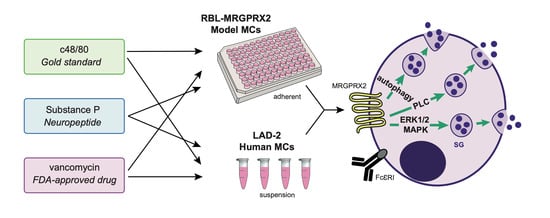
Authentic and Ectopically Expressed MRGPRX2 Elicit Similar Mechanisms to Stimulate Degranulation of Mast Cells
Abstract
:1. Introduction
2. Materials and Methods
2.1. Antibodies and Reagents
2.2. Cell Culture
2.3. Cell Transfection
2.4. Isolation of Mouse Peritoneal MCs
2.5. Mast Cell Activation
2.6. β-Hexosaminidase Release Assay
2.7. Histamine Release Assay
2.8. Flow Cytometry Analysis
2.9. Immunostaining and Laser Confocal Microscopy Analysis
2.10. Western Blot Analyses
2.11. RNA Purification and Quantitative Real-Time PCR
2.12. Statistical Analysis
3. Results
3.1. Dose and Time Dependence of Endogenous and Ectopic MRGPRX2 Responses
3.2. ERK1/2 Signaling Is Linked with Secretion in LAD-2 and RBL-MRGPRX2 Cells
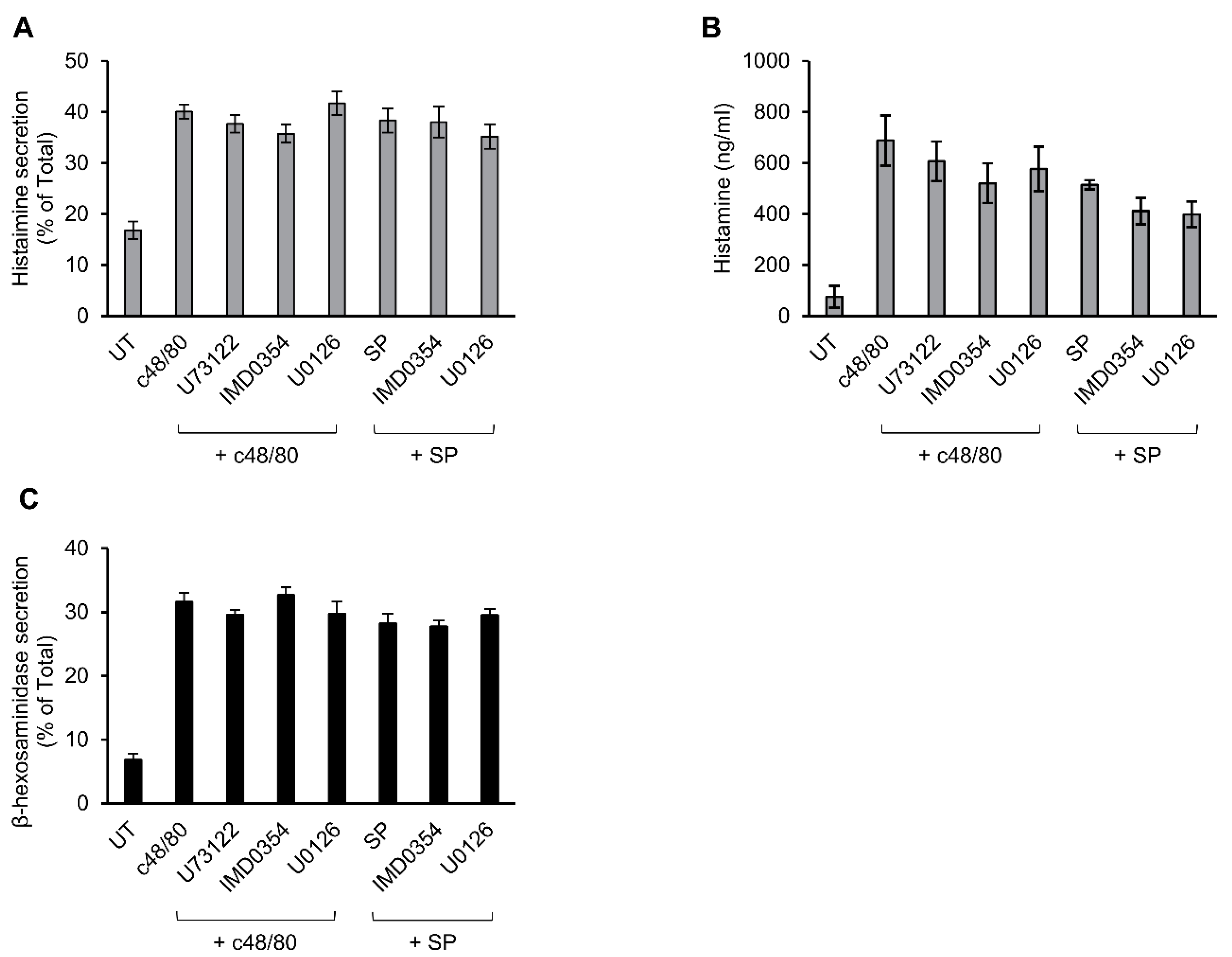
3.3. Inhibitor Profiling of MRGPRX2-Induced Secretion from LAD-2 and RBL-MRGPRX2 Cells
3.4. Inhibitor Profiling of MRGPRX2-Stimulated ERK1/2 Phosphorylation in LAD-2 and RBL-MRGPRX2 Cells
3.5. Secretion from Murine Peritoneal MCs Is Resistant to Inhibitors That Abrogate MRGPRX2-stimulated Secretion
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Shelburne, C.P.; Abraham, S.N. The mast cell in innate and adaptive immunity. Adv. Exp. Med. Biol. 2011, 716, 162–185. [Google Scholar] [CrossRef] [PubMed]
- Agier, J.; Pastwinska, J.; Brzezinska-Blaszczyk, E. An overview of mast cell pattern recognition receptors. Inflamm. Res. Off. J. Eur. Histamine Res. Soc. 2018. [Google Scholar] [CrossRef] [Green Version]
- Redegeld, F.A.; Yu, Y.; Kumari, S.; Charles, N.; Blank, U. Non-IgE mediated mast cell activation. Immunol. Rev. 2018, 282, 87–113. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, H.; Gupta, K.; Lee, D.; Bayir, A.K.; Ahn, H.; Ali, H. beta-Defensins activate human mast cells via Mas-related gene X2. J. Immunol. 2013, 191, 345–352. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lagunoff, D.; Martin, T.W.; Read, G. Agents that release histamine from mast cells. Annu. Rev. Pharmacol. Toxicol. 1983, 23, 331–351. [Google Scholar] [CrossRef] [PubMed]
- Ferry, X.; Brehin, S.; Kamel, R.; Landry, Y. G protein-dependent activation of mast cell by peptides and basic secretagogues. Peptides 2002, 23, 1507–1515. [Google Scholar] [CrossRef]
- McNeil, B.D.; Pundir, P.; Meeker, S.; Han, L.; Undem, B.J.; Kulka, M.; Dong, X. Identification of a mast-cell-specific receptor crucial for pseudo-allergic drug reactions. Nature 2015, 519, 237–241. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mousli, M.; Bronner, C.; Bueb, J.L.; Tschirhart, E.; Gies, J.P.; Landry, Y. Activation of rat peritoneal mast cells by substance P and mastoparan. J. Pharmacol. Exp. Ther. 1989, 250, 329–335. [Google Scholar] [PubMed]
- Subramanian, H.; Gupta, K.; Ali, H. Roles of Mas-related G protein-coupled receptor X2 on mast cell-mediated host defense, pseudoallergic drug reactions, and chronic inflammatory diseases. J. Allergy Clin. Immunol. 2016, 138, 700–710. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alysandratos, K.-D.; Asadi, S.; Angelidou, A.; Zhang, B.; Sismanopoulos, N.; Yang, H.; Critchfield, A.; Theoharides, T.C. Neurotensin and CRH interactions augment human mast cell activation. PLoS ONE 2012, 7, e48934. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Holgate, S.T. The role of mast cells and basophils in inflammation. Clin. Exp. Allergy J. Br. Soc. Allergy Clin. Immunol. 2000, 30, 28–32. [Google Scholar] [CrossRef] [PubMed]
- Schwartz, L.B.; Austen, K.F. Enzymes of the mast cell granule. J. Investig. Dermatol. 1980, 74, 349–353. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moon, T.C.; Befus, A.D.; Kulka, M. Mast cell mediators: Their differential release and the secretory pathways involved. Front. Immunol. 2014, 5, 569. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gordon, J.R.; Galli, S.J. Mast cells as a source of both preformed and immunologically inducible TNF-alpha/cachectin. Nature 1990, 346, 274–276. [Google Scholar] [CrossRef] [PubMed]
- Kulinski, J.M.; Muñoz-Cano, R.; Olivera, A. Sphingosine-1-phosphate and other lipid mediators generated by mast cells as critical players in allergy and mast cell function. Eur. J. Pharmacol. 2016, 778, 56–67. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Theoharides, T.C.; Kandere, K. Mast cell involvement in neurogenic inflammation. In Migraine: A Neuroinflammatory Disease? Spierings, E.L.H., Sánchez del Río, M., Eds.; Birkhäuser Basel: Basel, Switzerland, 2002; pp. 115–132. [Google Scholar] [CrossRef]
- Kulka, M.; Sheen, C.H.; Tancowny, B.P.; Grammer, L.C.; Schleimer, R.P. Neuropeptides activate human mast cell degranulation and chemokine production. Immunology 2008, 123, 398–410. [Google Scholar] [CrossRef]
- Ali, H. Mas-related G protein coupled receptor-X2: A potential new target for modulating mast cell-mediated allergic and inflammatory diseases. J. Immunobiol. 2016, 1, 115. [Google Scholar] [CrossRef] [Green Version]
- Tatemoto, K.; Nozaki, Y.; Tsuda, R.; Konno, S.; Tomura, K.; Furuno, M.; Ogasawara, H.; Edamura, K.; Takagi, H.; Iwamura, H.; et al. Immunoglobulin E-independent activation of mast cell is mediated by Mrg receptors. Biochem. Biophys. Res. Commun. 2006, 349, 1322–1328. [Google Scholar] [CrossRef] [PubMed]
- Babina, M. The pseudo-allergic/neurogenic route of mast cell activation via MRGPRX2: Discovery, functional programs, regulation, relevance to disease, and relation with allergic stimulation. Itch 2020, 5, e32. [Google Scholar] [CrossRef]
- Subramanian, H.; Gupta, K.; Guo, Q.; Price, R.; Ali, H. Mas-related gene X2 (MrgX2) is a novel G protein-coupled receptor for the antimicrobial peptide LL-37 in human mast cells: Resistance to receptor phosphorylation, desensitization, and internalization. J. Biol. Chem. 2011, 286, 44739–44749. [Google Scholar] [CrossRef] [Green Version]
- Zylka, M.J.; Dong, X.; Southwell, A.L.; Anderson, D.J. Atypical expansion in mice of the sensory neuron-specific Mrg G protein-coupled receptor family. Proc. Natl. Acad. Sci. USA 2003, 100, 10043–10048. [Google Scholar] [CrossRef] [Green Version]
- van der Kleij, H.P.; Ma, D.; Redegeld, F.A.; Kraneveld, A.D.; Nijkamp, F.P.; Bienenstock, J. Functional expression of neurokinin 1 receptors on mast cells induced by IL-4 and stem cell factor. J. Immunol. 2003, 171, 2074–2079. [Google Scholar] [CrossRef] [Green Version]
- Subramanian, H.; Kashem, S.W.; Collington, S.J.; Qu, H.; Lambris, J.D.; Ali, H. PMX-53 as a dual CD88 antagonist and an agonist for Mas-related gene 2 (MrgX2) in human mast cells. Mol. Pharmacol. 2011, 79, 1005–1013. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Olszewski, M.B.; Groot, A.J.; Dastych, J.; Knol, E.F. TNF Trafficking to Human Mast Cell Granules: Mature Chain-Dependent Endocytosis. J. Immunol. 2007, 178, 5701–5709. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Klein, O.; Krier-Burris, R.A.; Lazki-Hagenbach, P.; Gorzalczany, Y.; Mei, Y.; Ji, P.; Bochner, B.S.; Sagi-Eisenberg, R. Mammalian diaphanous-related formin 1 (mDia1) coordinates mast cell migration and secretion through its actin-nucleating activity. J. Allergy Clin. Immunol. 2019, 144, 1074–1090. [Google Scholar] [CrossRef] [PubMed]
- Shefler, I.; Taube, Z.; Medalia, O.; Sagi-Eisenberg, R. Basic secretagogues activate protein tyrosine phosphorylation and release of arachidonic acid in mast cells via a novel protein kinase C and phosphatidylinositol 3-kinase-dependent mechanism. Eur. J. Immunol. 1998, 28, 3468–3478. [Google Scholar] [CrossRef]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef] [PubMed]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- BOLTE, S.; CORDELIÈRES, F.P. A guided tour into subcellular colocalization analysis in light microscopy. J. Microsc. 2006, 224, 213–232. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Spandidos, A.; Wang, X.; Wang, H.; Seed, B. PrimerBank: A resource of human and mouse PCR primer pairs for gene expression detection and quantification. Nucleic Acids Res. 2009, 38, D792–D799. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Spandidos, A.; Wang, X.; Wang, H.; Dragnev, S.; Thurber, T.; Seed, B. A comprehensive collection of experimentally validated primers for Polymerase Chain Reaction quantitation of murine transcript abundance. BMC Genom. 2008, 9, 633. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, X.; Seed, B. A PCR primer bank for quantitative gene expression analysis. Nucleic Acids Res. 2003, 31, e154. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sahid, M.N.A.; Liu, S.; Mogi, M.; Maeyama, K. Tachykinin-1 receptor antagonism suppresses substance-P- and compound 48/80-induced mast cell activation from rat mast cells expressing functional mas-related GPCR B3. Inflamm. Res. 2020, 69, 289–298. [Google Scholar] [CrossRef] [PubMed]
- Kiseljak-Vassiliades, K.; Xu, M.; Mills, T.S.; Smith, E.E.; Silveira, L.J.; Lillehei, K.O.; Kerr, J.M.; Kleinschmidt-DeMasters, B.K.; Wierman, M.E. Differential somatostatin receptor (SSTR) 1-5 expression and downstream effectors in histologic subtypes of growth hormone pituitary tumors. Mol. Cell. Endocrinol. 2015, 417, 73–83. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, E.K.; Song, J.; Seo, Y.; Koh, E.M.; Kim, S.-H.; Jung, K.J. Inhibitory Effects of AF-343, a Mixture of Cassia tora L., Ulmus pumila L., and Taraxacum officinale, on Compound 48/80-Mediated Allergic Responses in RBL-2H3 Cells. Molecules 2020, 25, 2434. [Google Scholar] [CrossRef]
- Petrosino, S.; Schiano Moriello, A.; Verde, R.; Allarà, M.; Imperatore, R.; Ligresti, A.; Mahmoud, A.M.; Peritore, A.F.; Iannotti, F.A.; Di Marzo, V. Palmitoylethanolamide counteracts substance P-induced mast cell activation in vitro by stimulating diacylglycerol lipase activity. J. NeuroInflamm. 2019, 16, 274. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Azouz, N.P.; Matsui, T.; Fukuda, M.; Sagi-Eisenberg, R. Decoding the regulation of mast cell exocytosis by networks of Rab GTPases. J. Immunol. 2012, 189, 2169–2180. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, M.; Baumgart, T.; Hammond, S.; Holowka, D.; Baird, B. Differential targeting of secretory lysosomes and recycling endosomes in mast cells revealed by patterned antigen arrays. J. Cell Sci. 2007, 120, 3147–3154. [Google Scholar] [CrossRef] [Green Version]
- Azouz, N.P.; Zur, N.; Efergan, A.; Ohbayashi, N.; Fukuda, M.; Amihai, D.; Hammel, I.; Rothenberg, M.E.; Sagi-Eisenberg, R. Rab5 Is a Novel Regulator of Mast Cell Secretory Granules: Impact on Size, Cargo, and Exocytosis. J. Immunol. 2014, 192, 4043–4053. [Google Scholar] [CrossRef] [Green Version]
- Paton, W.D. Compound 48/80: A potent histamine liberator. Br. J. Pharmacol. Chemother. 1951, 6, 499–508. [Google Scholar] [CrossRef] [Green Version]
- Green, D.P.; Limjunyawong, N.; Gour, N.; Pundir, P.; Dong, X. A Mast-Cell-Specific Receptor Mediates Neurogenic Inflammation and Pain. Neuron 2019, 101, 412–420. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Navinés-Ferrer, A.; Serrano-Candelas, E.; Lafuente, A.; Muñoz-Cano, R.; Martín, M.; Gastaminza, G. MRGPRX2-mediated mast cell response to drugs used in perioperative procedures and anaesthesia. Sci. Rep. 2018, 8, 11628. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beaven, M.A.; Ozawa, K. Role of calcium, protein kinase C and MAP kinase in the activation of mast cells. Allergol. Int. 1996, 45, 73–84. [Google Scholar] [CrossRef] [Green Version]
- Jabril-Cuenod, B.; Zhang, C.; Scharenberg, A.M.; Paolini, R.; Numerof, R.; Beaven, M.A.; Kinet, J.P. Syk-dependent phosphorylation of Shc. A potential link between FcepsilonRI and the Ras/mitogen-activated protein kinase signaling pathway through SOS and Grb2. J. Biol. Chem. 1996, 271, 16268–16272. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Santini, F.; Beaven, M.A. Tyrosine phosphorylation of a mitogen-activated protein kinase-like protein occurs at a late step in exocytosis. Studies with tyrosine phosphatase inhibitors and various secretagogues in rat RBL-2H3 cells. J. Biol. Chem. 1993, 268, 22716–22722. [Google Scholar] [CrossRef]
- Zhang, C.; Baumgartner, R.A.; Yamada, K.; Beaven, M.A. Mitogen-activated protein (MAP) kinase regulates production of tumor necrosis factor-alpha and release of arachidonic acid in mast cells. Indications of communication between p38 and p42 MAP kinases. J. Biol. Chem. 1997, 272, 13397–13402. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, M.-S.; Rådinger, M.; Gilfillan, A.M. The multiple roles of phosphoinositide 3-kinase in mast cell biology. Trends Immunol. 2008, 29, 493–501. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peng, Y.; Power, M.R.; Li, B.; Lin, T.-J. Inhibition of IKK down-regulates antigen + IgE-induced TNF production by mast cells: A role for the IKK-IκB-NF-κB pathway in IgE-dependent mast cell activation. J. Leukoc. Biol. 2005, 77, 975–983. [Google Scholar] [CrossRef]
- Hwang, S.L.; Lu, Y.; Li, X.; Kim, Y.D.; Cho, Y.S.; Jahng, Y.; Son, J.K.; Lee, Y.J.; Kang, W.; Taketomi, Y.; et al. ERK1/2 antagonize AMPK-dependent regulation of FcεRI-mediated mast cell activation and anaphylaxis. J. Allergy Clin. Immunol. 2014, 134, 714–721. [Google Scholar] [CrossRef]
- Shefler, I.; Seger, R.; Sagi-Eisenberg, R. Gi-mediated activation of mitogen-activated protein kinase (MAPK) pathway by receptor mimetic basic secretagogues of connective tissue-type mast cells: Bifurcation of arachidonic acid-induced release upstream of MAPK. J. Pharmacol. Exp. Ther. 1999, 289, 1654–1661. [Google Scholar]
- Lobingier, B.T.; von Zastrow, M. When trafficking and signaling mix: How subcellular location shapes G protein-coupled receptor activation of heterotrimeric G proteins. Traffic 2019, 20, 130–136. [Google Scholar] [CrossRef] [Green Version]
- Kimata, M.; Inagaki, N.; Kato, T.; Miura, T.; Serizawa, I.; Nagai, H. Roles of mitogen-activated protein kinase pathways for mediator release from human cultured mast cells. Biochem. Pharmacol. 2000, 60, 589–594. [Google Scholar] [CrossRef]
- Hu Frisk, J.M.; Kjellén, L.; Melo, F.R.; Öhrvik, H.; Pejler, G. Mitogen-Activated Protein Kinase Signaling Regulates Proteoglycan Composition of Mast Cell Secretory Granules. Front. Immunol. 2018, 9, 1670. [Google Scholar] [CrossRef] [PubMed]
- Arifuzzaman, M.; Mobley, Y.R.; Choi, H.W.; Bist, P.; Salinas, C.A.; Brown, Z.D.; Chen, S.L.; Staats, H.F.; Abraham, S.N. MRGPR-mediated activation of local mast cells clears cutaneous bacterial infection and protects against reinfection. Sci. Adv. 2019, 5, eaav0216. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chahdi, A.; Mousli, M.; Landry, Y. Substance P-related inhibitors of mast cell exocytosis act on G-proteins or on the cell surface. Eur. J. Pharmacol. 1998, 341, 329–335. [Google Scholar] [CrossRef]
- Suzuki, K.; Verma, I.M. Phosphorylation of SNAP-23 by IkappaB kinase 2 regulates mast cell degranulation. Cell 2008, 134, 485–495. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roget, K.; Ben-Addi, A.; Mambole-Dema, A.; Gantke, T.; Yang, H.-T.; Janzen, J.; Morrice, N.; Abbott, D.; Ley, S.C. IκB kinase 2 regulates TPL-2 activation of extracellular signal-regulated kinases 1 and 2 by direct phosphorylation of TPL-2 serine 400. Mol. Cell. Biol. 2012, 32, 4684–4690. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ushio, H.; Ueno, T.; Kojima, Y.; Komatsu, M.; Tanaka, S.; Yamamoto, A.; Ichimura, Y.; Ezaki, J.; Nishida, K.; Komazawa-Sakon, S.; et al. Crucial role for autophagy in degranulation of mast cells. J. Allergy Clin. Immunol. 2011, 127, 1267–1276. [Google Scholar] [CrossRef] [PubMed]
- Nakano, H.; Ushio, H. An unexpected role for autophagy in degranulation of mast cells. Autophagy 2011, 7, 657–659. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martinez-Lopez, N.; Athonvarangkul, D.; Mishall, P.; Sahu, S.; Singh, R. Autophagy proteins regulate ERK phosphorylation. Nat. Commun. 2013, 4, 2799. [Google Scholar] [CrossRef] [PubMed]







| Inhibitor | Target | Company | Cat. No. |
|---|---|---|---|
| Go6976 | PKCα/β1, PKD | A.G. Scientific (San Diego, CA, USA) | G-1017 |
| GF109203X | PKC | A.G. Scientific | G-1063 |
| U73122 | PLC (+PLD) | TOCRIS (Minneapolis, MN, USA) | 1268 |
| LY294002 | PI3K | TOCRIS | 1130 |
| wortmannin | PI3K + PI4K | A.G. Scientific | W-1022 |
| U0126 | MEK1, MEK2 | A.G. Scientific | U-1026 |
| MRT68921 | ULK1/2 | TOCRIS | 5780 |
| IMD0354 | IKKβ | Cayman Chemical (Ann Arbor, MI, USA) | 17290 |
| 3-MA | PI3K type III | Sigma-Aldrich | M9281 |
| EGTA | Ca2+ chelator | Sigma-Aldrich | E4378 |
| hMRGPRX2 (PrimerBank ID40255006c1) [32,33,34] | fw 5′-CTGGTAGGAAACGGGTTTGTG-3′ rv 5′-GCTGAGGACGTAGACAGAGAAG-3′ |
| rMRGPRB3 [35] | fw 5′-CCCCTGGAATGTTCTTTTGTGTAG-3′ rv 5′-ACAGTGAAAAATGCAGGAACTTGG-3′ |
| hGAPDH (cat #: HP205798, sequence from Origene, ordered from Hy Laboratories Ltd., Rehovot, Israel) | fw 5′-GTCTCCTCTGACTTCAACAGCG-3′ rv 5′-ACCACCCTGTTGCTGTAGCCAA-3′ |
| rGAPDH [36] | fw 5′-TGGAGTCTACTGGCGTCT-3′ rv 5′-TGTCATATTTVTCGTGGT-3′ |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lazki-Hagenbach, P.; Ali, H.; Sagi-Eisenberg, R. Authentic and Ectopically Expressed MRGPRX2 Elicit Similar Mechanisms to Stimulate Degranulation of Mast Cells. Cells 2021, 10, 376. https://doi.org/10.3390/cells10020376
Lazki-Hagenbach P, Ali H, Sagi-Eisenberg R. Authentic and Ectopically Expressed MRGPRX2 Elicit Similar Mechanisms to Stimulate Degranulation of Mast Cells. Cells. 2021; 10(2):376. https://doi.org/10.3390/cells10020376
Chicago/Turabian StyleLazki-Hagenbach, Pia, Hydar Ali, and Ronit Sagi-Eisenberg. 2021. "Authentic and Ectopically Expressed MRGPRX2 Elicit Similar Mechanisms to Stimulate Degranulation of Mast Cells" Cells 10, no. 2: 376. https://doi.org/10.3390/cells10020376
APA StyleLazki-Hagenbach, P., Ali, H., & Sagi-Eisenberg, R. (2021). Authentic and Ectopically Expressed MRGPRX2 Elicit Similar Mechanisms to Stimulate Degranulation of Mast Cells. Cells, 10(2), 376. https://doi.org/10.3390/cells10020376