAGCTGGAGGTCTTAT | 60 |
| AFP | AGCTTGGTGGTGGATGAAAC | CCCTCTTCAGCAAAGCAGAC | 60 |
| NCAM | ATGGAAACTCTATTAAAGTGAACCTG | TAGACCTCATACTCAGCATTCCAGT | 60 |
| NESTIN | GCGTTGGAACAGAGGTTGGA | TGGGAGCAAAGATCCAAGAC | 60 |
| XIST | CGGTACGTTGAAGTTAGGGAATG | GTGCTGTCTAATCCAATGGGTAG | 60 |
| XACT | TGAATGAAGGCACATCTGA | CTCTCCCAGCCTATTTGTGG | 60 |
| ZFX | ATGGAAGAAGCAGATGTGTC | TTGAGGCTGAAGTAATGTCA | 60 |
| RNF12 | ACCGATTGGATCGAGAAGAAGC | TGTAGTCGTCTCAGCAACTCT | 60 |
| EZH2 | ACCGGTTGTGGGCTGCACAC | TGCAGCGGCATCCCGGAAAG | 60 |
| MECP2 | CCAGGACTTGAGCAGCAGCG | CGGGAAGCTTTGTCAGAGCCC | 60 |
| SOX17 | GAGCCAAGGGCGAGTCCCGTA | CCTTCCACGACTTGCCCAGCAT | 65 |
| TFAP2C (AP-2γ) | CGCTCATGTGACTCTCCTGACATCC | TGGGCCGCCAATAGCATGTTCT | 60 |
| PRDM14 | CTACCGAGCCCGAGTGGCCTAC | TAGAGCCATCCCGGGACCGCA | 60 |
| NANOS3 | CCCGAAACTCGGCAGGCAAGA | AAGGCTCAGACTTCCCGGCAC | 60 |
| DPPA3 (STELLA) | ACGCCGATGGACCCATCACAGTTT | TCTCGGAGGAGATTTGAGAGGCCC | 60 |
| DAZL | TGGCCCTTCTTTCAGTGACTTC | GACCCTAGGGGGCACTAGTAA | 60 |
| DDX4 (VASA) | TTCTTCACAAGCTCCCAATCCA | TTCTTCTCTGCATCAAAACCACA | 60 |
| DNMT1 | GGGCTACCTGGCTAAAGTCAA | CTGCCATTCCCACTCTACGG | 60 |
| DNMT3A | TGGGATTCATCCAGACTCATGC | AAAGTGAGAAACTGGGCCTGAA | 60 |
| DNMT3B | TAACTGGAGCCACGACGTAAC | GCATCCGTCATCTTTCAGCCTA | 60 |
| TET1 | GCTGCTGTCAGGGAAATCAT | ACCATCACAGCAGTTGGACA | 60 |
| TET2 | CCAATAGGACATGATCCAGG | TCTGGATGAGCTCTCTCAGG | 60 |
| TET3 | TCGGAGACACCCTCTACCAG | CTTGCAGCCGTTGAAGTACA | 60 |
| IGF2 | GGGAGTTCTGGGGTAGGAAG | GAAAAATGCCCCAAGAAACA | 60 |
| IGF2R | CTTGACTGGGGCAATGATTT | GACGCATCGGGTGTAACTTT | 60 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
de Souza, A.F.; Bressan, F.F.; Pieri, N.C.G.; Botigelli, R.C.; Revay, T.; Haddad, S.K.; Covas, D.T.; Ramos, E.S.; King, W.A.; Meirelles, F.V. Generation of Primordial Germ Cell-like Cells from iPSCs Derived from Turner Syndrome Patients. Cells 2021, 10, 3099. https://doi.org/10.3390/cells10113099
de Souza AF, Bressan FF, Pieri NCG, Botigelli RC, Revay T, Haddad SK, Covas DT, Ramos ES, King WA, Meirelles FV. Generation of Primordial Germ Cell-like Cells from iPSCs Derived from Turner Syndrome Patients. Cells. 2021; 10(11):3099. https://doi.org/10.3390/cells10113099
Chicago/Turabian Stylede Souza, Aline Fernanda, Fabiana Fernandes Bressan, Naira Caroline Godoy Pieri, Ramon Cesar Botigelli, Tamas Revay, Simone Kashima Haddad, Dimas Tadeu Covas, Ester Silveira Ramos, Willian Allan King, and Flavio Vieira Meirelles. 2021. "Generation of Primordial Germ Cell-like Cells from iPSCs Derived from Turner Syndrome Patients" Cells 10, no. 11: 3099. https://doi.org/10.3390/cells10113099
APA Stylede Souza, A. F., Bressan, F. F., Pieri, N. C. G., Botigelli, R. C., Revay, T., Haddad, S. K., Covas, D. T., Ramos, E. S., King, W. A., & Meirelles, F. V. (2021). Generation of Primordial Germ Cell-like Cells from iPSCs Derived from Turner Syndrome Patients. Cells, 10(11), 3099. https://doi.org/10.3390/cells10113099