The SC-35 Splicing Factor Interacts with RNA Pol II and A-Type Lamin Depletion Weakens This Interaction
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Cultivation and Treatment
2.2. Immunofluorescence Staining
2.3. Western Blot
2.4. Laser Scanning Confocal Microscopy
2.5. FLIM-FRET Technique
2.6. FRAP Analysis
2.7. Statistical Analysis
3. Results
3.1. The SC-35 Protein Was Localized in Nuclear Blebs, and Inhibitors of RNA Polymerases Changed Its Level
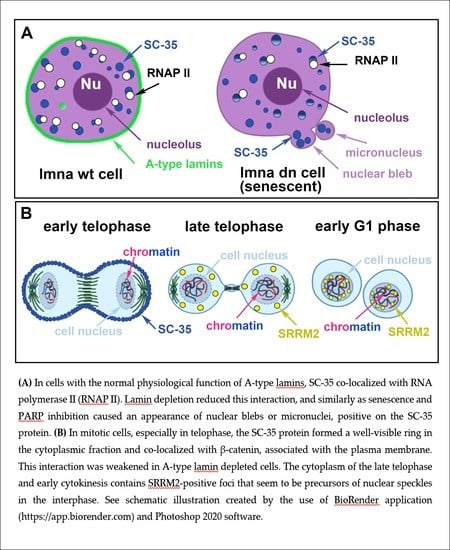
3.2. The Highest Level of SC-35 Was Accompanied by Depletion of A-Type Lamins in Distinct Cell Types
3.3. A Spatial Link of SC-35 Positive Nuclear Speckles to DNA Repair Proteins and Nucleoli
3.4. The SC-35 Protein Decorates the Plasma Membrane in Mitotic Cells and the Degree of Its Colocalization with PCNA Is Enhanced in the Late S-phase of the Cell Cycle
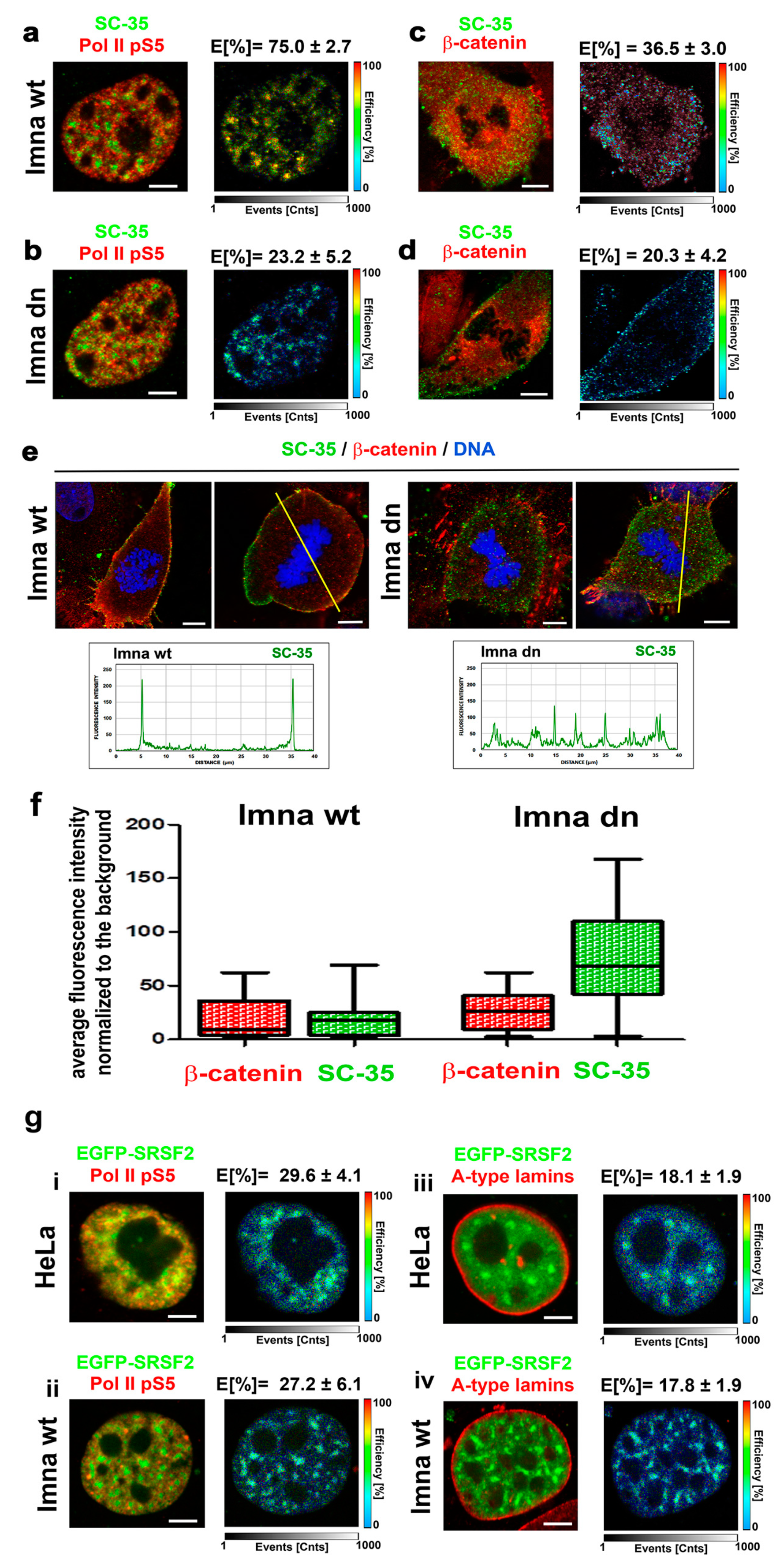
3.5. A-Type Lamin-Dependent Interaction between the SC-35 Protein and RNA Polymerase II in Interphase Cells, or SC-35 and β-catenin Interaction in Mitosis
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Misteli, T.; Spector, D.L. RNA polymerase II targets pre-mRNA splicing factors to transcription sites in vivo. Mol. Cell 1999, 3, 697–705. [Google Scholar] [CrossRef]
- Misteli, T.; Caceres, J.F.; Spector, D.L. The dynamics of a pre-mRNA splicing factor in living cells. Nature 1997, 387, 523–527. [Google Scholar] [CrossRef] [PubMed]
- Fakan, S. Perichromatin fibrils are in situ forms of nascent transcripts. Trends Cell Biol. 1994, 4, 86–90. [Google Scholar] [CrossRef]
- Lamond, A.I.; Spector, D.L. Nuclear speckles: A model for nuclear organelles. Nat. Rev. Mol. Cell Biol. 2003, 4, 605–612. [Google Scholar] [CrossRef] [PubMed]
- Spector, D.L. Nuclear organization of pre-mRNA processing. Curr. Opin. Cell Biol. 1993, 5, 442–447. [Google Scholar] [CrossRef]
- Prasanth, K.V.; Sacco-Bubulya, P.A.; Prasanth, S.G.; Spector, D.L. Sequential entry of components of the gene expression machinery into daughter nuclei. Mol. Biol. Cell 2003, 14, 1043–1057. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, K.; Parnaik, V.K. Differential dynamics of splicing factor SC35 during the cell cycle. J. Biosci. 2008, 33, 345–354. [Google Scholar] [CrossRef]
- Dorn, R.; Reuter, G.; Loewendorf, A. Transgene analysis proves mRNA trans-splicing at the complex mod(mdg4) locus in Drosophila. Proc. Natl. Acad. Sci. USA 2001, 98, 9724–9729. [Google Scholar] [CrossRef]
- Horiuchi, T.; Giniger, E.; Aigaki, T. Alternative trans-splicing of constant and variable exons of a Drosophila axon guidance gene, lola. Genes Dev. 2003, 17, 2496–2501. [Google Scholar] [CrossRef]
- McManus, C.J.; Duff, M.O.; Eipper-Mains, J.; Graveley, B.R. Global analysis of trans-splicing in Drosophila. Proc. Natl. Acad. Sci. USA 2010, 107, 12975–12979. [Google Scholar] [CrossRef]
- Li, H.; Wang, J.; Ma, X.; Sklar, J. Gene fusions and RNA trans-splicing in normal and neoplastic human cells. Cell Cycle 2009, 8, 218–222. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.; Heere-Ress, E.; Boucher, B.; Defesche, J.C.; Kastelein, J.; Lavoie, M.A.; Genest, J., Jr. Familial hypercholesterolemia. Acceptor splice site (G-->C) mutation in intron 7 of the LDL-R gene: Alternate RNA editing causes exon 8 skipping or a premature stop codon in exon 8. LDL-R(Honduras-1) [LDL-R1061(-1) G-->C]. Atherosclerosis 1999, 146, 125–131. [Google Scholar] [CrossRef]
- Reid, D.W.; Nicchitta, C.V. Diversity and selectivity in mRNA translation on the endoplasmic reticulum. Nat. Rev. Mol. Cell Biol. 2015, 16, 221–231. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Liu, J.; Huang, B.O.; Xu, Y.M.; Li, J.; Huang, L.F.; Lin, J.; Zhang, J.; Min, Q.H.; Yang, W.M.; et al. Mechanism of alternative splicing and its regulation. Biomed. Rep. 2015, 3, 152–158. [Google Scholar] [CrossRef] [PubMed]
- Luco, R.F.; Pan, Q.; Tominaga, K.; Blencowe, B.J.; Pereira-Smith, O.M.; Misteli, T. Regulation of alternative splicing by histone modifications. Science 2010, 327, 996–1000. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.Y.; Zhu, Y.R.; Dai, D.J.; Wang, X.; Jin, H.C. Epigenetic regulation of alternative splicing. Am. J. Cancer Res. 2018, 8, 2346–2358. [Google Scholar]
- Luco, R.F.; Misteli, T. More than a splicing code: Integrating the role of RNA, chromatin and non-coding RNA in alternative splicing regulation. Curr. Opin. Genet. Dev. 2011, 21, 366–372. [Google Scholar] [CrossRef]
- Adhikari, S.; Xiao, W.; Zhao, Y.L.; Yang, Y.G. m(6)A: Signaling for mRNA splicing. RNA Biol. 2016, 13, 756–759. [Google Scholar] [CrossRef]
- Kim, M.; Patel, B.; Schroeder, K.E.; Raza, A.; Dejong, J. Organization and transcriptional output of a novel mRNA-like piRNA gene (mpiR) located on mouse chromosome 10. RNA 2008, 14, 1005–1011. [Google Scholar] [CrossRef][Green Version]
- Gruenbaum, Y.; Foisner, R. Lamins: Nuclear intermediate filament proteins with fundamental functions in nuclear mechanics and genome regulation. Annu. Rev. Biochem. 2015, 84, 131–164. [Google Scholar] [CrossRef]
- Ilik, I.A.; Malszycki, M.; Lubke, A.K.; Schade, C.; Meierhofer, D.; Aktas, T. SON and SRRM2 are essential for nuclear speckle formation. eLife 2020, 9. [Google Scholar] [CrossRef] [PubMed]
- Rein, I.D.; Landsverk, K.S.; Micci, F.; Patzke, S.; Stokke, T. Replication-induced DNA damage after PARP inhibition causes G2 delay, and cell line-dependent apoptosis, necrosis and multinucleation. Cell Cycle 2015, 14, 3248–3260. [Google Scholar] [CrossRef] [PubMed]
- Singh, M.; Hunt, C.R.; Pandita, R.K.; Kumar, R.; Yang, C.R.; Horikoshi, N.; Bachoo, R.; Serag, S.; Story, M.D.; Shay, J.W.; et al. Lamin A/C depletion enhances DNA damage-induced stalled replication fork arrest. Mol. Cell Biol. 2013, 33, 1210–1222. [Google Scholar] [CrossRef] [PubMed]
- Bartova, E.; Malyskova, B.; Komurkova, D.; Legartova, S.; Suchankova, J.; Krejci, J.; Kozubek, S. Function of heterochromatin protein 1 during DNA repair. Protoplasma 2017, 254, 1233–1240. [Google Scholar] [CrossRef] [PubMed]
- Essers, J.; Theil, A.F.; Baldeyron, C.; van Cappellen, W.A.; Houtsmuller, A.B.; Kanaar, R.; Vermeulen, W. Nuclear dynamics of PCNA in DNA replication and repair. Mol. Cell Biol. 2005, 25, 9350–9359. [Google Scholar] [CrossRef]
- Stixova, L.; Sehnalova, P.; Legartova, S.; Suchankova, J.; Hruskova, T.; Kozubek, S.; Sorokin, D.V.; Matula, P.; Raska, I.; Kovarik, A.; et al. HP1beta-dependent recruitment of UBF1 to irradiated chromatin occurs simultaneously with CPDs. Epigenetics Chromatin 2014, 7, 39. [Google Scholar] [CrossRef]
- McCrea, P.D.; Turck, C.W.; Gumbiner, B. A homolog of the armadillo protein in Drosophila (plakoglobin) associated with E-cadherin. Science 1991, 254, 1359–1361. [Google Scholar] [CrossRef]
- Sullivan, T.; Escalante-Alcalde, D.; Bhatt, H.; Anver, M.; Bhat, N.; Nagashima, K.; Stewart, C.L.; Burke, B. Loss of A-type lamin expression compromises nuclear envelope integrity leading to muscular dystrophy. J. Cell Biol. 1999, 147, 913–920. [Google Scholar] [CrossRef]
- Stixova, L.; Matula, P.; Kozubek, S.; Gombitova, A.; Cmarko, D.; Raska, I.; Bartova, E. Trajectories and nuclear arrangement of PML bodies are influenced by A-type lamin deficiency. Biol. Cell 2012, 104, 418–432. [Google Scholar] [CrossRef]
- Bartova, E.; Pachernik, J.; Harnicarova, A.; Kovarik, A.; Kovarikova, M.; Hofmanova, J.; Skalnikova, M.; Kozubek, M.; Kozubek, S. Nuclear levels and patterns of histone H3 modification and HP1 proteins after inhibition of histone deacetylases. J. Cell Sci. 2005, 118, 5035–5046. [Google Scholar] [CrossRef]
- Bartova, E.; Krejci, J.; Harnicarova, A.; Kozubek, S. Differentiation of human embryonic stem cells induces condensation of chromosome territories and formation of heterochromatin protein 1 foci. Differentiation 2008, 76, 24–32. [Google Scholar] [CrossRef] [PubMed]
- Bartova, E.; Harnicarova, A.; Pachernik, J.; Kozubek, S. Nuclear topography and expression of the BCR/ABL fusion gene and its protein level influenced by cell differentiation and RNA interference. Leuk. Res. 2005, 29, 901–913. [Google Scholar] [CrossRef] [PubMed]
- Lukasova, E.; Kovarˇík, A.; Bac ikova, A.; Falk, M.; Kozubek, S. Loss of lamin B receptor is necessary to induce cellular senescence. Biochem. J. 2017, 474, 281–300. [Google Scholar] [CrossRef] [PubMed]
- Krejci, J.; Harnicarova, A.; Kurova, J.; Uhlirova, R.; Kozubek, S.; Legartova, S.; Hajek, R.; Bartova, E. Nuclear organization of PML bodies in leukaemic and multiple myeloma cells. Leuk. Res. 2008, 32, 1866–1877. [Google Scholar] [CrossRef] [PubMed]
- Ismail, I.H.; Andrin, C.; McDonald, D.; Hendzel, M.J. BMI1-mediated histone ubiquitylation promotes DNA double-strand break repair. J. Cell Biol. 2010, 191, 45–60. [Google Scholar] [CrossRef]
- Svobodova Kovarikova, A.; Stixova, L.; Kovarik, A.; Komurkova, D.; Legartova, S.; Fagherazzi, P.; Bartova, E. N(6)-Adenosine Methylation in RNA and a Reduced m3G/TMG Level in Non-Coding RNAs Appear at Microirradiation-Induced DNA Lesions. Cells 2020, 9, 360. [Google Scholar] [CrossRef]
- Suchankova, J.; Legartova, S.; Ruckova, E.; Vojtesek, B.; Kozubek, S.; Bartova, E. Mutations in the TP53 gene affected recruitment of 53BP1 protein to DNA lesions, but level of 53BP1 was stable after gamma-irradiation that depleted MDC1 protein in specific TP53 mutants. Histochem. Cell Biol. 2017, 148, 239–255. [Google Scholar] [CrossRef]
- Dundr, M.; Hoffmann-Rohrer, U.; Hu, Q.; Grummt, I.; Rothblum, L.I.; Phair, R.D.; Misteli, T. A kinetic framework for a mammalian RNA polymerase in vivo. Science 2002, 298, 1623–1626. [Google Scholar] [CrossRef]
- Bartova, E.; Sustackova, G.; Stixova, L.; Kozubek, S.; Legartova, S.; Foltankova, V. Recruitment of Oct4 protein to UV-damaged chromatin in embryonic stem cells. PLoS ONE 2011, 6, e27281. [Google Scholar] [CrossRef]
- Eriksson, S.; Kim, S.K.; Kubista, M.; Norden, B. Binding of 4’,6-diamidino-2-phenylindole (DAPI) to AT regions of DNA: Evidence for an allosteric conformational change. Biochemistry 1993, 32, 2987–2998. [Google Scholar] [CrossRef]
- Legartova, S.; Jugova, A.; Stixova, L.; Kozubek, S.; Fojtova, M.; Zdrahal, Z.; Lochmanova, G.; Bartova, E. Epigenetic aspects of HP1 exchange kinetics in apoptotic chromatin. Biochimie 2013, 95, 167–179. [Google Scholar] [CrossRef] [PubMed]
- Legartova, S.; Lochmanova, G.; Zdrahal, Z.; Kozubek, S.; Sponer, J.; Krepl, M.; Pokorna, P.; Bartova, E. DNA Damage Changes Distribution Pattern and Levels of HP1 Protein Isoforms in the Nucleolus and Increases Phosphorylation of HP1beta-Ser88. Cells 2019, 8, 1097. [Google Scholar] [CrossRef] [PubMed]
- Edmond, V.; Moysan, E.; Khochbin, S.; Matthias, P.; Brambilla, C.; Brambilla, E.; Gazzeri, S.; Eymin, B. Acetylation and phosphorylation of SRSF2 control cell fate decision in response to cisplatin. EMBO J. 2011, 30, 510–523. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Alibhai, D.; Margineanu, A.; Laine, R.; Kennedy, G.; McGinty, J.; Warren, S.; Kelly, D.; Alexandrov, Y.; Munro, I.; et al. FLIM FRET technology for drug discovery: Automated multiwell-plate high-content analysis, multiplexed readouts and application in situ. Chemphyschem 2011, 12, 609–626. [Google Scholar] [CrossRef] [PubMed]
- Lakowicz, J.R.; Gryczynski, I.I.; Gryczynski, Z. High Throughput Screening with Multiphoton Excitation. J. Biomol. Screen 1999, 4, 355–362. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Sillen, A.; Engelborghs, Y. The correct use of “average” fluorescence parameters. Photochem. Photobiol. 1998, 67, 475–486. [Google Scholar] [CrossRef]
- Daubner, G.M.; Clery, A.; Jayne, S.; Stevenin, J.; Allain, F.H. A syn-anti conformational difference allows SRSF2 to recognize guanines and cytosines equally well. EMBO J. 2012, 31, 162–174. [Google Scholar] [CrossRef]
- Legartova, S.; Sehnalova, P.; Malyskova, B.; Kuntziger, T.; Collas, P.; Cmarko, D.; Raska, I.; Sorokin, D.V.; Kozubek, S.; Bartova, E. Localized Movement and Levels of 53BP1 Protein Are Changed by gamma-irradiation in PML Deficient Cells. J. Cell Biochem. 2016, 117, 2583–2596. [Google Scholar] [CrossRef] [PubMed]
- Bubulya, P.A.; Prasanth, K.V.; Deerinck, T.J.; Gerlich, D.; Beaudouin, J.; Ellisman, M.H.; Ellenberg, J.; Spector, D.L. Hypophosphorylated SR splicing factors transiently localize around active nucleolar organizing regions in telophase daughter nuclei. J. Cell Biol. 2004, 167, 51–63. [Google Scholar] [CrossRef] [PubMed]
- Spector, D.L.; Lamond, A.I. Nuclear speckles. Cold Spring Harb. Perspect. Biol. 2011, 3. [Google Scholar] [CrossRef]
- Galganski, L.; Urbanek, M.O.; Krzyzosiak, W.J. Nuclear speckles: Molecular organization, biological function and role in disease. Nucleic Acids Res. 2017, 45, 10350–10368. [Google Scholar] [CrossRef] [PubMed]
- Fu, X.D.; Maniatis, T. The 35-kDa mammalian splicing factor SC35 mediates specific interactions between U1 and U2 small nuclear ribonucleoprotein particles at the 3′ splice site. Proc. Natl. Acad. Sci. USA 1992, 89, 1725–1729. [Google Scholar] [CrossRef] [PubMed]
- Neugebauer, K.M. On the importance of being co-transcriptional. J. Cell Sci. 2002, 115, 3865–3871. [Google Scholar] [CrossRef]
- Shimi, T.; Pfleghaar, K.; Kojima, S.; Pack, C.G.; Solovei, I.; Goldman, A.E.; Adam, S.A.; Shumaker, D.K.; Kinjo, M.; Cremer, T.; et al. The A- and B-type nuclear lamin networks: Microdomains involved in chromatin organization and transcription. Genes Dev. 2008, 22, 3409–3421. [Google Scholar] [CrossRef] [PubMed]
- Bercht Pfleghaar, K.; Taimen, P.; Butin-Israeli, V.; Shimi, T.; Langer-Freitag, S.; Markaki, Y.; Goldman, A.E.; Wehnert, M.; Goldman, R.D. Gene-rich chromosomal regions are preferentially localized in the lamin B deficient nuclear blebs of atypical progeria cells. Nucleus 2015, 6, 66–76. [Google Scholar] [CrossRef] [PubMed]
- Funkhouser, C.M.; Sknepnek, R.; Shimi, T.; Goldman, A.E.; Goldman, R.D.; Olvera de la Cruz, M. Mechanical model of blebbing in nuclear lamin meshworks. Proc. Natl. Acad. Sci. USA 2013, 110, 3248–3253. [Google Scholar] [CrossRef]
- Gonzalez-Suarez, I.; Redwood, A.B.; Gonzalo, S. Loss of A-type lamins and genomic instability. Cell Cycle 2009, 8, 3860–3865. [Google Scholar] [CrossRef]
- Caruso, R.A.; Fedele, F.; Crisafulli, C.; Paparo, D.; Parisi, A.; Luciano, R.; Cavallari, V. Abnormal nuclear structures (micronuclei, nuclear blebs, strings, and pockets) in a case of anaplastic giant cell carcinoma of the thyroid: An immunohistochemical and ultrastructural study. Ultrastruct. Pathol. 2011, 35, 14–18. [Google Scholar] [CrossRef]
- Utani, K.; Okamoto, A.; Shimizu, N. Generations of micronuclei during interphase by coupling between cytoplasmic membrane blebbing and nuclear budding. PLoS ONE 2011, 6, e27233. [Google Scholar] [CrossRef]
- Rai, A.K.; Chen, J.X.; Selbach, M.; Pelkmans, L. Kinase-controlled phase transition of membraneless organelles in mitosis. Nature 2018, 559, 211–216. [Google Scholar] [CrossRef]
- Dundr, M.; Misteli, T. Biogenesis of nuclear bodies. Cold Spring Harb. Perspect. Biol. 2010, 2, a000711. [Google Scholar] [CrossRef] [PubMed]








| Cell Line | Cell Culture Medium | Reference |
|---|---|---|
| A549 (human lung cancer) (#ATCC® CCL-185™, Germany) | DMEM medium supplemented with 10% FCS | [30] |
| HL60 (human acute promyelocytic leukemia) (#ATCC® CCL-240™, Germany) | IMDM medium supplemented with 10% FCS | [32] |
| MCF7 (human adenocarcinoma) (#ATCC® HTB-22™, Germany) | EMEM medium supplemented with 0.01 mg/mL human recombinant insulin and 10% FCS | [33] |
| MOLP8 (human multiple myeloma) (#ACC 569, DSMZ, Germany) | RPMI 1640 medium supplemented with 20% FCS | [34] |
| U2OS (human osteosarcoma) collaboration with the Institute of Biology and Medical Genetics, Charles University in Prague) | DMEM medium supplemented with 10% FCS | [35] |
| U937 (human histiocytic lymphoma) (#ATCC® CRL-1593.2™, Germany) | RPMI 1640 medium supplemented with 10% FCS | [34] |
| HaCaT (human keratinocytes) (#300493, CLS, Germany) | DMEM medium supplemented with 10% FCS | [36] |
| IMR90 (human lung fibroblast) (#ATCC® CCL-186™, Germany) | EMEM medium supplemented with 10% FCS, 1% non-essential amino acids (NEAA), and 2mM glutamine | [37] |
| QYCy3 (DONOR) | ECCy5(M−1cm−1) | QYCy5 (ACCEPTOR) | J(λ) (*1 × 1015 M−1cm−1nm4) | R0(Å) | R0 × QYCy5 |
| 0.15 | 250,000 | 0.30 | 7.6 | 49.73 | 14.92 |
| QYEGFP (DONOR) | EC AF594 * (M−1cm−1) | QYAF594 (ACCEPTOR) | J(λ) (*1 × 1015 M−1cm−1nm4) | R0(Å) | R0 × QYAF594 |
| 0.60 | 92,000 | 0.66 | 1.80 | 49.30 | 32.54 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Legartová, S.; Fagherazzi, P.; Stixová, L.; Kovařík, A.; Raška, I.; Bártová, E. The SC-35 Splicing Factor Interacts with RNA Pol II and A-Type Lamin Depletion Weakens This Interaction. Cells 2021, 10, 297. https://doi.org/10.3390/cells10020297
Legartová S, Fagherazzi P, Stixová L, Kovařík A, Raška I, Bártová E. The SC-35 Splicing Factor Interacts with RNA Pol II and A-Type Lamin Depletion Weakens This Interaction. Cells. 2021; 10(2):297. https://doi.org/10.3390/cells10020297
Chicago/Turabian StyleLegartová, Soňa, Paolo Fagherazzi, Lenka Stixová, Aleš Kovařík, Ivan Raška, and Eva Bártová. 2021. "The SC-35 Splicing Factor Interacts with RNA Pol II and A-Type Lamin Depletion Weakens This Interaction" Cells 10, no. 2: 297. https://doi.org/10.3390/cells10020297
APA StyleLegartová, S., Fagherazzi, P., Stixová, L., Kovařík, A., Raška, I., & Bártová, E. (2021). The SC-35 Splicing Factor Interacts with RNA Pol II and A-Type Lamin Depletion Weakens This Interaction. Cells, 10(2), 297. https://doi.org/10.3390/cells10020297