Hundreds of LncRNAs Display Circadian Rhythmicity in Zebrafish Larvae
Abstract
:1. Introduction
2. Materials and Methods
2.1. Fish Husbandry and Embryo Production
2.2. Deep Sequencing-Based Transcriptome Analysis
2.3. Zebrafish Larval RNA-Seq Datasets under DD and LL Conditions
2.4. Identification of Zebrafish Larval lncRNAs
2.5. Collection of Zebrafish Pineal Gland and Testis lncRNAs
2.6. Identification of Zebrafish Larval lncRNAs Displaying Circadian Expression
2.7. Investigating Circadian Regulation of Zebrafish Larval lncRNAs Mediated by E-Box, D-Box and RORE Regulatory Motifs
2.8. GO, COG, and KEGG Enrichment and Annotation of Circadianly Expressed Zebrafish Larval lncRNAs
2.9. Principal Component Analysis (PCA) of Circadian Zebrafish Larval lncRNAs
2.10. Predicting Orthologs of Zebrafish Larval lncRNAs with NCBI BLAST
2.11. Uncovering 3D Models and Functions of lncRNA-Encoded Peptides
3. Results
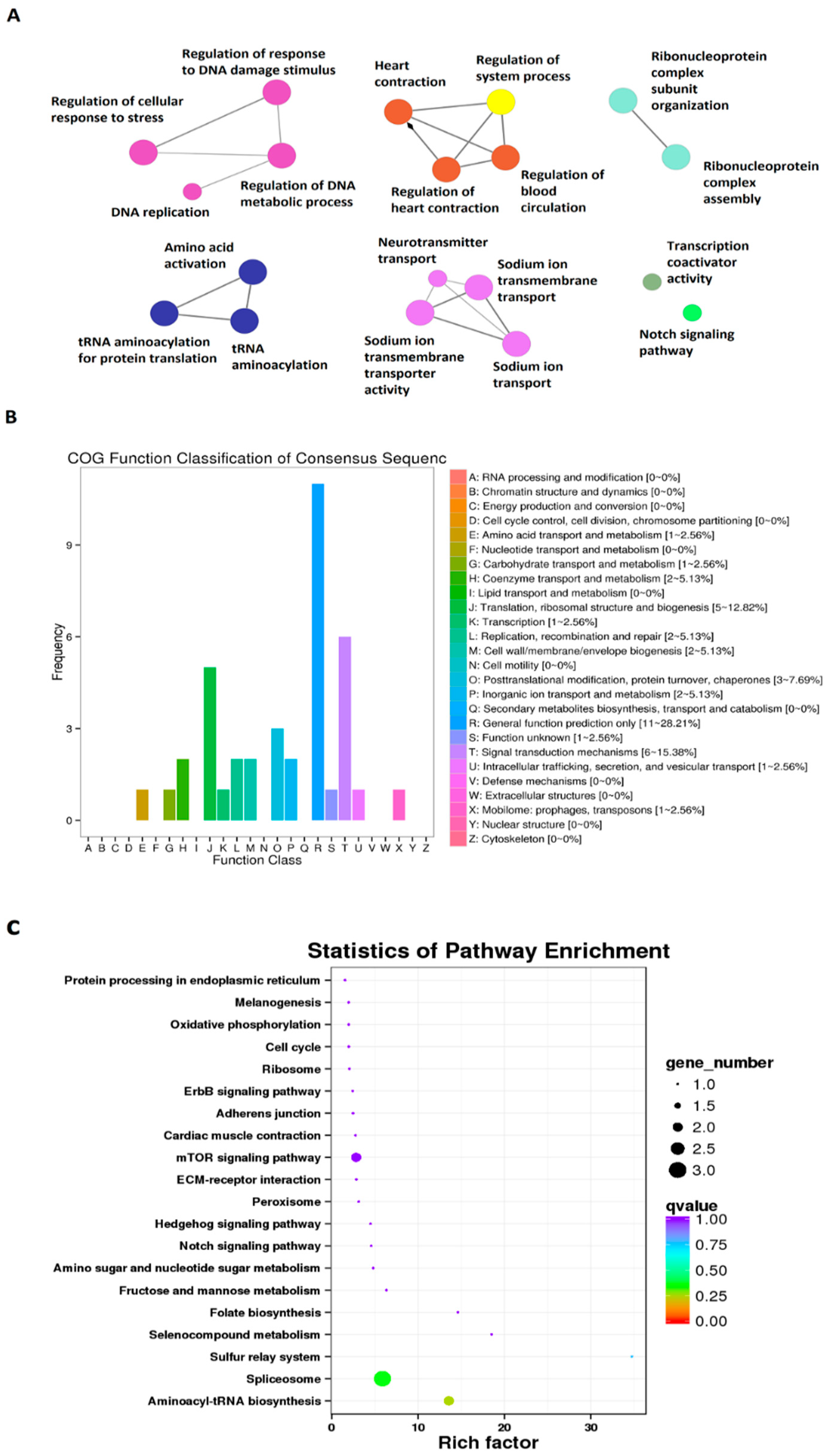
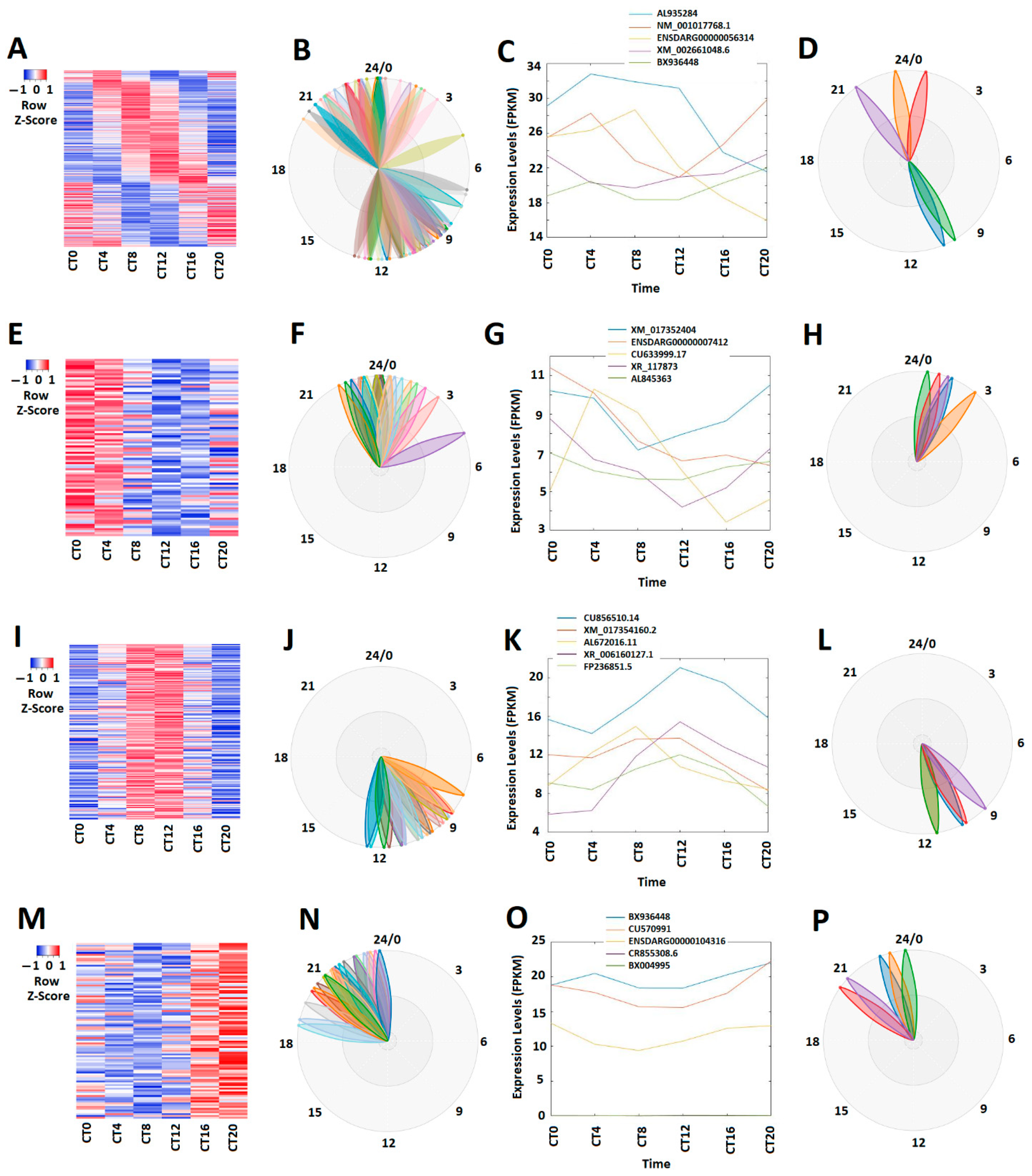
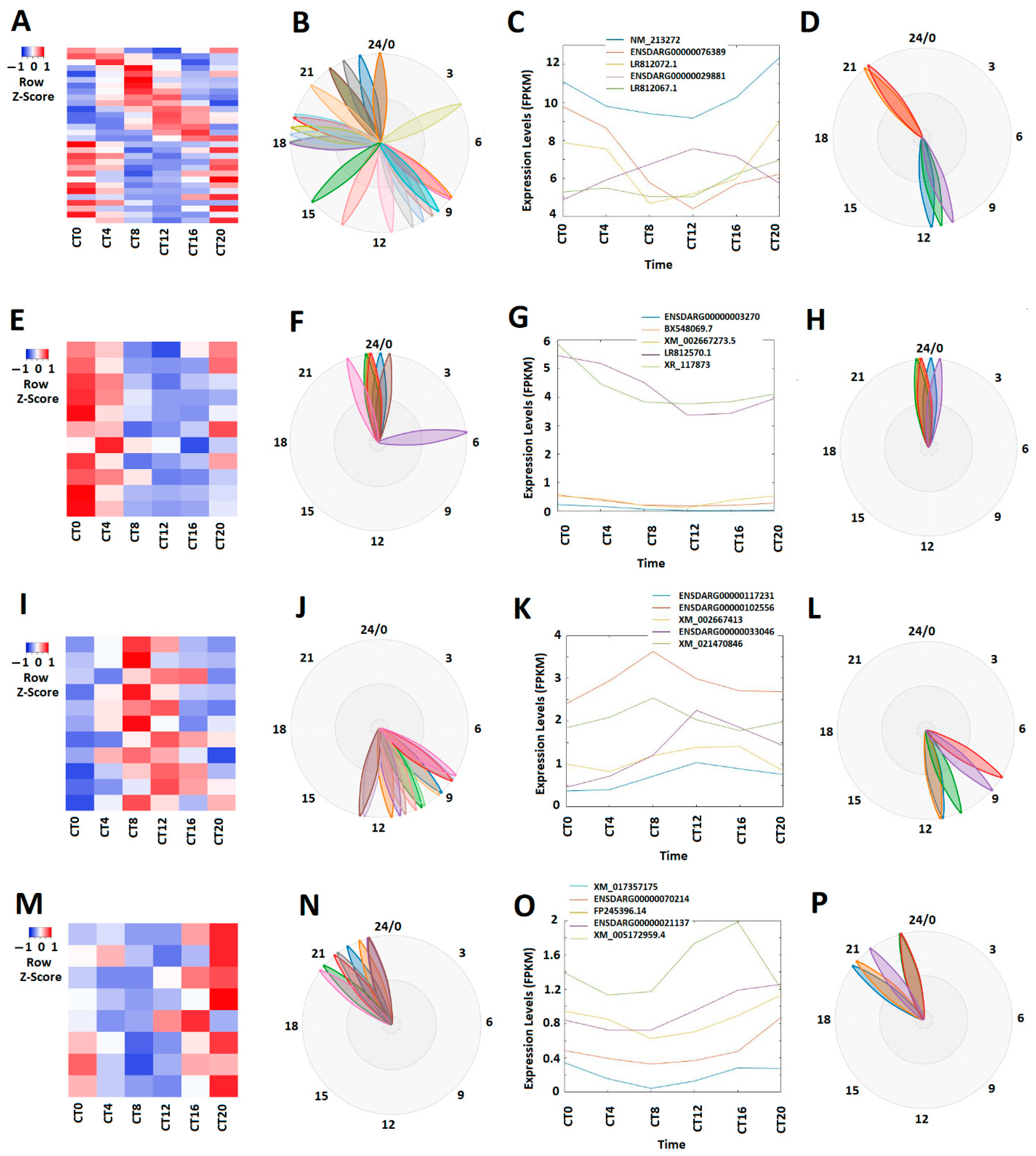
3.1. Circadianly Expressed Zebrafish Larval lncRNAs under Constant Darkness (DD) Condition and Their GO, COG, and KEGG Analyses
3.2. Circadianly Expressed Zebrafish Larval lncRNAs under Constant Light (LL) Condition and Their GO, COG, and KEGG Analyses
3.3. Circadianly Expressed Zebrafish Larval lncRNAs Are Coexpressed under Both Constant Darkness (DD) and Constant Light (LL) Condition and Their GO, COG, and KEGG Analyses
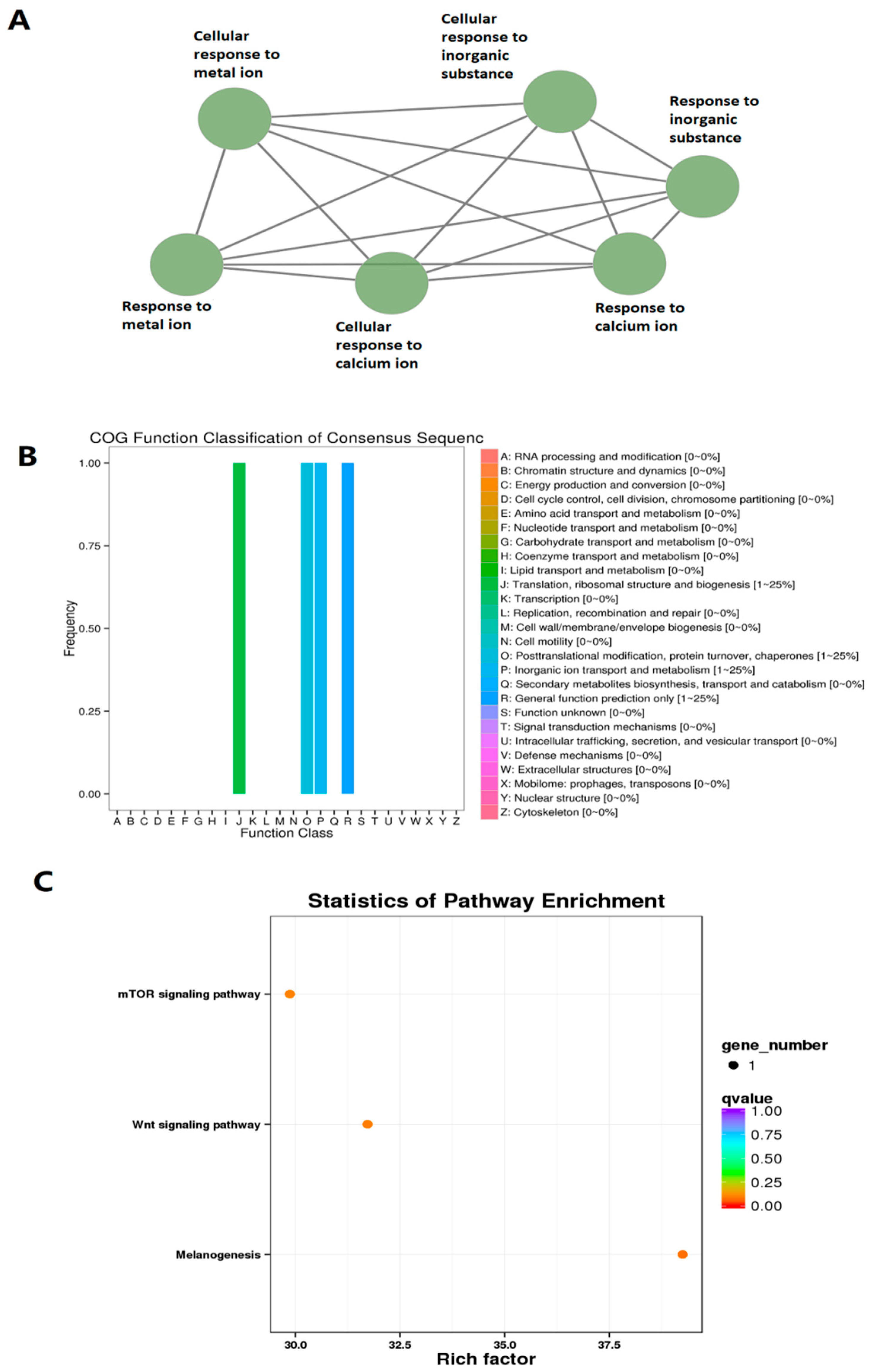
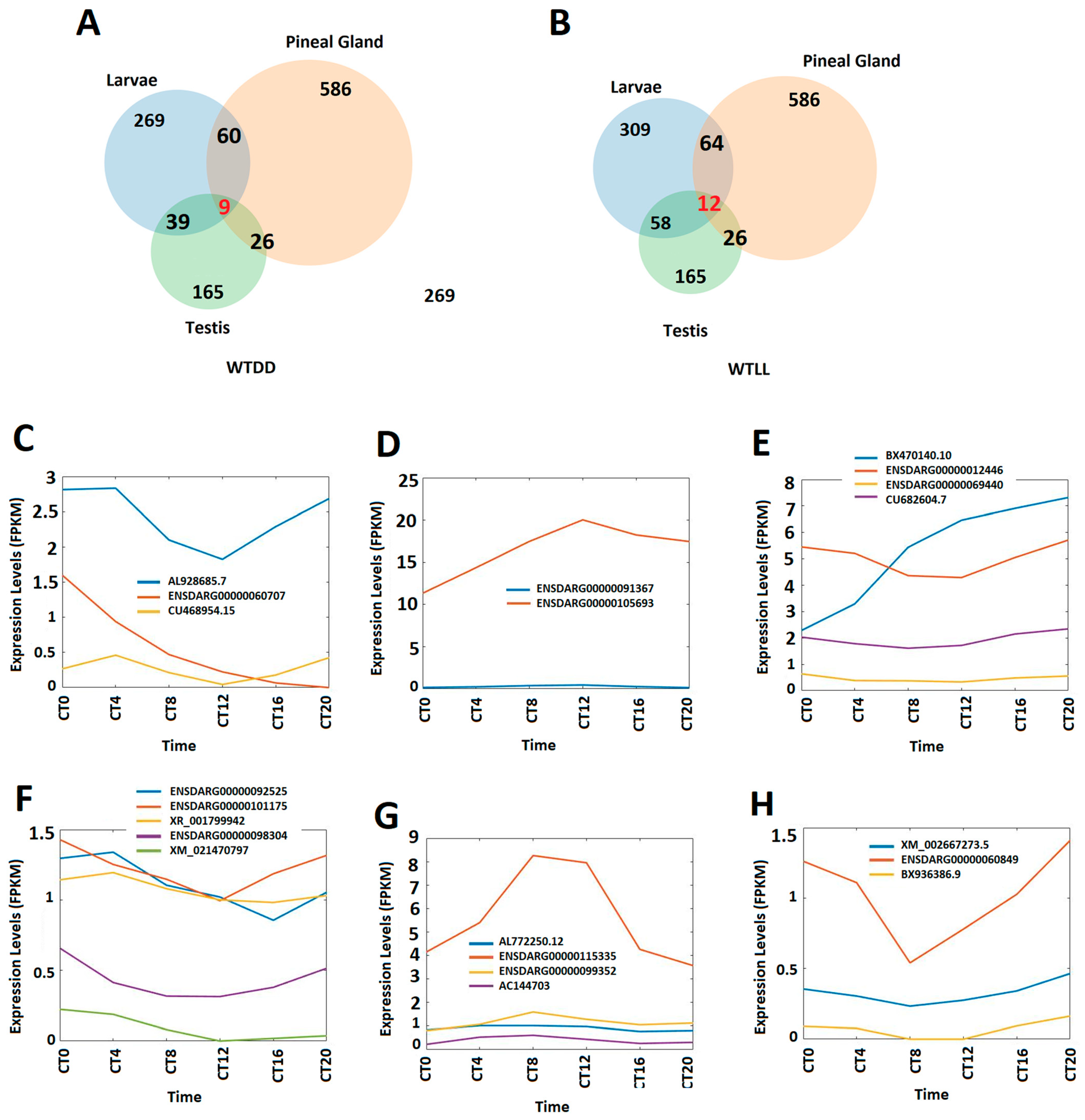
3.4. Circadianly Expressed Zebrafish Larval lncRNAs Are Coexpressed in the Pineal Gland and the Testis
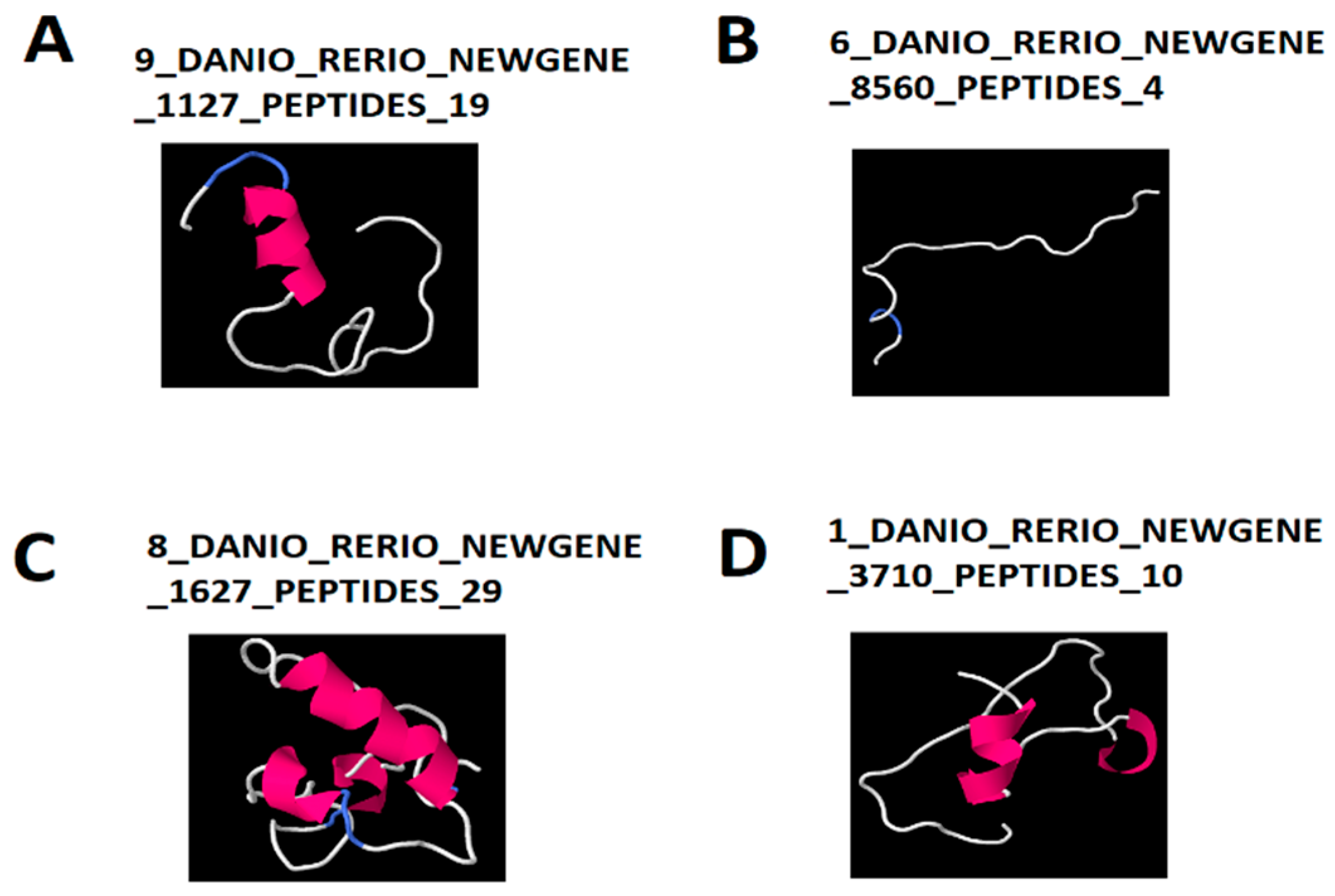
3.5. Uncovering Coexpressing lncRNA-Encoded Peptides and Their 3D Structures
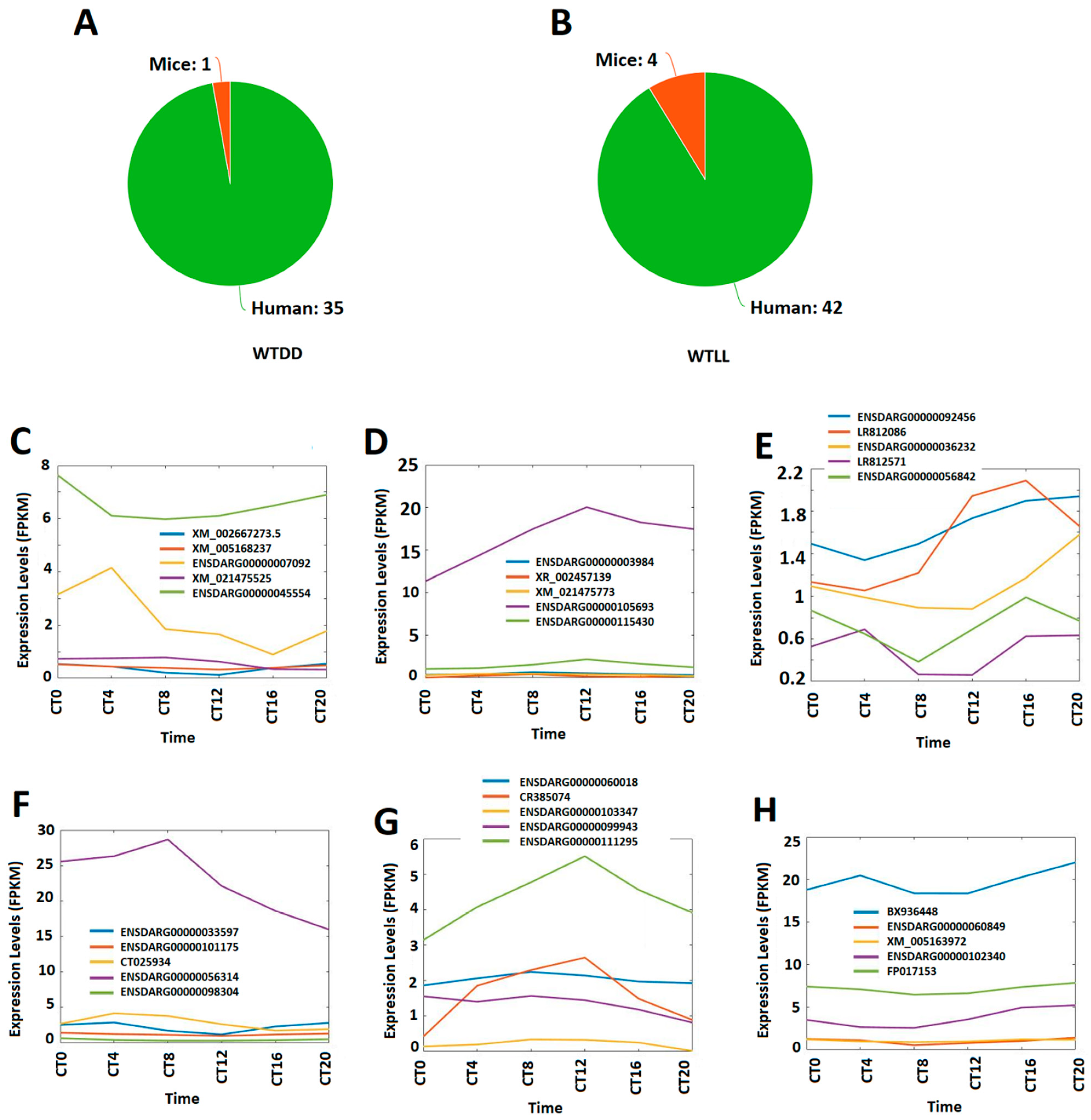
3.6. Conservation of Circadianly Expressed Zebrafish Larval lncRNAs under DD and LL Conditions with Mice and Humans
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Young, M.W. Life’s 24-hour clock: Molecular control of circadian rhythms in animal cells. Trends Biochem. Sci. 2000, 25, 601–606. [Google Scholar] [CrossRef]
- Segers, A.; Depoortere, I. Circadian clocks in the digestive system. Nat. Rev. Gastroenterol. Hepatol. 2021, 18, 239–251. [Google Scholar] [CrossRef]
- Sancar, A.; Lindsey-Boltz, L.A.; Gaddameedhi, S.; Selby, C.P.; Ye, R.; Chiou, Y.-Y.; Kemp, M.G.; Hu, J.; Lee, J.H.; Ozturk, N. Circadian clock, cancer, and chemotherapy. Biochemistry 2015, 54, 110–123. [Google Scholar] [CrossRef] [PubMed]
- Vatine, G.; Vallone, D.; Gothilf, Y.; Foulkes, N.S. It’s time to swim! Zebrafish and the circadian clock. FEBS Lett. 2011, 585, 1485–1494. [Google Scholar] [CrossRef]
- Beer, K.; Helfrich-Förster, C. Model and Non-model Insects in Chronobiology. Front. Behav. Neurosci. 2020, 14, 221. [Google Scholar] [CrossRef]
- Tataroglu, O.; Emery, P. Studying circadian rhythms in Drosophila melanogaster. Methods 2014, 68, 140–150. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ben-Moshe, Z.; Foulkes, N.S.; Gothilf, Y. Functional Development of the Circadian Clock in the Zebrafish Pineal Gland. BioMed Res. Int. 2014, 2014, 235781. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mishra, S.K.; Liu, T.; Wang, H. Identification of Rhythmically Expressed LncRNAs in the Zebrafish Pineal Gland and Testis. Int. J. Mol. Sci. 2021, 22, 7810. [Google Scholar] [CrossRef]
- Zhong, Z.; Wang, M.; Huang, G.; Zhang, S.; Wang, H. Molecular Genetic and Genomic Analyses of Zebrafish Circadian Rhythmicity; Springer: New Delhi, India, 2017; pp. 193–209. [Google Scholar] [CrossRef]
- Huang, G.; Zhang, F.; Ye, Q.; Wang, H. The circadian clock regulates autophagy directly through the nuclear hormone receptor Nr1d1/Reverbα and indirectly via Cebpb/(C/ebpβ) in zebrafish. Autophagy 2016, 2, 1292–1309. [Google Scholar] [CrossRef] [Green Version]
- Huang, J.; Zhong, Z.; Wang, M.; Chen, X.; Tan, Y.; Zhang, S.; He, W.; He, X.; Huang, G.; Lu, H.; et al. Circadian Modulation of Dopamine Levels and Dopaminergic Neuron Development Contributes to Attention Deficiency and Hyperactive Behavior. J. Neurosci. 2015, 35, 2572–2587. [Google Scholar] [CrossRef] [Green Version]
- Wang, M.; Zhong, Z.; Zhong, Y.; Zhang, W.; Wang, H. The Zebrafish Period2 Protein Positively Regulates the Circadian Clock through Mediation of Retinoic Acid Receptor (RAR)-related Orphan Receptor α (Rorα). J. Biol. Chem. 2015, 290, 4367–4382. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.; Zhou, Q.; Kesinger, J.W.; Norris, C.; Valdez, C. Heme Regulates Exocrine Peptidase Precursor Genes in Zebrafish. Exp. Biol. Med. 2007, 232, 1170–1180. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, H.; Kesinger, J.W.; Zhou, Q.; Wren, J.D.; Martin, G.; Turner, S.; Tang, Y.; Frank, M.B.; Centola, M. Identification and characterization of zebrafish ocular formation genes. Genome 2008, 51, 222–235. [Google Scholar] [CrossRef]
- Kelua, J.J.; Pipaliaa, T.G.; Hughes, S.M. Circadian regulation of muscle growth independent of locomotor activity. Proc. Natl. Acad. Sci. USA 2020, 117, 31208–31218. [Google Scholar] [CrossRef] [PubMed]
- The FANTOM Consortium; Carninci, P.; Kasukawa, T.; Katayama, S.; Gough, J.; Frith, M.C.; Maeda, N.; Oyama, R.; Ravasi, T.; Lenhard, B.; et al. The Transcriptional Landscape of the Mammalian Genome. Science 2005, 309, 1559–1563. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cui, M.; Zheng, M.; Sun, B.; Wang, Y.; Ye, L.; Zhang, X. A Long Noncoding RNA Perturbs the Circadian Rhythm of Hepatoma Cells to Facilitate Hepatocarcinogenesis. Neoplasia 2015, 17, 79–88. [Google Scholar] [CrossRef] [Green Version]
- Fan, Z.; Zhao, M.; Joshi, P.D.; Li, P.; Zhang, Y.; Guo, W.; Xu, Y.; Wang, H.; Zhao, Z.; Yan, J. A class of circadian long non-coding RNAs mark enhancers modulating long-range circadian gene regulation. Nucleic Acids Res. 2017, 45, 5720–5738. [Google Scholar] [CrossRef] [Green Version]
- Coon, S.L.; Munson, P.J.; Cherukuri, P.F.; Sugden, D.; Rath, M.; Møller, M.; Clokie, S.; Fu, C.; Olanich, M.E.; Rangel, Z.; et al. Circadian changes in long noncoding RNAs in the pineal gland. Proc. Natl. Acad. Sci. USA 2012, 109, 13319–13324. [Google Scholar] [CrossRef] [Green Version]
- Bittman, E.L. Timing in the Testis. J. Biol. Rhythm. 2016, 31, 12–36. [Google Scholar] [CrossRef] [Green Version]
- Morse, D.; Cermakian, N.; Brancorsini, S.; Parvinen, M.; Sassone-Corsi, P. No Circadian Rhythms in Testis: Period1 Expression Is Clock Independent and Developmentally Regulated in the Mouse. Mol. Endocrinol. 2003, 17, 141–151. [Google Scholar] [CrossRef] [Green Version]
- Alvarez, J.D.; Sehgal, A. The Thymus Is Similar to the Testis in Its Pattern of Circadian Clock Gene Expression. J. Biol. Rhythm. 2005, 20, 111–121. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Wang, Y.; Wang, C.; Hu, J.-F.; Li, W. LncRNA Functions as a New Emerging Epigenetic Factor in Determining the Fate of Stem Cells. Front. Genet. 2020, 11, 277. [Google Scholar] [CrossRef] [PubMed]
- Zhao, T.; Xu, J.; Liu, L.; Bai, J.; Wang, L.; Xiao, Y.; Li, X.; Zhang, L. Computational identification of epigenetically regulated lncRNAs and their associated genes based on integrating genomic data. FEBS Lett. 2015, 589, 521–531. [Google Scholar] [CrossRef] [Green Version]
- Ørom, U.A.; Derrien, T.; Beringer, M.; Gumireddy, K.; Gardini, A.; Bussotti, G.; Lai, F.; Zytnicki, M.; Notredame, C.; Huang, Q.; et al. Long noncoding rnas with enhancer-like function in human cells. Cell 2010, 143, 46–58. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guttman, M.; Amit, I.; Garber, M.; French, C.; Lin, M.F.; Feldser, D.; Huarte, M.; Zuk, O.; Carey, B.W.; Cassady, J.P.; et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 2009, 458, 223–227. [Google Scholar] [CrossRef]
- Yunusov, D.; Anderson, L.; DaSilva, L.F.; Wysocka, J.; Ezashi, T.R.; Roberts, M.; Verjovski-Almeida, S. HIPSTR and thousands of lncRNAs are heterogeneously expressed in human embryos, primordial germ cells and stable cell lines. Sci. Rep. 2016, 6, 32753. [Google Scholar] [CrossRef] [Green Version]
- Derrien, T.; Johnson, R.; Bussotti, G.; Tanzer, A.; Djebali, S.; Tilgner, H.; Guernec, G.; Merkel, A.; Gonzalez, D.; Lagarde, J.; et al. The GENCODE v7 Catalogue of Human Long Non-Coding RNAs: Analysis of Their Structure, Evolution and Expression. Genome Res. 2012, 22, 1775–1789. [Google Scholar] [CrossRef] [Green Version]
- Hu, X.; Chen, W.; Li, J.; Huang, S.; Xu, X.; Zhang, X.; Xiang, S.; Liu, C. ZFLNC: A comprehensive and well-annotated database for zebrafish lncRNA. Database 2018, 2018, bay114. [Google Scholar] [CrossRef]
- Niu, L.; Lou, F.; Sun, Y.; Sun, L.; Cai, X.; Liu, Z.; Zhou, H.; Wang, H.; Wang, Z.; Bai, J.; et al. A micropeptide encoded by lncRNA MIR155HG suppresses autoimmune inflammation via modulating antigen presentation. Sci. Adv. 2020, 6, eaaz2059. [Google Scholar] [CrossRef]
- Lin, Y.; Xiao, M.; Chen, H.; Meng, Y.; Zhao, N.; Yang, L.; Tang, H.; Wang, J.; Liu, X.; Zhu, Y.; et al. A novel mitochondrial micropeptide MPM enhances mitochondrial respiratory activity and promotes myogenic differentiation. Cell Death Dis. 2019, 10, 528. [Google Scholar] [CrossRef]
- Sousa, M.E.; Farkas, M.H. Micropeptide. PLoS Genet. 2018, 14, e1007764. [Google Scholar] [CrossRef] [PubMed]
- Xing, J.; Liu, H.; Jiang, W.; Wang, L. LncRNA-Encoded Peptide: Functions and Predicting Methods. Front. Oncol. 2021, 10, 3071. [Google Scholar] [CrossRef] [PubMed]
- Reinier, A.B.; Jaé, N.; Holdt, L.; Dimmeler, S. Long Noncoding RNAs From Clinical Genetics to Therapeutic Targets? J. Am. Coll. Cardiol. 2016, 67, 1214–1226. [Google Scholar]
- Anderson, D.M.; Anderson, K.M.; Chang, C.-L.; Makarewich, C.A.; Nelson, B.R.; McAnally, J.R.; Kasaragod, P.; Shelton, J.M.; Liou, J.; Bassel-Duby, R.; et al. A Micropeptide Encoded by a Putative Long Noncoding RNA Regulates Muscle Performance. Cell 2015, 160, 595–606. [Google Scholar] [CrossRef] [Green Version]
- Li, X.L.; Pongor, L.; Tang, W.; Das, S.; Muys, B.R.; Jones, M.F.; Lazar, S.B.; Dangelmaier, A.E.; Hartford, C.C.; Grammatikakis, I.; et al. A small protein encoded by a putative lncRNA regulates apoptosis and tumorigenicity in human colorectal cancer cells. eLife 2020, 9, e53734. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wu, S.; Zhu, X.; Zhang, L.; Deng, J.; Li, F.; Guo, B.; Zhang, S.; Wu, R.; Zhang, Z.; et al. LncRNA-encoded polypeptide ASRPS inhibits triple-negative breast cancer angiogenesis. J. Exp. Med. 2019, 217, jem.20190950. [Google Scholar] [CrossRef]
- Anfossi, S.; Calin, G.A. When non-coding is not enough. J. Exp. Med. 2020, 217, jem.20192009. [Google Scholar] [CrossRef]
- Mishra, S.K.; Wang, H. Computational Analysis Predicts Hundreds of Coding lncRNAs in Zebrafish. Biology 2021, 10, 371. [Google Scholar] [CrossRef]
- Westerfield, M. The Zebrafish Book: A Guide for the Laboratory Use of Zebrafish (Brachydanio rerio), 2nd ed.; University of Oregon Press: Eugene, OR, USA, 1993; 300p. [Google Scholar]
- Wang, L.; Park, H.J.; Dasari, S.; Wang, S.; Kocher, J.-P.; Li, W. CPAT: Coding-Potential Assessment Tool using an alignment-free logistic regression model. Nucleic Acids Res. 2013, 41, e74. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.; Anafi, R.C.; Hughes, M.E.; Kornacker, K.; Hogenesch, J.B. MetaCycle: An integrated R package to evaluate periodicity in large scale data. Bioinformatics 2016, 32, 3351–3353. [Google Scholar] [CrossRef] [Green Version]
- Zieliński, T.; Moore, A.M.; Troup, E.; Halliday, K.; Millar, A.J. Strengths and Limitations of Period Estimation Methods for Circadian Data. PLoS ONE 2014, 9, e96462. [Google Scholar] [CrossRef] [Green Version]
- Leach, L.J.; Zhang, Z.; Lu, C.; Kearsey, M.J.; Luo, Z. The Role of Cis-Regulatory Motifs and Genetical Control of Expression in the Divergence of Yeast Duplicate Genes. Mol. Biol. Evol. 2007, 24, 2556–2565. [Google Scholar] [CrossRef] [PubMed]
- González-Barrios, M.; Fierro-González, J.C.; Krpelanova, E.; Mora-Lorca, J.A.; Pedrajas, J.R.; Peñate, X.; Chavez, S. Cis- and Trans-Regulatory Mechanisms of Gene Expression in the ASJ Sensory Neuron of Caenorhabditis elegans. Genetics 2015, 200, 123–134. [Google Scholar] [CrossRef] [Green Version]
- Barik, S. Molecular Interactions between Pathogens and the Circadian Clock. Int. J. Mol. Sci. 2019, 20, 5824. [Google Scholar] [CrossRef] [Green Version]
- Thiriet, M. Control of Cell Fate in the Circulatory and Ventilatory Systems. In Biomathematical and Biomechanical Modeling of the Circulatory and Ventilatory Systems; Springer: New York, NY, USA, 2012; Volume 2. [Google Scholar]
- Doherty, C.J.; Kay, S.A. Circadian Control of Global Gene Expression Patterns. Annu. Rev. Genet. 2010, 44, 419–444. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fang, G.; Bhardwaj, N.; Robilotto, R.; Gerstein, M.B. Getting Started in Gene Orthology and Functional Analysis. PLoS Comput. Biol. 2010, 6, e1000703. [Google Scholar] [CrossRef] [Green Version]
- Mudunuri, U.; Che, A.; Yi, M.; Stephens, R.M. bioDBnet: The biological database network. Bioinformatics 2009, 25, 555–556. [Google Scholar] [CrossRef] [Green Version]
- Grant, C.E.; Bailey, T.L.; Noble, W.S. FIMO: Scanning for occurrences of a given motif. Bioinformatics 2011, 27, 1017–1018. [Google Scholar] [CrossRef] [Green Version]
- Fornes, O.; Castro-Mondragon, J.A.; Khan, A.; Van Der Lee, R.; Zhang, X.; Richmond, A.P.; Modi, B.; Correard, S.; Gheorghe, M.; Baranašić, D.; et al. JASPAR 2020: Update of the open-access database of transcription factor binding profiles. Nucleic Acids Res. 2019, 48, D87–D92. [Google Scholar] [CrossRef]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene Ontology: Tool for the unification of biology. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef] [Green Version]
- Galperin, M.; Kristensen, D.M.; Makarova, K.S.; Wolf, Y.; Koonin, E.V. Microbial genome analysis: The COG approach. Briefings Bioinform. 2017, 20, 1063–1070. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Götz, S.; García-Gómez, J.M.; Terol, J.; Williams, T.D.; Nagaraj, S.H.; Nueda, M.J.; Robles, M.; Talón, M.; Dopazo, J.; Conesa, A. High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res. 2008, 36, 3420–3435. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID Bioinformatics Resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef] [PubMed]
- Lever, J.; Krzywinski, M.; Altman, N. Principal component analysis. Nat. Methods 2017, 14, 641–642. [Google Scholar] [CrossRef] [Green Version]
- Gasteiger, E.; Gattiker, A.; Hoogland, C.; Ivanyi, I.; Ron, D.A.; Bairoch, A. ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003, 31, 3784–3788. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, C.-C.; Hwang, J.-K.; Yang, J.-M. (PS)2: Protein structure prediction server. Nucleic Acids Res. 2006, 34, W152–W157. [Google Scholar] [CrossRef] [Green Version]
- Chen, C.-C.; Hwang, J.-K.; Yang, J.-M. (PS)2-v2: Template-based protein structure prediction server. BMC Bioinform. 2009, 10, 366. [Google Scholar] [CrossRef] [Green Version]
- Echeazarra, L.; Hortigón-Vinagre, M.P.; Casis, O.; Gallego, M. Adult and Developing Zebrafish as Suitable Models for Cardiac Electrophysiology and Pathology in Research and Industry. Front. Physiol. 2021, 11, 607860. [Google Scholar] [CrossRef] [PubMed]
- Singleman, C.; Holtzman, N.G. Growth and Maturation in the Zebrafish, Danio Rerio: A Staging Tool for Teaching and Research. Zebrafish 2014, 11, 396–406. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, Y.; Liu, N.; Wang, J.P.; Wang, Y.Q.; Yu, X.L.; Wang, Z.B.; Cheng, X.C.; Zou, Q. Regulatory long non-coding rna and its functions. J. Physiol. Biochem. 2012, 68, 611–618. [Google Scholar] [CrossRef]
- Tovin, A.; Alon, S.; Ben-Moshe, Z.; Mracek, P.; Vatine, G.; Foulkes, N.S.; Jacob-Hirsch, J.; Rechavi, G.; Toyama, R.; Coon, S.L.; et al. Systematic Identification of Rhythmic Genes Reveals camk1gb as a New Element in the Circadian Clockwork. PLoS Genet. 2012, 8, e1003116. [Google Scholar] [CrossRef]
- Ye, M.; Zhang, J.; Wei, M.; Liu, B.; Dong, K. Emerging role of long noncoding RNA-encoded micropeptides in cancer. Cancer Cell Int. 2020, 20, 506. [Google Scholar] [CrossRef]
- Ozsolak, F.; Milos, P.M. RNA sequencing: Advances, challenges and opportunities. Nat. Rev. Genet. 2011, 12, 87–98. [Google Scholar] [CrossRef] [PubMed]
- Yu, F.; Zhang, Y.; Cheng, C.; Wang, W.; Zhou, Z.; Rang, W.; Yu, H.; Wei, Y.; Wu, Q.; Zhang, Y. Poly(A)-seq: A method for direct sequencing and analysis of the transcriptomic poly(A)-tails. PLoS ONE 2020, 15, e0234696. [Google Scholar] [CrossRef] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mishra, S.K.; Zhong, Z.; Wang, H. Hundreds of LncRNAs Display Circadian Rhythmicity in Zebrafish Larvae. Cells 2021, 10, 3173. https://doi.org/10.3390/cells10113173
Mishra SK, Zhong Z, Wang H. Hundreds of LncRNAs Display Circadian Rhythmicity in Zebrafish Larvae. Cells. 2021; 10(11):3173. https://doi.org/10.3390/cells10113173
Chicago/Turabian StyleMishra, Shital Kumar, Zhaomin Zhong, and Han Wang. 2021. "Hundreds of LncRNAs Display Circadian Rhythmicity in Zebrafish Larvae" Cells 10, no. 11: 3173. https://doi.org/10.3390/cells10113173
APA StyleMishra, S. K., Zhong, Z., & Wang, H. (2021). Hundreds of LncRNAs Display Circadian Rhythmicity in Zebrafish Larvae. Cells, 10(11), 3173. https://doi.org/10.3390/cells10113173





