IgLON5 Regulates the Adhesion and Differentiation of Myoblasts
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Neutralization of IgLON5
2.3. Gene Knockdown
2.4. Cell Adhesion and Proliferation Assays
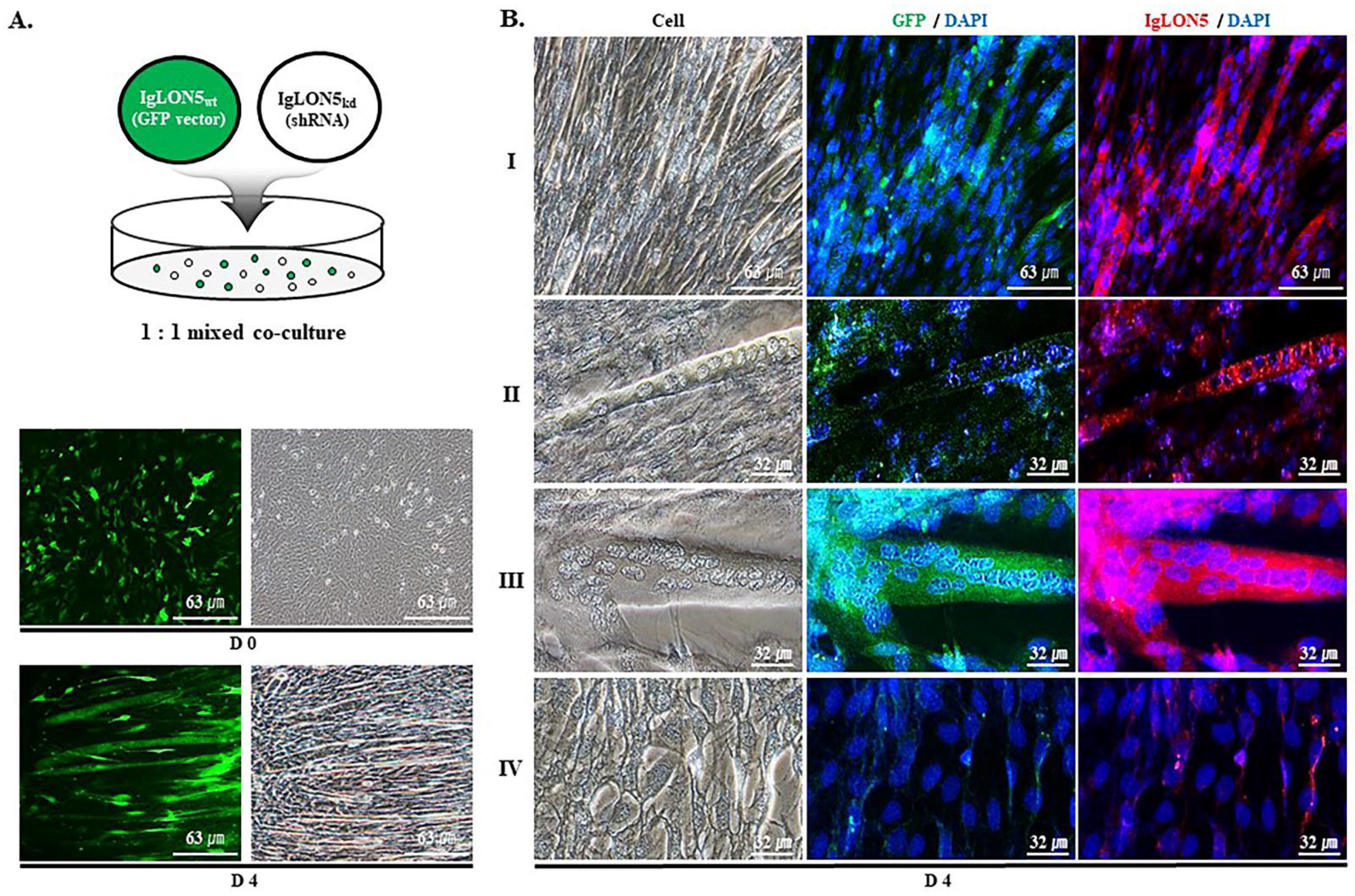
2.5. Co-Culture of IgLON5wt and IgLON5kd Cells
2.6. Cell Adhesion Analysis Using Centrifugation
2.7. Fusion and Maturation Indices
2.8. Cell Cycle Analysis
2.9. Metabolite Analysis
2.10. Animal Experiment
2.11. Immunocytochemistry
2.12. Total RNA Extraction, cDNA Synthesis, and Real-Time RT-PCR
2.13. Western Blot Analysis
2.14. Immunohistochemistry and Immunofluorescence
2.15. Statistical Analysis
3. Results
3.1. IgLON5 Regulated Myoblast Differentiation
3.2. IgLON5 Expression and Myotube Formation
3.3. The roles of IgLON5 during Adhesion and Proliferation
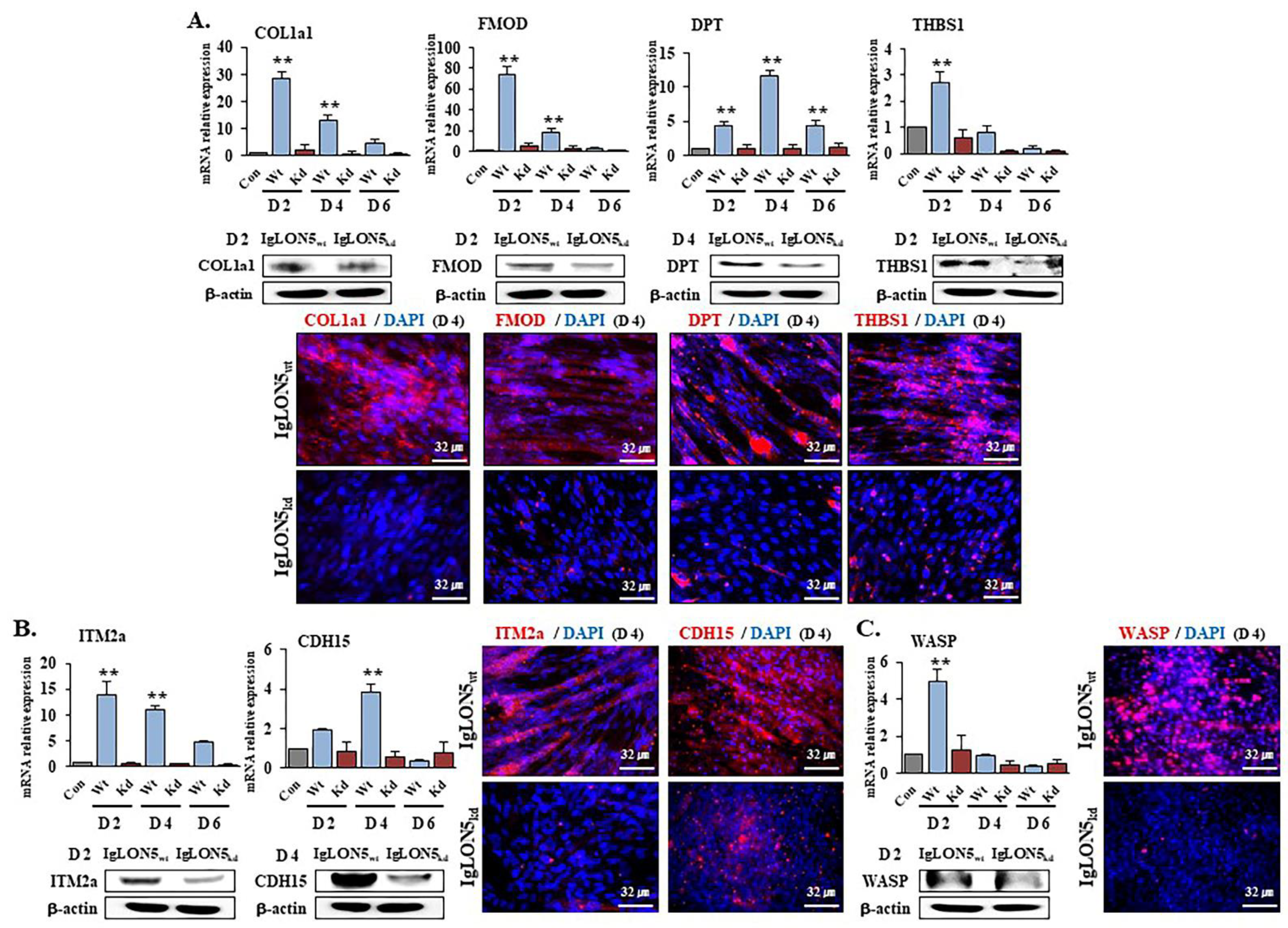
3.4. Gene Expression Profiles in IgLON5wt and IgLON5kd Cells
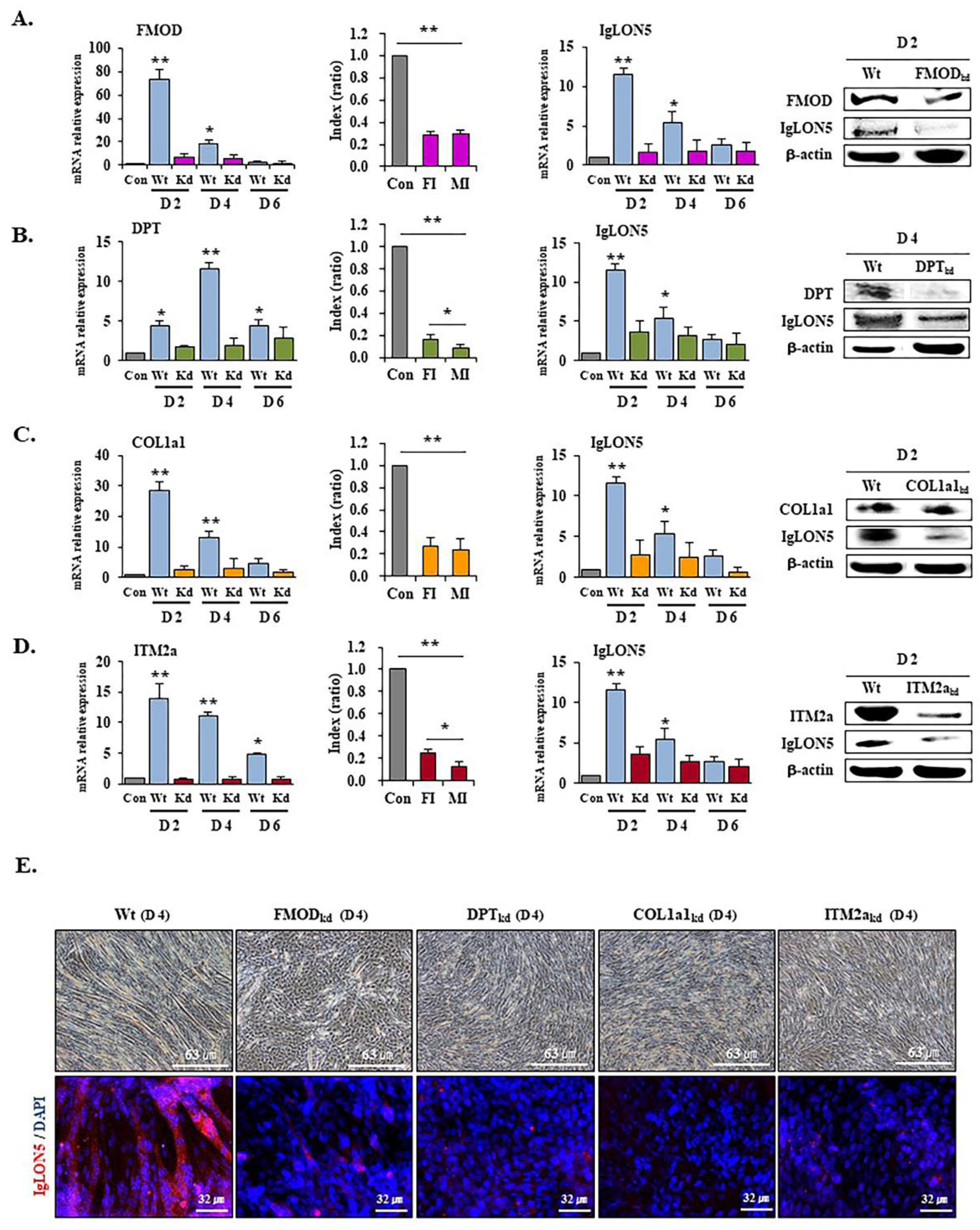
3.5. IgLON5 Expressions in FMOD, DPT, COL1a1, and ITM2a Knockdown Cells
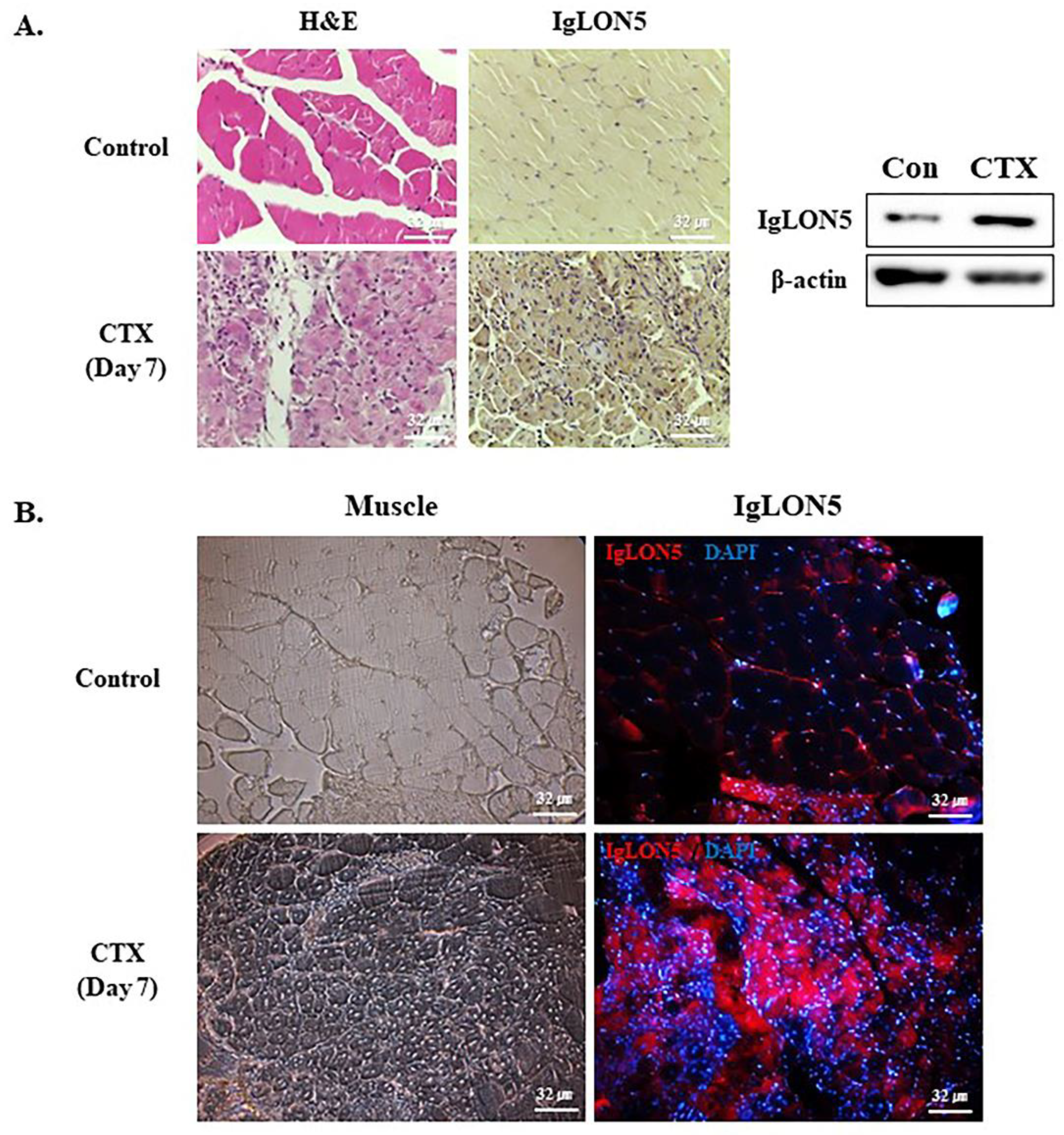
3.6. IgLON5 Expression during Muscle Regeneration
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| MSCs | myogenic stem (satellite) cells |
| ECM | Extracellular matrix |
| IgSF | Immunoglobulin superfamily |
| OPCML | Opioid-binding cell adhesion molecule |
| GPI | Glycosylphosphatidylinositol |
| LSAMP | limbic system-associated membrane protein, |
| IgLON5 | Immunoglobulin-like cell adhesion molecule 5 |
| MYOG | Myogenin |
| MYOD | Myoblast determination protein |
| MYL2 | Myosin light chain 2 |
| PSP | Progressive supranuclear palsy |
| FMOD | Fibromodulin |
| DPT | Dermatopontin |
| Itm2A | Integral membrane protein 2A |
| THBS1 | Thrombospondin-1 |
| COL1a1 | Collagen type 1 alpha 1 |
| WASP | Wiskott–Aldrich syndrome protein |
| CDH15 | Cadherin 15 |
| CTX | Cardiotoxin |
References
- Kim, K.M.; Jang, H.C.; Lim, S. Differences among skeletal muscle mass indices derived from height-, weight-, and body mass index-adjusted models in assessing sarcopenia. Korean J. Intern. Med. 2016, 31, 643–650. [Google Scholar] [CrossRef] [PubMed]
- Yin, H.; Price, F.; Rudnicki, M.A. Satellite cells and the muscle stem cell niche. Physiol Rev 2013, 93, 23–67. [Google Scholar] [CrossRef] [PubMed]
- Lee, E.J.; Jan, A.T.; Baig, M.H.; Ashraf, J.M.; Nahm, S.S.; Kim, Y.W.; Park, S.Y.; Choi, I. Fibromodulin: A master regulator of myostatin controlling progression of satellite cells through a myogenic program. FASEB J. 2016, 30, 2708–2719. [Google Scholar] [CrossRef]
- Lee, E.J.; Lee, H.J.; Kamli, M.R.; Pokharel, S.; Bhat, A.R.; Lee, Y.H.; Choi, B.H.; Chun, T.; Kang, S.W.; Lee, Y.S.; et al. Depot-specific gene expression profiles during differentiation and transdifferentiation of bovine muscle satellite cells, and differentiation of preadipocytes. Genomics 2012, 100, 195–202. [Google Scholar] [CrossRef]
- Blau, H.M.; Cosgrove, B.D.; Ho, A.T. The central role of muscle stem cells in regenerative failure with aging. Nat. Med. 2015, 21, 854–862. [Google Scholar] [CrossRef]
- Ahmad, K.; Lee, E.J.; Moon, J.S.; Park, S.Y.; Choi, I. Multifaceted Interweaving Between Extracellular Matrix, Insulin Resistance, and Skeletal Muscle. Cells 2018, 7, 148. [Google Scholar] [CrossRef]
- Charge, S.B.; Rudnicki, M.A. Cellular and molecular regulation of muscle regeneration. Physiol. Rev. 2004, 84, 209–238. [Google Scholar] [CrossRef]
- Lee, E.J.; Jan, A.T.; Baig, M.H.; Ahmad, K.; Malik, A.; Rabbani, G.; Kim, T.; Lee, I.K.; Lee, Y.H.; Park, S.Y.; et al. Fibromodulin and regulation of the intricate balance between myoblast differentiation to myocytes or adipocyte-like cells. FASEB J. 2018, 32, 768–781. [Google Scholar] [CrossRef] [PubMed]
- Baig, M.H.; Jan, A.T.; Rabbani, G.; Ahmad, K.; Ashraf, J.M.; Kim, T.; Min, H.S.; Lee, Y.H.; Cho, W.K.; Ma, J.Y.; et al. Methylglyoxal and Advanced Glycation End products: Insight of the regulatory machinery affecting the myogenic program and of its modulation by natural compounds. Sci. Rep. 2017, 7, 5916. [Google Scholar] [CrossRef]
- Frantz, C.; Stewart, K.M.; Weaver, V.M. The extracellular matrix at a glance. J. Cell. Sci. 2010, 123, 4195–4200. [Google Scholar] [CrossRef]
- Yue, B. Biology of the extracellular matrix: An overview. J. Glaucoma. 2014, 23, S20–S23. [Google Scholar] [CrossRef] [PubMed]
- Willander, H.; Hermansson, E.; Johansson, J.; Presto, J. BRICHOS domain associated with lung fibrosis, dementia and cancer--a chaperone that prevents amyloid fibril formation? FEBS J. 2011, 278, 3893–3904. [Google Scholar] [CrossRef] [PubMed]
- Van den Plas, D.; Merregaert, J. Constitutive overexpression of the integral membrane protein Itm2A enhances myogenic differentiation of C2C12 cells. Cell Biol. Int. 2004, 28, 199–207. [Google Scholar] [CrossRef] [PubMed]
- Kim, T.; Ahmad, K.; Shaikh, S.; Jan, A.T.; Seo, M.G.; Lee, E.J.; Choi, I. Dermatopontin in Skeletal Muscle Extracellular Matrix Regulates Myogenesis. Cells 2019, 8, 332. [Google Scholar] [CrossRef]
- Okamoto, O.; Fujiwara, S. Dermatopontin, a novel player in the biology of the extracellular matrix. Connect Tissue Res. 2006, 47, 177–189. [Google Scholar] [CrossRef]
- Adams, J.C.; Lawler, J. The thrombospondins. Cold Spring Harb Perspect Biol. 2011, 3, a009712. [Google Scholar] [CrossRef]
- Stenina, O.I.; Krukovets, I.; Wang, K.; Zhou, Z.; Forudi, F.; Penn, M.S.; Topol, E.J.; Plow, E.F. Increased expression of thrombospondin-1 in vessel wall of diabetic Zucker rat. Circulation 2003, 107, 3209–3215. [Google Scholar] [CrossRef] [PubMed]
- Tan, R.P.A.; Leshchyns’ka, I.; Sytnyk, V. Glycosylphosphatidylinositol-Anchored Immunoglobulin Superfamily Cell Adhesion Molecules and Their Role in Neuronal Development and Synapse Regulation. Front. Mol. Neurosci. 2017, 10, 378. [Google Scholar] [CrossRef] [PubMed]
- Sterky, F.H.; Trotter, J.H.; Lee, S.J.; Recktenwald, C.V.; Du, X.; Zhou, B.; Zhou, P.; Schwenk, J.; Fakler, B.; Sudhof, T.C. Carbonic anhydrase-related protein CA10 is an evolutionarily conserved pan-neurexin ligand. Proc. Natl. Acad. Sci. USA 2017, 114, E1253–E1262. [Google Scholar] [CrossRef] [PubMed]
- Sellar, G.C.; Watt, K.P.; Rabiasz, G.J.; Stronach, E.A.; Li, L.; Miller, E.P.; Massie, C.E.; Miller, J.; Contreras-Moreira, B.; Scott, D.; et al. OPCML at 11q25 is epigenetically inactivated and has tumor-suppressor function in epithelial ovarian cancer. Nat. Genet. 2003, 34, 337–343. [Google Scholar] [CrossRef]
- Pimenta, A.F.; Levitt, P. Characterization of the genomic structure of the mouse limbic system-associated membrane protein (Lsamp) gene. Genomics 2004, 83, 790–801. [Google Scholar] [CrossRef] [PubMed]
- Karagogeos, D. Neural GPI-anchored cell adhesion molecules. Front. Biosci. 2003, 8, s1304–s1320. [Google Scholar] [CrossRef]
- Ranaivoson, F.M.; Turk, L.S.; Ozgul, S.; Kakehi, S.; von Daake, S.; Lopez, N.; Trobiani, L.; De Jaco, A.; Denissova, N.; Demeler, B.; et al. A Proteomic Screen of Neuronal Cell-Surface Molecules Reveals IgLONs as Structurally Conserved Interaction Modules at the Synapse. Structure 2019, 27, 893–906.e9. [Google Scholar] [CrossRef] [PubMed]
- Sanz, R.; Ferraro, G.B.; Fournier, A.E. IgLON cell adhesion molecules are shed from the cell surface of cortical neurons to promote neuronal growth. J. Biol. Chem. 2015, 290, 4330–4342. [Google Scholar] [CrossRef] [PubMed]
- Sabater, L.; Gaig, C.; Gelpi, E.; Bataller, L.; Lewerenz, J.; Torres-Vega, E.; Contreras, A.; Giometto, B.; Compta, Y.; Embid, C.; et al. A novel non-rapid-eye movement and rapid-eye-movement parasomnia with sleep breathing disorder associated with antibodies to IgLON5: A case series, characterisation of the antigen, and post-mortem study. Lancet Neurol. 2014, 13, 575–586. [Google Scholar] [CrossRef]
- Leypoldt, F.; Armangue, T.; Dalmau, J. Autoimmune encephalopathies. Ann. N Y Acad. Sci. 2015, 1338, 94–114. [Google Scholar] [CrossRef]
- Minhas, H.M.; Pescosolido, M.F.; Schwede, M.; Piasecka, J.; Gaitanis, J.; Tantravahi, U.; Morrow, E.M. An unbalanced translocation involving loss of 10q26.2 and gain of 11q25 in a pedigree with autism spectrum disorder and cerebellar juvenile pilocytic astrocytoma. Am. J. Med. Genet. A 2013, 161A, 787–791. [Google Scholar] [CrossRef] [PubMed]
- Lee, E.J.; Pokharel, S.; Jan, A.T.; Huh, S.; Galope, R.; Lim, J.H.; Lee, D.M.; Choi, S.W.; Nahm, S.S.; Kim, Y.W.; et al. Transthyretin: A Transporter Protein Essential for Proliferation of Myoblast in the Myogenic Program. Int. J. Mol. Sci. 2017, 18, 115. [Google Scholar] [CrossRef]
- Lagha, M.; Mayeuf-Louchart, A.; Chang, T.; Montarras, D.; Rocancourt, D.; Zalc, A.; Kormish, J.; Zaret, K.S.; Buckingham, M.E.; Relaix, F. Itm2a is a Pax3 target gene, expressed at sites of skeletal muscle formation in vivo. PLoS ONE 2013, 8, e63143. [Google Scholar] [CrossRef] [PubMed]
- Mandai, S.; Furukawa, S.; Kodaka, M.; Hata, Y.; Mori, T.; Nomura, N.; Ando, F.; Mori, Y.; Takahashi, D.; Yoshizaki, Y.; et al. Loop diuretics affect skeletal myoblast differentiation and exercise-induced muscle hypertrophy. Sci. Rep. 2017, 7, 46369. [Google Scholar] [CrossRef]
- Ratkevicius, A.; Joyson, A.; Selmer, I.; Dhanani, T.; Grierson, C.; Tommasi, A.; DeVries, A.; Rauchhaus, P.; Crowther, D.; Alesci, S. Serum concentrations of myostatin and myostatin-interacting proteins do not differ between young and sarcopenic elderly men. J. Gerontol. Ser. A: Biomed. Sci. Med. Sci. 2011, 66, 620–626. [Google Scholar] [CrossRef]
- Feige, P.; Brun, C.E.; Ritso, M.; Rudnicki, M.A. Orienting Muscle Stem Cells for Regeneration in Homeostasis, Aging, and Disease. Cell Stem Cell 2018, 23, 653–664. [Google Scholar] [CrossRef]
- Collins, C.A.; Olsen, I.; Zammit, P.S.; Heslop, L.; Petrie, A.; Partridge, T.A.; Morgan, J.E. Stem cell function, self-renewal, and behavioral heterogeneity of cells from the adult muscle satellite cell niche. Cell 2005, 122, 289–301. [Google Scholar] [CrossRef]
- Kook, S.H.; Choi, K.C.; Son, Y.O.; Lee, K.Y.; Hwang, I.H.; Lee, H.J.; Chang, J.S.; Choi, I.H.; Lee, J.C. Satellite cells isolated from adult Hanwoo muscle can proliferate and differentiate into myoblasts and adipose-like cells. Mol. Cells 2006, 22, 239–245. [Google Scholar] [PubMed]
- Cohen, S.; Nathan, J.A.; Goldberg, A.L. Muscle wasting in disease: Molecular mechanisms and promising therapies. Nat. Rev. Drug Discov. 2015, 14, 58–74. [Google Scholar] [CrossRef]
- McNamee, C.J.; Reed, J.E.; Howard, M.R.; Lodge, A.P.; Moss, D.J. Promotion of neuronal cell adhesion by members of the IgLON family occurs in the absence of either support or modification of neurite outgrowth. J. Neurochem. 2002, 80, 941–948. [Google Scholar] [CrossRef] [PubMed]
- Malek, M.H.; Olfert, I.M. Global deletion of thrombospondin-1 increases cardiac and skeletal muscle capillarity and exercise capacity in mice. Exp. Physiol. 2009, 94, 749–760. [Google Scholar] [CrossRef]
- Svensson, L.; Aszódi, A.; Reinholt, F.P.; Fässler, R.; Heinegård, D.; Oldberg, A. Fibromodulin-null mice have abnormal collagen fibrils, tissue organization, and altered lumican deposition in tendon. J. Biol. Chem. 1999, 274, 9636–9647. [Google Scholar] [CrossRef] [PubMed]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lim, J.H.; Beg, M.M.A.; Ahmad, K.; Shaikh, S.; Ahmad, S.S.; Chun, H.J.; Choi, D.; Lee, W.-J.; Jin, J.-O.; Kim, J.; et al. IgLON5 Regulates the Adhesion and Differentiation of Myoblasts. Cells 2021, 10, 417. https://doi.org/10.3390/cells10020417
Lim JH, Beg MMA, Ahmad K, Shaikh S, Ahmad SS, Chun HJ, Choi D, Lee W-J, Jin J-O, Kim J, et al. IgLON5 Regulates the Adhesion and Differentiation of Myoblasts. Cells. 2021; 10(2):417. https://doi.org/10.3390/cells10020417
Chicago/Turabian StyleLim, Jeong Ho, Mirza Masroor Ali Beg, Khurshid Ahmad, Sibhghatulla Shaikh, Syed Sayeed Ahmad, Hee Jin Chun, Dukhwan Choi, Woo-Jong Lee, Jun-O Jin, Jihoe Kim, and et al. 2021. "IgLON5 Regulates the Adhesion and Differentiation of Myoblasts" Cells 10, no. 2: 417. https://doi.org/10.3390/cells10020417
APA StyleLim, J. H., Beg, M. M. A., Ahmad, K., Shaikh, S., Ahmad, S. S., Chun, H. J., Choi, D., Lee, W.-J., Jin, J.-O., Kim, J., Jan, A. T., Lee, E. J., & Choi, I. (2021). IgLON5 Regulates the Adhesion and Differentiation of Myoblasts. Cells, 10(2), 417. https://doi.org/10.3390/cells10020417







