MicroRNA-27b-3p Targets the Myostatin Gene to Regulate Myoblast Proliferation and Is Involved in Myoblast Differentiation
Abstract
:1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. Experimental Animals and Tissues
2.3. Cell Culture
2.4. Total RNA Extraction and Quality Detection, Complementary DNA (cDNA) Synthesis, and Quantitative Real-Time PCR (qRT-PCR)
2.5. Primers for Quantitative Real-Time PCR (qRT-PCR)
2.6. RNA Oligonucleotides and Plasmids Construction
2.7. Cell Transfection
2.8. Dual-Luciferase Reporter Assay
2.9. CCK-8 Assay
2.10. EdU Assay
2.11. Cell Cycle was Detected by Flow Cytometry
2.12. Immunofluorescence
2.13. Western Blot Assay
2.14. Statistical Analysis
3. Results
3.1. Expression of miR-27b-3p and MSTN in Tissues of Chicken
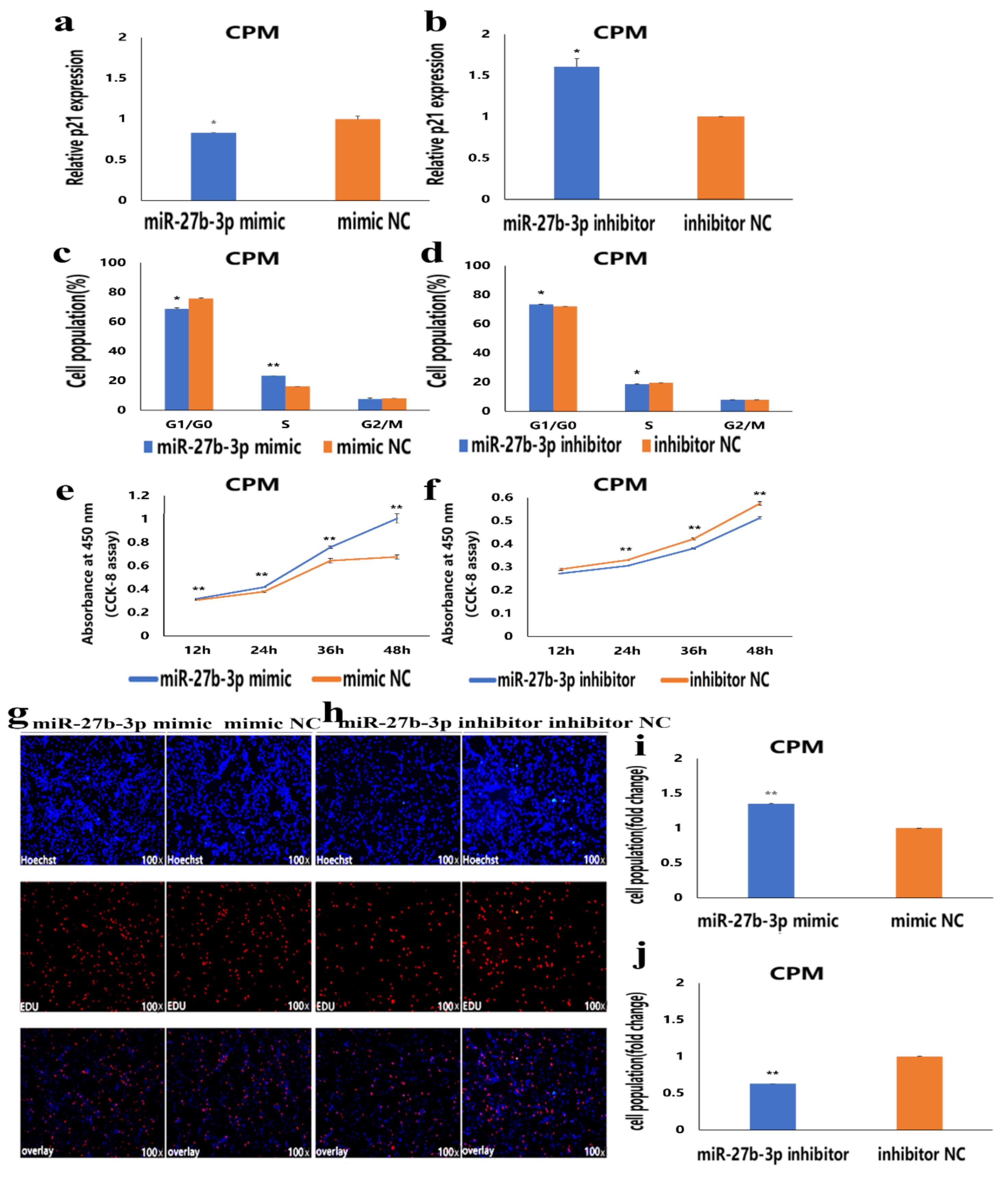
3.2. MiR-27b-3p Promotes CPMs Proliferation
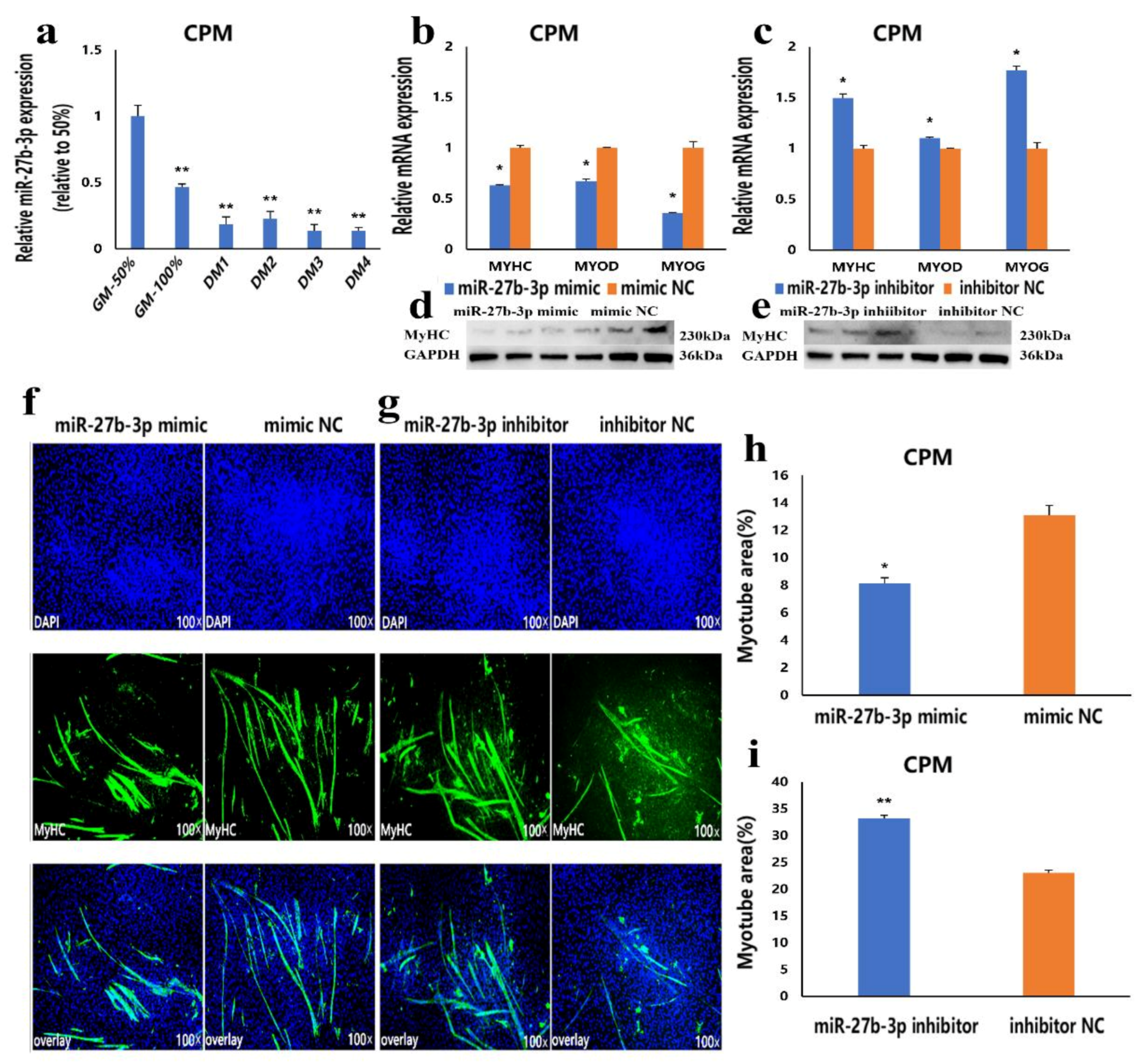
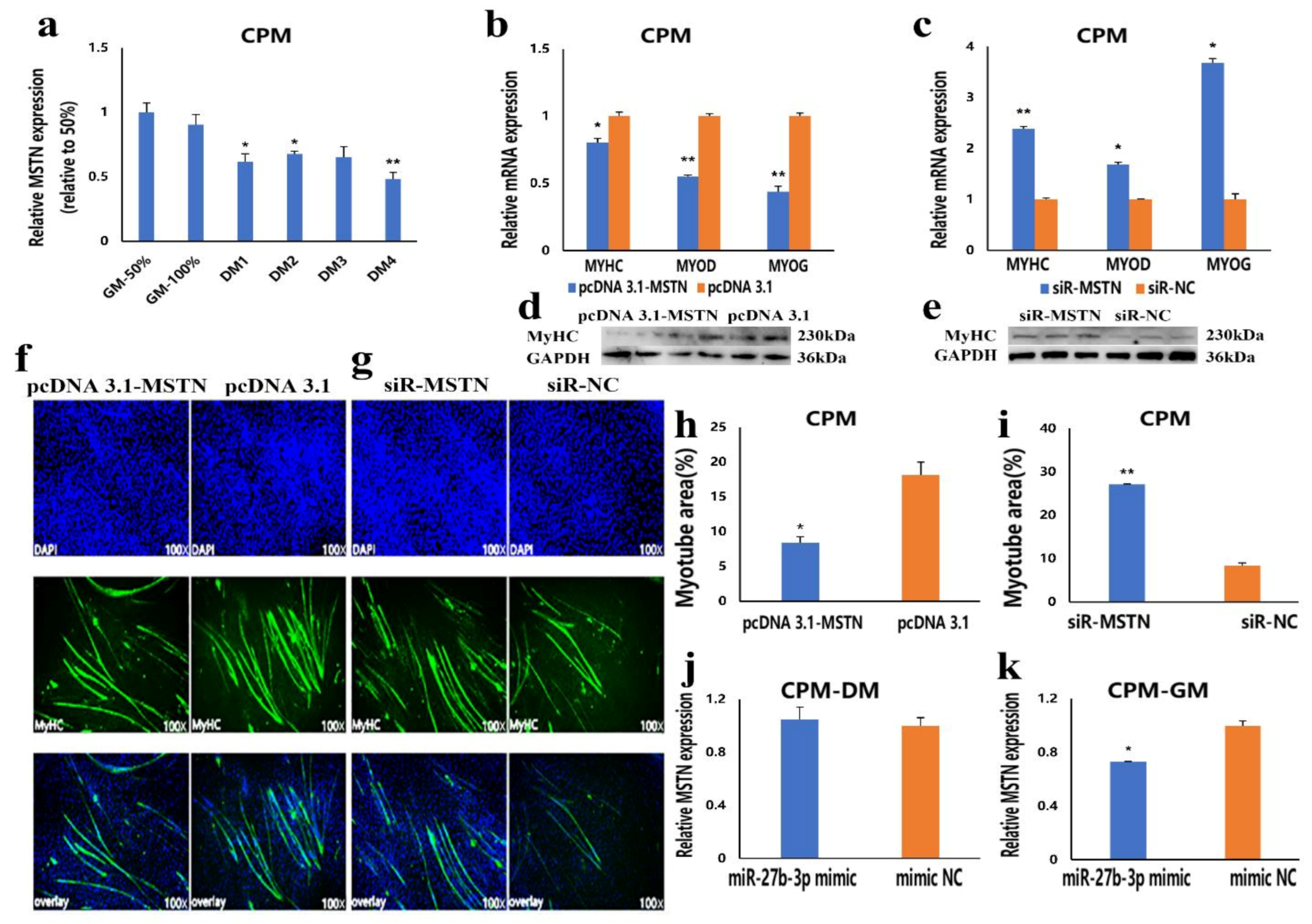
3.3. MiR-27b-3p Inhibits CPMs Differentiation
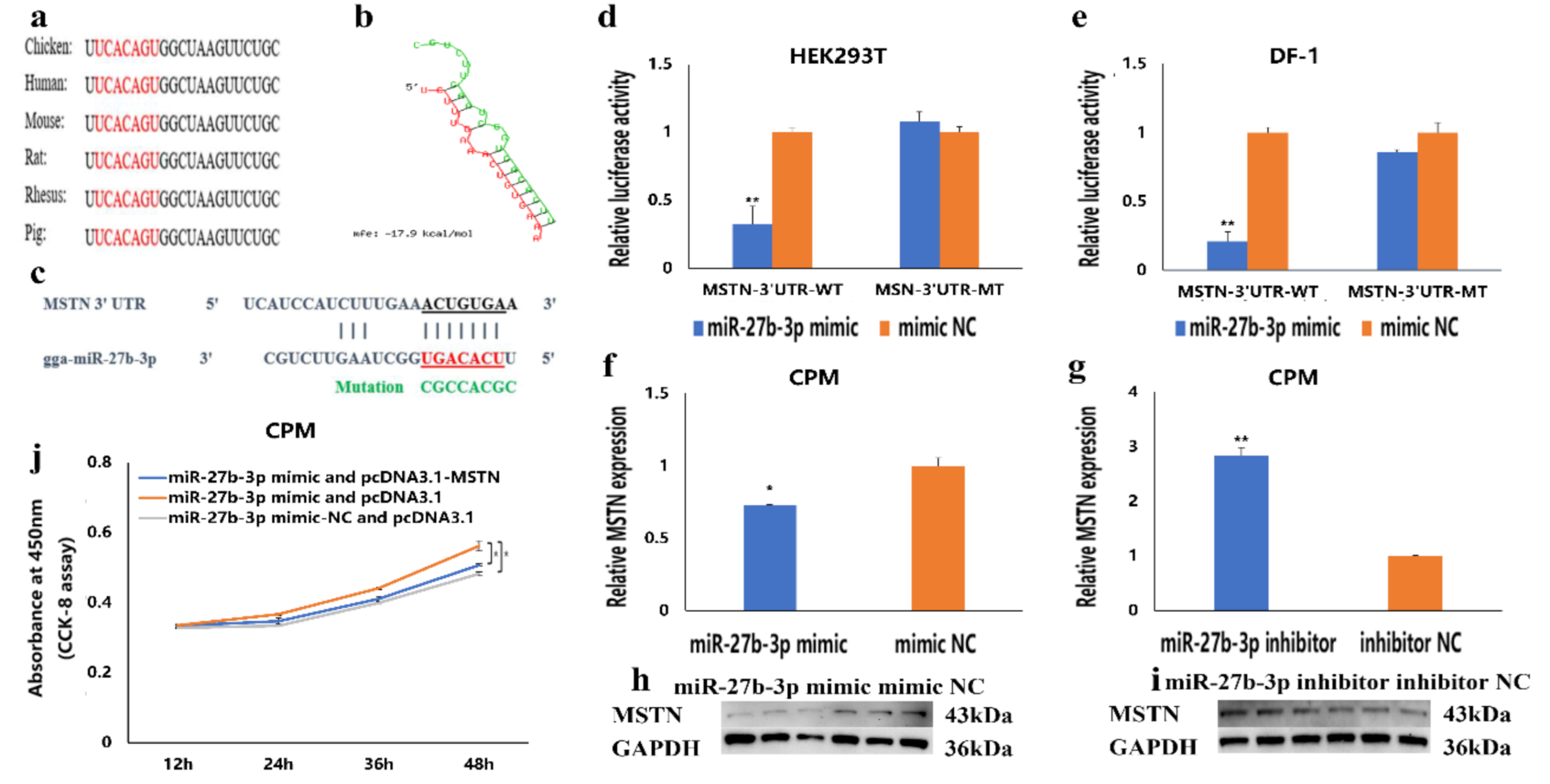
3.4. MSTN Is a Target Gene of miR-27b-3p
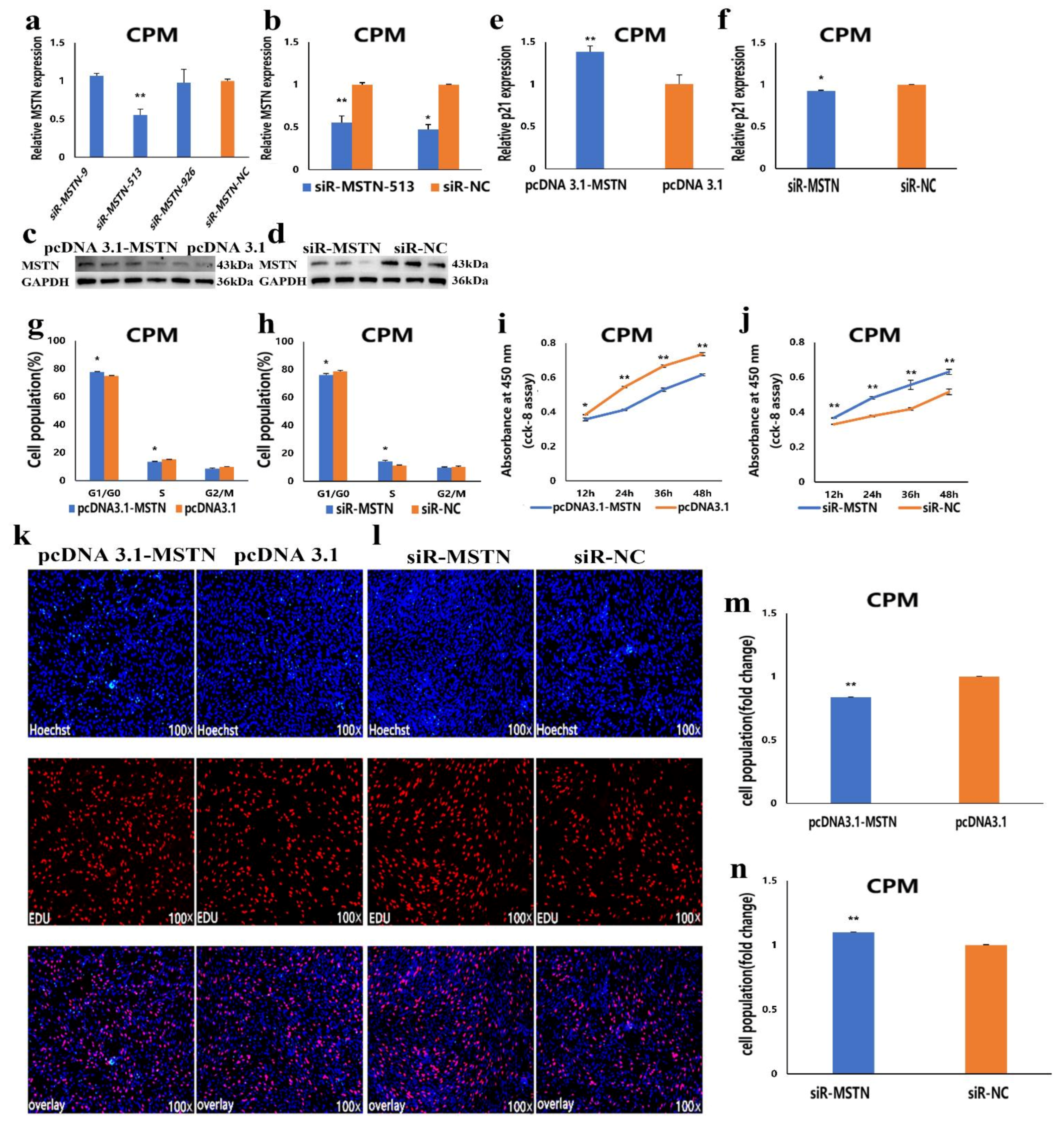
3.5. The Facilitation of miR-27b-3p on CPMs Proliferation was Achieved by Its Target Gene MSTN
3.6. The Inhibition of miR-27b-3p on MSTN Is Different between CPMs Proliferation and Differentiation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Lau, N.C.; Lim, L.P.; Weinstein, E.G.; Bartel, D.P. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science 2001, 294, 858–862. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Lian, L.; Zhang, D.; Qu, L.; Yang, N. gga-miR-26a targets NEK6 and suppresses Marek’s disease lymphoma cell prolif-eration. Poult. Sci. 2014, 93, 1097–1105. [Google Scholar] [CrossRef] [PubMed]
- Wahid, F.; Shehzad, A.; Khan, T.; Kim, Y.Y. MicroRNAs: Synthesis, mechanism, function, and recent clinical trials. Biochim. Biophys. Acta BBA Bioenerg. 2010, 1803, 1231–1243. [Google Scholar] [CrossRef] [Green Version]
- Cui, M.; Yao, X.; Lin, Y.; Zhang, D.; Cui, R.; Zhang, X. Interactive functions of microRNAs in the miR-23a-27a-24-2 cluster and the potential for targeted therapy in cancer. J. Cell Physiol. 2020, 235, 6–16. [Google Scholar] [CrossRef]
- Guo, H.; Ingolia, N.T.; Weissman, J.S.; Bartel, D.P. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nat. Cell Biol. 2010, 466, 835–840. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, G.; Wang, Y.; Li, Y.; Cui, L.; Zhao, Y.; Zhao, B.; Li, K. MiR-206, a Key Modulator of Skeletal Muscle Development and Disease. Int. J. Biol. Sci. 2015, 11, 345–352. [Google Scholar] [CrossRef]
- Sempere, L.F.; Freemantle, S.; Pitha-Rowe, I.; Moss, E.; Dmitrovsky, E.; Ambros, V. Expression profiling of mammalian mi-croRNAs uncovers a subset of brain-expressed microRNAs with possible roles in murine and human neuronal differentiation. Genome Biol. 2004, 5, R13. [Google Scholar] [CrossRef] [Green Version]
- Chen, J.F.; Mandel, E.M.; Thomson, J.M.; Wu, Q.; Callis, T.E.; Hammond, S.M.; Conlon, F.L.; Wang, D.Z. The role of mi-croRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat. Genet. 2006, 38, 228–233. [Google Scholar] [CrossRef] [PubMed]
- Luo, W.; Wu, H.; Ye, Y.; Li, Z.; Hao, S.; Kong, L.; Zheng, X.; Lin, S.; Nie, Q.; Zhang, X. The transient expression of miR-203 and its inhibiting effects on skeletal muscle cell proliferation and differentiation. Cell Death Dis. 2014, 5, e1347. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luo, W.; Nie, Q.; Zhang, X. MicroRNAs Involved in Skeletal Muscle Differentiation. J. Genet. Genom. 2013, 40, 107–116. [Google Scholar] [CrossRef]
- Chen, Y.; Chen, G.; Zhang, B.; Liu, C.; Yu, Y.; Jin, Y. miR-27b-3p suppresses cell proliferation, migration and invasion by tar-geting LIMK1 in colorectal cancer. Int. J. Clin. Exp. Pathol. 2017, 10, 9251–9261. [Google Scholar]
- Allen, D.L.; Loh, A.S. Posttranscriptional mechanisms involving microRNA-27a and b contribute to fast-specific and gluco-corticoid-mediated myostatin expression in skeletal muscle. Am. J. Physiol. Cell Physiol. 2011, 300, C124–C137. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lui, W.-O.; Pourmand, N.; Patterson, B.K.; Fire, A. Patterns of Known and Novel Small RNAs in Human Cervical Cancer. Cancer Res. 2007, 67, 6031–6043. [Google Scholar] [CrossRef] [Green Version]
- Michael, M.Z.; Connor, S.M.O.; Pellekaan, N.G.V.H.; Young, G.P.; James, R.J. Reduced accumulation of specific microRNAs in colorectal neoplasia. Mol. Cancer Res. 2003, 1, 882–891. [Google Scholar] [PubMed]
- Tao, J.; Zhi, X.; Zhang, X.; Fu, M.; Huang, H.; Fan, Y.; Guan, W.; Zou, C. miR-27b-3p suppresses cell proliferation through targeting receptor tyrosine kinase like orphan receptor 1 in gastric cancer. J. Exp. Clin. Cancer Res. 2015, 34, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, N.; Tang, Y.; Liu, B.; Cong, W.; Liu, C.; Xiao, J. Retinoid acid-induced microRNA-27b-3p impairs C2C12 myoblast prolifer-ation and differentiation by suppressing alpha-dystrobrevin. Exp. Cell Res. 2017, 350, 301–311. [Google Scholar] [CrossRef]
- Langley, B.; Thomas, M.; Bishop, A.; Sharma, M.; Gilmour, S.; Kambadur, R. Myostatin Inhibits Myoblast Differentiation by Down-regulating MyoD Expression. J. Biol. Chem. 2002, 277, 49831–49840. [Google Scholar] [CrossRef] [Green Version]
- McPherron, A.C.; Lee, S.-J. Double muscling in cattle due to mutations in the myostatin gene. Proc. Natl. Acad. Sci. USA 1997, 94, 12457–12461. [Google Scholar] [CrossRef] [Green Version]
- Mott, I.; Ivarie, R. Expression of myostatin is not altered in lines of poultry exhibiting myofiber hyper- and hypoplasia. Poult. Sci. 2002, 81, 799–804. [Google Scholar] [CrossRef]
- Ji, S.; Losinski, R.L.; Cornelius, S.G.; Frank, G.R.; Willis, G.M.; Gerrard, D.E.; Depreux, F.F.S.; Spurlock, M.E. Myostatin expression in porcine tissues: Tissue specificity and developmental and postnatal regulation. Am. J. Physiol. Content 1998, 275, R1265–R1273. [Google Scholar] [CrossRef] [PubMed]
- McFarland, D.C.; Velleman, S.G.; Pesall, J.E.; Liu, C. The role of myostatin in chicken (Gallus domesticus) myogenic satellite cell proliferation and differentiation. Gen. Comp. Endocrinol. 2007, 151, 351–357. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Cai, B.; Ma, M.; Chen, B.; Li, Z.; Abdalla, B.A.; Nie, Q.; Zhang, X. MiR-16-5p targets SESN1 to regulate the p53 signaling pathway, affecting myoblast proliferation and apoptosis, and is involved in myoblast differentiation. Cell Death Dis. 2018, 9, 1–15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Darnell, D.K.; Kaur, S.; Stanislaw, S.; Konieczka, J.K.; Yatskievych, T.A.; Antin, P.B. MicroRNA expression during chick embryo development. Dev. Dyn. 2006, 235, 3156–3165. [Google Scholar] [CrossRef]
- Lie, S.; Morrison, J.L.; Williams-Wyss, O.; Suter, C.M.; Humphreys, D.T.; Ozanne, S.E.; Zhang, S.; MacLaughlin, S.M.; Kleemann, D.O.; Walker, S.K. Impact of periconceptional and preimplantation undernutrition on factors regulating myo-genesis and protein synthesis in muscle of singleton and twin fetal sheep. Physiol. Rep. 2015, 3, e12495. [Google Scholar] [CrossRef]
- Li, W.; Chang, N.; Tian, L.; Yang, J.; Ji, X.; Xie, J.; Yang, L.; Li, L. miR-27b-3p, miR-181a-1-3p, and miR-326-5p are involved in the inhibition of macrophage activation in chronic liver injury. J. Mol. Med. 2017, 95, 1091–1105. [Google Scholar] [CrossRef]
- Cheng, X.; Kan, P.; Ma, Z.; Wang, Y.; Song, W.; Huang, C.; Zhang, B. Exploring the potential value of miR-148b-3p, miR-151b and miR-27b-3p as biomarkers in acute ischemic stroke. Biosci. Rep. 2018, 38, 38. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, T.; Zhang, G.; Wu, P.; Duan, L.; Li, G.; Liu, Q.; Wang, J. Dissection of Myogenic Differentiation Signatures in Chickens by RNA-Seq Analysis. Genes 2018, 9, 34. [Google Scholar] [CrossRef] [Green Version]
- Gu, S.; Jin, L.; Zhang, F.; Sarnow, P.; Kay, M.A. Biological basis for restriction of microRNA targets to the 3′ untranslated region in mammalian mRNAs. Nat. Struct. Mol. Biol. 2009, 16, 144–150. [Google Scholar] [CrossRef] [Green Version]
- Yin, H.; He, H.; Shen, X.; Zhao, J.; Cao, X.; Han, S.; Cui, C.; Chen, Y.; Wei, Y.; Xia, L.; et al. miR-9-5p Inhibits Skeletal Muscle Satellite Cell Proliferation and Differentiation by Targeting IGF2BP3 through the IGF2-PI3K/Akt Signaling Pathway. Int. J. Mol. Sci. 2020, 21, 1655. [Google Scholar] [CrossRef] [Green Version]
- Sun, Y.; Xu, T.; Cao, Y.-W.; Ding, X.-Q. Antitumor effect of miR-27b-3p on lung cancer cells via targeting Fzd7. Eur. Rev. Med Pharmacol. Sci. 2017, 21, 4113–4123. [Google Scholar] [PubMed]
- Chen, D.; Si, W.; Shen, J.; Du, C.; Lou, W.; Bao, C.; Zheng, H.; Pan, J.; Zhong, G.; Xu, L.; et al. miR-27b-3p inhibits proliferation and potentially reverses multi-chemoresistance by targeting CBLB/GRB2 in breast cancer cells. Cell Death Dis. 2018, 9, 1–13. [Google Scholar] [CrossRef]
- Zhang, C.; Zou, Y.; Dai, D.-Q. Downregulation of microRNA-27b-3p via aberrant DNA methylation contributes to malignant behavior of gastric cancer cells by targeting GSPT1. Biomed. Pharmacother. 2019, 119, 109417. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Chen, F.; Wu, P.; Li, T.; He, M.; Yin, X.; Shi, H.; Duan, Y.; Zhang, T.; Wang, J.; et al. MicroRNA-7 Targets the KLF4 Gene to Regulate the Proliferation and Differentiation of Chicken Primary Myoblasts. Front. Genet. 2020, 11, 842. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, Y.; Yulin, B.; Tang, B.; Wang, M.; Zhang, C.; Zhang, W.; Jin, J.; Li, T.; Zhao, R.; et al. CRISPR/Cas9-mediated sheep MSTN gene knockout and promote sSMSCs differentiation. J. Cell. Biochem. 2019, 120, 1794–1806. [Google Scholar] [CrossRef]
- Taylor, W.E.; Bhasin, S.; Artaza, J.; Byhower, F.; Azam, M.; Willard, D.H., Jr.; Kull, F.C., Jr.; Gonzalez-Cadavid, N. Myostatin inhibits cell proliferation and protein synthesis in C2C12 muscle cells. Am. J. Physiol. Metab. 2001, 280, E221–E228. [Google Scholar] [CrossRef] [Green Version]
- Ríos, R.; Carneiro, I.; Arce, V.M.; Devesa, J.; Rios, R. Myostatin is an inhibitor of myogenic differentiation. Am. J. Physiol. Physiol. 2002, 282, C993–C999. [Google Scholar] [CrossRef] [Green Version]
- Berkes, C.A.; Tapscott, S.J. MyoD and the transcriptional control of myogenesis. Semin. Cell Dev. Biol. 2005, 16, 585–595. [Google Scholar] [CrossRef]
- Edmondson, D.G.; Olson, E.N. A gene with homology to the myc similarity region of MyoD1 is expressed during myogenesis and is sufficient to activate the muscle differentiation program. Genes Dev. 1990, 4, 1450. [Google Scholar] [CrossRef] [Green Version]
- Toniolo, L.; Maccatrozzo, L.; Patruno, M.; Pavan, E.; Caliaro, F.; Rossi, R.; Rinaldi, C.; Canepari, M.; Reggiani, C.; Mascarello, F. Fiber types in canine muscles: Myosin isoform expression and functional characterization. Am. J. Physiol. Physiol. 2007, 292, C1915–C1926. [Google Scholar] [CrossRef] [PubMed]
- Brennecke, J.; Stark, A.; Russell, R.B.; Cohen, S.M. Principles of microRNA-target recognition. PLoS Biol. 2005, 3, e85. [Google Scholar] [CrossRef] [PubMed]

| Gene | Primer Sequence (5′-3′) | Annealing Temperature (°C) |
|---|---|---|
| Gga-miR-27b-3p | F:GCGCGTTCACAGTGGCTAAG | 60 |
| R:AGTGCAGGGTCCGAGGTATT | ||
| Stem-loop primer | GTCGTATCCAGTGCAGGGTCCGAGG | |
| TATTCGCACTGGATACGACGCAGAA | ||
| U6 | F:GTCACTTCTGGTGGCGGTAA | 60 |
| R:GTTCAGTAGAGGGTCAAA |
| Gene | Primer Sequence (5′-3′) | Product Size (bp) | Annealing Temperature (°C) | Accession Number |
|---|---|---|---|---|
| MSTN | F:GCTTTTACCCAAAGCTCCTCCAC | 179 | 60 | NM_001001461.1 |
| R:AGCAACATTTTGGTTTTCCCTCC | ||||
| P21 | F:GAGATGCTGAAGGAGATCAATGAG | 102 | 60 | NM_204396.1 |
| R:GTGGTCAGTCCGAGCCTTTT | ||||
| MYOD1 | F:GCTACTACACGGAATCACCAAAT | 200 | 60 | NM_204214.2 |
| R:CTGGGCTCCACTGTCACTCA | ||||
| MYHC | F:CTCCTCACGCTTTGGTAA | 213 | 60 | NM_001319304.1 |
| MYOG | R:TGATAGTCGTATGGGTTGGT | 320 | 60 | NM_204184.1 |
| F:CGGAGGCTGAAGAAGGTGAA | ||||
| R:CGGTCCTCTGCCTGGTCAT | ||||
| β-actin | F:CAGCCATCTTTCTTGGGTAT | 169 | 60 | NM_205518.1 |
| R:CTGTGATCTCCTTCTGCATCC |
| Fragment Name | Sequence (5′-3′) |
|---|---|
| miR-27b-3p mimic | UUCACAGUGGCUAAGUUCUGC |
| AGAACUUAGCCACUGUGAAUU | |
| mimic-NC | UUCUCCGAACGUGUCACGUTT |
| ACGUGACACGUUCGGAGAATT | |
| miR-27b-3p inhibitor | GCAGAACUUAGCCACUGUGAA |
| inhibitor-NC | CAGUACUUUUGUGUAGUACAA |
| siR-MSTN | GCAGAUCCUGAGACUCAUUTT |
| AAUGAGUCUCAGGAUCUGCTT | |
| siR-NC | UUCUCCGAACGUGUCACGUTT |
| ACGUGACACGUUCGGAGAATT |
| Primer Name | Primer Sequence (5′-3′) | Product Size (bp) | Annealing Temperature (°C) |
|---|---|---|---|
| MSTN-3′ UTR-WT | F:AAAGATCCTTTATTAAGCTTCTGTCGTGAGATCCACCATT | 298 | 62 |
| R:ATAGGCCGGCATAGACGCGTCCCATTTGTTAAATCCGGTG | |||
| MSTN-3′ UTR-MT | F:TTGAACGCCACGCATTACGTACGCTAGGCATTGCC | 6742 | 68 |
| R: GTAATGCGTGGCGTTCAAAGATGGATGAGGGGATATAG |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, G.; He, M.; Wu, P.; Zhang, X.; Zhou, K.; Li, T.; Zhang, T.; Xie, K.; Dai, G.; Wang, J. MicroRNA-27b-3p Targets the Myostatin Gene to Regulate Myoblast Proliferation and Is Involved in Myoblast Differentiation. Cells 2021, 10, 423. https://doi.org/10.3390/cells10020423
Zhang G, He M, Wu P, Zhang X, Zhou K, Li T, Zhang T, Xie K, Dai G, Wang J. MicroRNA-27b-3p Targets the Myostatin Gene to Regulate Myoblast Proliferation and Is Involved in Myoblast Differentiation. Cells. 2021; 10(2):423. https://doi.org/10.3390/cells10020423
Chicago/Turabian StyleZhang, Genxi, Mingliang He, Pengfei Wu, Xinchao Zhang, Kaizhi Zhou, Tingting Li, Tao Zhang, Kaizhou Xie, Guojun Dai, and Jinyu Wang. 2021. "MicroRNA-27b-3p Targets the Myostatin Gene to Regulate Myoblast Proliferation and Is Involved in Myoblast Differentiation" Cells 10, no. 2: 423. https://doi.org/10.3390/cells10020423






