Statins as a Therapeutic Approach for the Treatment of Pseudoxanthoma Elasticum Patients: Evaluation of the Spectrum Efficacy of Atorvastatin In Vitro
Abstract
:1. Introduction
2. Materials and Methods
2.1. Experimental Design
2.2. Cell Culture
2.3. Delipidation of FCS
2.4. Nucleic Acid Isolation
2.5. Gene Expression Analysis
2.6. Immunoassays for Evaluation of SASP Factors in Cell Culture Supernatants
2.7. Evaluation of ENPP1 Activity in Cell Culture Supernatants
2.8. Statistical Analysis
3. Results
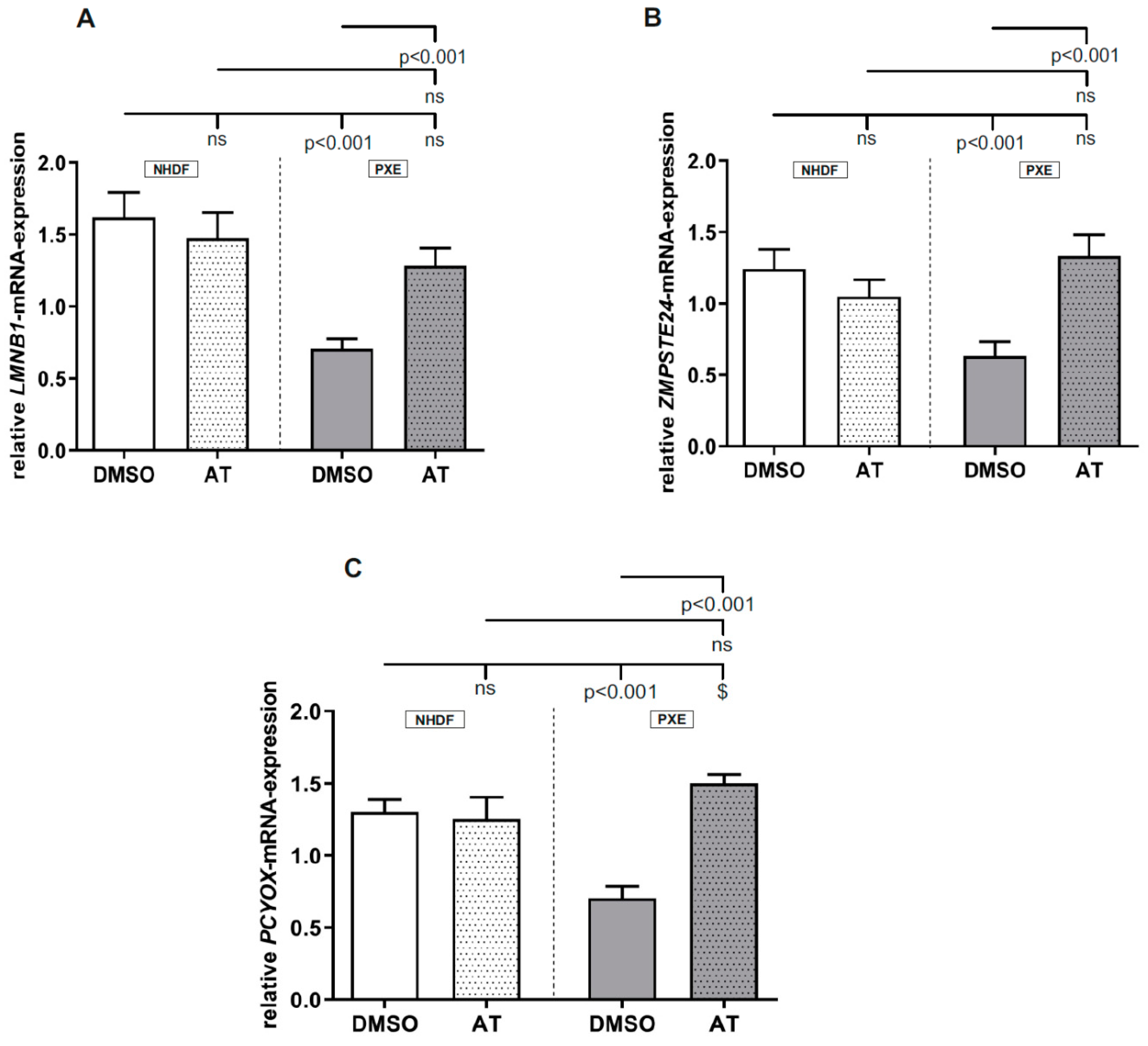
3.1. Atorvastatin Has Positive Effects on Gene Expression of Targets Linked to Cholesterol Biosynthesis
3.2. Atorvastatin Has Positive Effects on Gene Expression of Factors Involved in Prenylation Processes
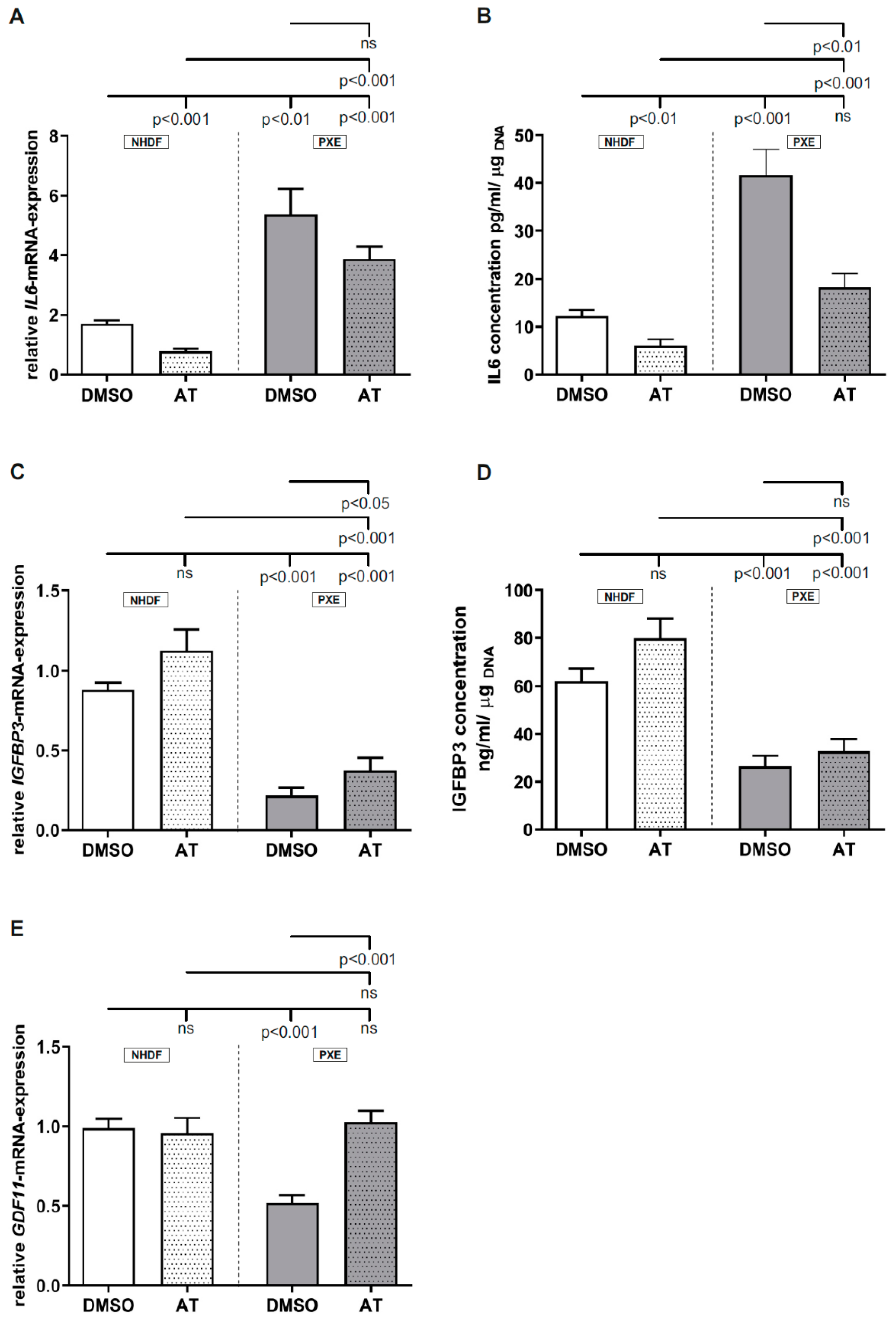
3.3. Limited Effect of Atorvastatin on Inflammatory Markers
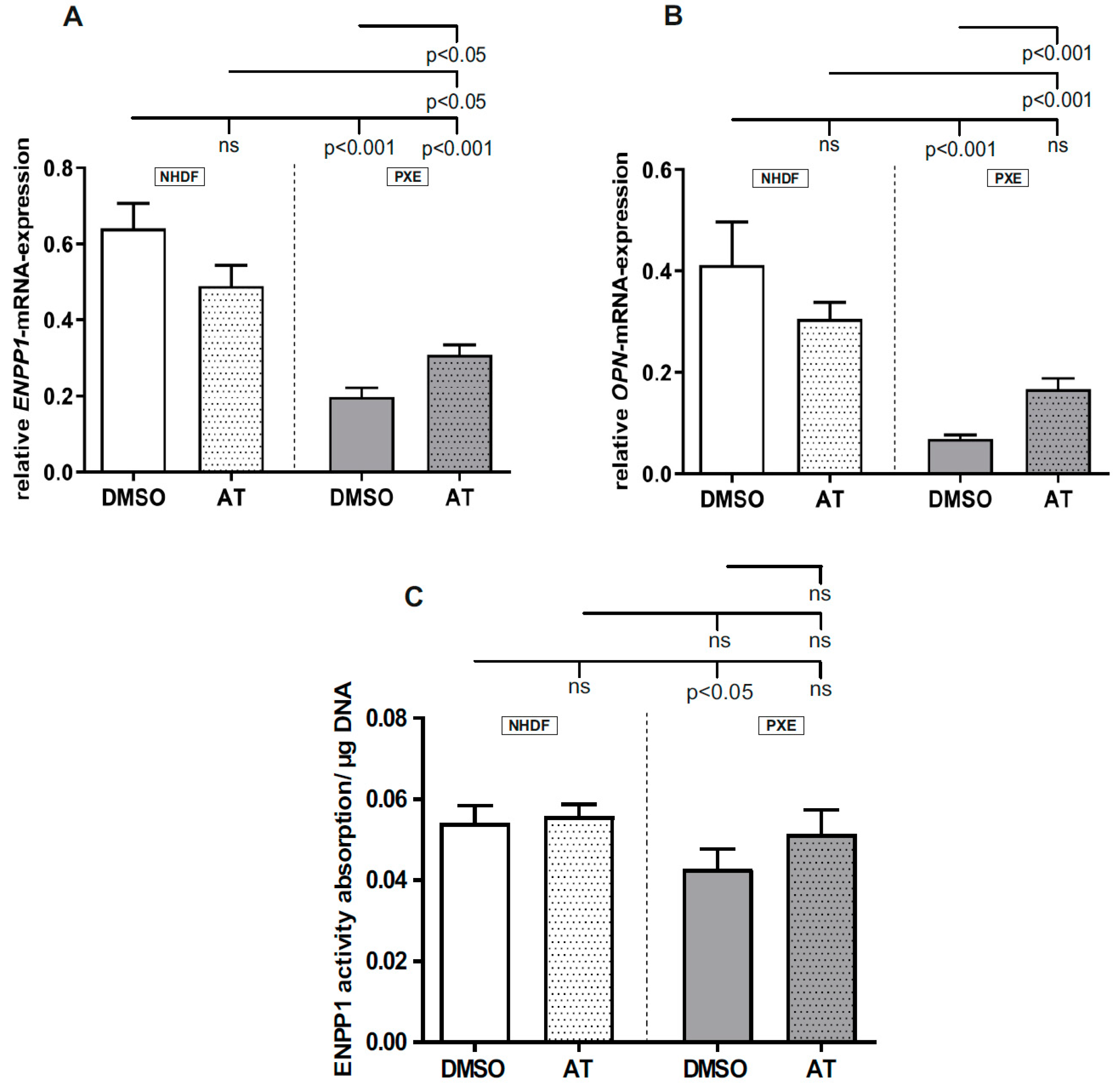
3.4. Limited Effect of Atorvastatin on Gene Expression of Calcification-Associated Factors
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Germain, D.P. Pseudoxanthoma Elasticum. Orphanet. J. Rare Dis. 2017, 12, 85. [Google Scholar] [CrossRef]
- Neldner, K.H. Pseudoxanthoma Elasticum. Clin. Derm. 1988, 6, 1–159. [Google Scholar] [CrossRef]
- Li, Q.; van de Wetering, K.; Uitto, J. Pseudoxanthoma Elasticum as a Paradigm of Heritable Ectopic Mineralization Disorders. Am. J. Pathol. 2019, 189, 216–225. [Google Scholar] [CrossRef] [Green Version]
- Miki, K.; Yuri, T.; Takeda, N.; Takehana, K.; Iwasaka, T.; Tsubura, A. An Autopsy Case of Pseudoxanthoma Elasticum: Histochemical Characteristics. Med. Mol. Morphol. 2007, 40, 172–177. [Google Scholar] [CrossRef] [PubMed]
- Mendelsohn, G.; Bulkley, B.H.; Hutchins, G.M. Cardiovascular Manifestations of Pseudoxanthoma Elasticum. Arch. Pathol. Lab. Med. 1978, 102, 298–302. [Google Scholar]
- Verschuere, S.; Van Gils, M.; Nollet, L.; Vanakker, O.M. From Membrane to Mineralization: The Curious Case of the ABCC6 Transporter. Febs. Lett. 2020, 594, 4109–4133. [Google Scholar] [CrossRef]
- Kornet, L.; Bergen, A.A.B.; Hoeks, A.P.G.; Cleutjens, J.P.; Oostra, R.-J.; Daemen, M.J.; van Soest, S.; Reneman, R.S. In Patients with Pseudoxanthoma Elasticum a Thicker and More Elastic Carotid Artery Is Associated with Elastin Fragmentation and Proteoglycans Accumulation. Ultrasound Med. Biol. 2004, 30, 1041–1048. [Google Scholar] [CrossRef] [PubMed]
- Lefthériotis, G.; Abraham, P.; Le Corre, Y.; Le Saux, O.; Henrion, D.; Ducluzeau, P.H.; Prunier, F.; Martin, L. Relationship between Ankle Brachial Index and Arterial Remodeling in Pseudoxanthoma Elasticum. J. Vasc. Surg. 2011, 54, 1390–1394. [Google Scholar] [CrossRef] [Green Version]
- Le Saux, O.; Urban, Z.; Tschuch, C.; Csiszar, K.; Bacchelli, B.; Quaglino, D.; Pasquali-Ronchetti, I.; Pope, F.M.; Richards, A.; Terry, S.; et al. Mutations in a Gene Encoding an ABC Transporter Cause Pseudoxanthoma Elasticum. Nat. Genet. 2000, 25, 223–227. [Google Scholar] [CrossRef] [PubMed]
- Legrand, A.; Cornez, L.; Samkari, W.; Mazzella, J.-M.; Venisse, A.; Boccio, V.; Auribault, K.; Keren, B.; Benistan, K.; Germain, D.P.; et al. Mutation Spectrum in the ABCC6 Gene and Genotype–Phenotype Correlations in a French Cohort with Pseudoxanthoma Elasticum. Genet. Med. 2017, 19, 909–917. [Google Scholar] [CrossRef] [PubMed]
- Jansen, R.S.; Kucukosmanoglu, A.; de Haas, M.; Sapthu, S.; Otero, J.A.; Hegman, I.E.M.; Bergen, A.A.B.; Gorgels, T.G.M.F.; Borst, P.; van de Wetering, K. ABCC6 Prevents Ectopic Mineralization Seen in Pseudoxanthoma Elasticum by Inducing Cellular Nucleotide Release. Proc. Natl. Acad. Sci. USA 2013, 110, 20206–20211. [Google Scholar] [CrossRef] [Green Version]
- Jansen, R.S.; Duijst, S.; Mahakena, S.; Sommer, D.; Szeri, F.; Váradi, A.; Plomp, A.; Bergen, A.A.; Oude Elferink, R.P.J.; Borst, P.; et al. ABCC6–Mediated ATP Secretion by the Liver Is the Main Source of the Mineralization Inhibitor Inorganic Pyrophosphate in the Systemic Circulation—Brief Report. Arter. Thromb. Vasc. Biol. 2014, 34, 1985–1989. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boraldi, F.; Annovi, G.; Bartolomeo, A.; Quaglino, D. Fibroblasts from Patients Affected by Pseudoxanthoma Elasticum Exhibit an Altered PPi Metabolism and Are More Responsive to Pro-Calcifying Stimuli. J. Derm. Sci. 2014, 74, 72–80. [Google Scholar] [CrossRef] [Green Version]
- Boraldi, F.; Annovi, G.; Vermeer, C.; Schurgers, L.J.; Trenti, T.; Tiozzo, R.; Guerra, D.; Quaglino, D. Matrix Gla Protein and Alkaline Phosphatase Are Differently Modulated in Human Dermal Fibroblasts from PXE Patients and Controls. J. Investig. Dermatol. 2013, 133, 946–954. [Google Scholar] [CrossRef] [Green Version]
- Salamat, M.; Dhar, P.K.; Neagu, D.L.; Lyon, J.B. Aortic Calcification in a Patient with Hutchinson–Gilford Progeria Syndrome. Pediatr. Cardiol. 2010, 31, 925–926. [Google Scholar] [CrossRef] [PubMed]
- Gonzalo, S.; Kreienkamp, R.; Askjaer, P. Hutchinson-Gilford Progeria Syndrome: A Premature Aging Disease Caused by LMNA Gene Mutations. Ageing Res. Rev. 2017, 33, 18–29. [Google Scholar] [CrossRef] [Green Version]
- Tiemann, J.; Wagner, T.; Vanakker, O.M.; van Gils, M.; Cabrera, J.-L.B.; Ibold, B.; Faust, I.; Knabbe, C.; Hendig, D. Cellular and Molecular Biomarkers Indicate Premature Aging in Pseudoxanthoma Elasticum Patients. Aging Dis. 2020, 11, 536. [Google Scholar] [CrossRef]
- Villa-Bellosta, R.; Rivera-Torres, J.; Osorio, F.G.; Acín-Pérez, R.; Enriquez, J.A.; López-Otín, C.; Andrés, V. Defective Extracellular Pyrophosphate Metabolism Promotes Vascular Calcification in a Mouse Model of Hutchinson-Gilford Progeria Syndrome That Is Ameliorated on Pyrophosphate Treatment. Circulation 2013, 127, 2442–2451. [Google Scholar] [CrossRef] [Green Version]
- Guo, H.; Li, Q.; Chou, D.W.; Uitto, J. Atorvastatin counteracts Aberrant Soft Tissue Mineralization in a Mouse Model of Pseudoxanthoma Elasticum (Abcc6−/−). J. Mol. Med. 2013, 91, 1177–1184. [Google Scholar] [CrossRef] [Green Version]
- Luft, F.C. Pseudoxanthoma Elasticum and Statin Prophylaxis. J. Mol. Med. 2013, 91, 1129–1130. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Istvan, E.S. Structural Mechanism for Statin Inhibition of HMG-CoA Reductase. Science 2001, 292, 1160–1164. [Google Scholar] [CrossRef] [Green Version]
- Rauch, U.; Osende, J.I.; Chesebro, J.H.; Fuster, V.; Vorchheimer, D.A.; Harris, K.; Harris, P.; Sandler, D.A.; Fallon, J.T.; Jayaraman, S.; et al. Statins and Cardiovascular Diseases: The Multiple Effects of Lipid-Lowering Therapy by Statins. Atherosclerosis 2000, 153, 181–189. [Google Scholar] [CrossRef]
- Wang, C.-Y.; Liu, P.-Y.; Liao, J.K. Pleiotropic Effects of Statin Therapy: Molecular Mechanisms and Clinical Results. Trends Mol. Med. 2008, 14, 37–44. [Google Scholar] [CrossRef] [Green Version]
- Maeda, T.; Kawane, T.; Horiuchi, N. Statins Augment Vascular Endothelial Growth Factor Expression in Osteoblastic Cells via Inhibition of Protein Prenylation. Endocrinology 2003, 144, 681–692. [Google Scholar] [CrossRef]
- Kuzaj, P.; Kuhn, J.; Dabisch-Ruthe, M.; Faust, I.; Götting, C.; Knabbe, C.; Hendig, D. ABCC6- a New Player in Cellular Cholesterol and Lipoprotein Metabolism? Lipids Health Dis. 2014, 13, 118. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gibson, K.M.; Hoffmann, G.; Schwall, A.; Broock, R.L.; Aramaki, S.; Sweetman, L.; Nyhan, W.L.; Brandt, I.K.; Wappner, R.S.; Lehnert, W. 3-Hydroxy-3-Methylglutaryl Coenzyme A Reductase Activity in Cultured Fibroblasts from Patients with Mevalonate Kinase Deficiency: Differential Response to Lipid Supplied by Fetal Bovine Serum in Tissue Culture Medium. J. Lipid Res. 1990, 31, 515–521. [Google Scholar] [CrossRef]
- Lau, W.M.; Doucet, M.; Stadel, R.; Huang, D.; Weber, K.L.; Kominsky, S.L. Enpp1: A Potential Facilitator of Breast Cancer Bone Metastasis. PLoS ONE 2013, 8, e66752. [Google Scholar] [CrossRef] [PubMed]
- Goldstein, J.L.; Brown, M.S. Regulation of the Mevalonate Pathway. Nature 1990, 343, 425–430. [Google Scholar] [CrossRef] [PubMed]
- McKnight, G.L.; Reasoner, J.; Gilbert, T.; Sundquist, K.O.; Hokland, B.; McKernan, P.A.; Champagne, J.; Johnson, C.J.; Bailey, M.C.; Holly, R. Cloning and Expression of a Cellular High Density Lipoprotein-Binding Protein That Is up-Regulated by Cholesterol Loading of Cells. J. Biol. Chem. 1992, 267, 12131–12141. [Google Scholar] [CrossRef]
- Chiu, D.S.; Oram, J.F.; LeBoeuf, R.C.; Alpers, C.E.; O’Brien, K.D. High-Density Lipoprotein-Binding Protein (HBP)/Vigilin Is Expressed in Human Atherosclerotic Lesions and Colocalizes With Apolipoprotein E. ATVB 1997, 17, 2350–2358. [Google Scholar] [CrossRef]
- Lacher, S.M.; Bruttger, J.; Kalt, B.; Berthelet, J.; Rajalingam, K.; Wörtge, S.; Waisman, A. HMG-CoA Reductase Promotes Protein Prenylation and Therefore Is Indispensible for T-Cell Survival. Cell Death Dis. 2017, 8, e2824. [Google Scholar] [CrossRef] [PubMed]
- Zineh, I.; Luo, X.; Welder, G.J.; Debella, A.E.; Wessel, T.R.; Arant, C.B.; Schofield, R.S.; Chegini, N. Modulatory Effects of AT on Endothelial Cell-Derived Chemokines, Cytokines, and Angiogenic Factors. Pharmacotherapy 2006, 26, 333–340. [Google Scholar] [CrossRef]
- Omoigui, S. The Interleukin-6 Inflammation Pathway from Cholesterol to Aging--Role of Statins, Bisphosphonates and Plant Polyphenols in Aging and Age-Related Diseases. Immun. Ageing 2007, 4, 1. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Narayanan, R.P.; Gittins, M.; Siddals, K.W.; Oliver, R.L.; Hudson, J.E.; White, A.; Durrington, P.; Davies, R.R.; Rutter, M.K.; Gibson, J.M. AT Administration Is Associated with Dose-Related Changes in IGF Bioavailability. Eur. J. Endocrinol. 2013, 168, 543–548. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oliver, W.T.; Rosenberger, J.; Lopez, R.; Gomez, A.; Cummings, K.K.; Fiorotto, M.L. The Local Expression and Abundance of Insulin-like Growth Factor (IGF) Binding Proteins in Skeletal Muscle Are Regulated by Age and Gender but Not Local IGF-I in Vivo. Endocrinology 2005, 146, 5455–5462. [Google Scholar] [CrossRef] [Green Version]
- Mei, W.; Xiang, G.; Li, Y.; Li, H.; Xiang, L.; Lu, J.; Xiang, L.; Dong, J.; Liu, M. GDF11 Protects against Endothelial Injury and Reduces Atherosclerotic Lesion Formation in Apolipoprotein E-Null Mice. Mol. Ther. 2016, 24, 1926–1938. [Google Scholar] [CrossRef] [Green Version]
- Dabisch-Ruthe, M.; Kuzaj, P.; Götting, C.; Knabbe, C.; Hendig, D. Pyrophosphates as a Major Inhibitor of Matrix Calcification in Pseudoxanthoma Elasticum. J. Derm. Sci. 2014, 75, 109–120. [Google Scholar] [CrossRef]
- Nam, H.K.; Liu, J.; Li, Y.; Kragor, A.; Hatch, N.E. Ectonucleotide Pyrophosphatase/Phosphodiesterase-1 (ENPP1) Protein Regulates Osteoblast Differentiation. J. Biol. Chem. 2011, 286, 39059–39071. [Google Scholar] [CrossRef] [Green Version]
- Fleisch, H.; Bisaz, S. Mechanism of Calcification: Inhibitory Role of Pyrophosphate. Nature 1962, 195, 911. [Google Scholar] [CrossRef]
- Lomashvili, K.A.; Narisawa, S.; Millán, J.L.; O’Neill, W.C. Vascular Calcification Is Dependent on Plasma Levels of Pyrophosphate. Kidney Int. 2014, 85, 1351–1356. [Google Scholar] [CrossRef] [Green Version]
- Boskey, A.L.; Maresca, M.; Ullrich, W.; Doty, S.B.; Butler, W.T.; Prince, C.W. Osteopontin-Hydroxyapatite Interactions in Vitro: Inhibition of Hydroxyapatite Formation and Growth in a Gelatin-Gel. Bone Min. 1993, 22, 147–159. [Google Scholar] [CrossRef]
- Giachelli, C.M.; Steitz, S. Osteopontin: A Versatile Regulator of Inflammation and Biomineralization. Matrix Biol. 2000, 19, 615–622. [Google Scholar] [CrossRef]

| Sample ID | Gender | Age 1 | Biopsy Source | ABCC6 Genotype 2 | Genotype Status | Phenodex Score 3 | |
|---|---|---|---|---|---|---|---|
| PXE Patients | |||||||
| P3M a | Male | 57 | Neck | c.3421C > T (p.(Arg1141*) | c.3883 − 6G > A (SSM) | cht | S3, V2; C0 |
| P128M a | Male | 51 | Neck | c.3769_3770insC (p.L1259fsX1277) | c.3769_3770insC (p.L1259fsX1277) | hm | S2; E2; G0; C1 |
| P255F a | Female | 48 | Arm | c.3421C > T (p.Arg1141*) | c.2787 + 1G > T | cht | S3; E2; G0; C0 |
| Healthy Controls | |||||||
| M57A b (AG13145) | Male | 57 | Arm | - | - | wt | Not applicable |
| M52A b (AG11482) | Male | 52 | Arm | - | - | wt | Not applicable |
| F48A b (AG14284) | Female | 48 | Arm | - | - | wt | Not applicable |
| Gene | Protein | 5′-3′ Sequence | Reference 1 | Annealing Temperature (°C) | Efficiency |
|---|---|---|---|---|---|
| β-ACTIN Beta-actin | ß-Actin | CGCGAGAAGATGACCC ATTGCCAATGGTGATGAC | NM_001101 | 59 | 2.0 |
| GAPDH Glycerinaldehyd-3-phosphat-dehydrogenase | GAPDH | AGGTCGGAGTCAACGGAT TCCTGGAAGATGGTGATG | NM_002046 | 59 | 1.8 |
| β2M Beta-2- microglobulin | ß2M | TGTGCTCGCGCTACTCTCTCTT CGGATGGATGAAACCCAGACA | NM_004048 | 59 | 2.0 |
| ENPP1 Ectonucleotide pyrophosphatase/phosphodiesterase 1 | ENPP1 | AATGCCCCTTTGGACACT CCCGTAACTTTTGGT | NM 006,208 | 59 | 1.8 |
| IL6 Interleukin 6 | IL6 | ACAGCCACTCACCTCTTCAG GTGCCTCTTTGCTGCTTTCAC | NM 000600.4 | 63 | 1.9 |
| LMNB1 Lamin B1 | Lamin B1 | GCAGACTTACCATGCCAAAC TCCCTTATTTCCGCCATCTC | NM 005573.3 | 63 | 1.9 |
| FDPS Farnesyldiphosphate synthase | FDPS | TTGCTCCTCCCTCAGAATGAACGTGCCTCCAATGGCATTGTACTC | NM_001135821.1 | 59 | 1.8 |
| HDLBP High-density lipoprotein-binding protein | HDLBP | GAACGCAGTTCACAGTACAG GTAACGGTGAAGGTCAAAGG | NM_005336 | 60 | 1.6 |
| HMGCR HMG-CoA Reduktase | HMGCR | AAGTTTGCCCTCAGTTCC ACTGACATGCAGCCAAAG | NM 000859.2 | 59 | 1.9 |
| IGFBP3 Insulin-like growth factor-binding protein 3 | IGFBP3 | GCGCCAGGAAATGCTAGTGA GGGAATGTGTACACCCCTGG | NM_001013398.1 | 63 | 1.8 |
| LDLR Low-density lipoprotein receptor | LDLR | CGACTGCAAGGACAAATCTG AGTCATATTCCCGGTACAC | NM_000527 | 60 | 1.6 |
| OPN Osteopontin | OPN | TGATGACCATGTGGACAG ACCATTCAACTCCTCGCT | NM 000,900 | 63 | 1.8 |
| PCYOX Prenylcysteinoxidase 1 | PCYOX | CACTTCAGCAGCCTATTACC CCAGTTGCTCTCAAATACC | NM 016,297 | 59 | 1.9 |
| ZMPSTE24 Zinc metalloprotease STE24 | ZMPSTE24 | ACCACCGGAGTTAGGACAGA CCAGGGACTGAGTGATCTCAT | NM_005857.5 | 66 | 1.9 |
| GDF11 Growth differentiation factor 11 | GDF11 | AGGCCATTGGCAGAGCATCGAC GTCCCAGCCGAAAGCCTCAAAG | NM_005811.3 | 63 | 2.0 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tiemann, J.; Lindenkamp, C.; Plümers, R.; Faust, I.; Knabbe, C.; Hendig, D. Statins as a Therapeutic Approach for the Treatment of Pseudoxanthoma Elasticum Patients: Evaluation of the Spectrum Efficacy of Atorvastatin In Vitro. Cells 2021, 10, 442. https://doi.org/10.3390/cells10020442
Tiemann J, Lindenkamp C, Plümers R, Faust I, Knabbe C, Hendig D. Statins as a Therapeutic Approach for the Treatment of Pseudoxanthoma Elasticum Patients: Evaluation of the Spectrum Efficacy of Atorvastatin In Vitro. Cells. 2021; 10(2):442. https://doi.org/10.3390/cells10020442
Chicago/Turabian StyleTiemann, Janina, Christopher Lindenkamp, Ricarda Plümers, Isabel Faust, Cornelius Knabbe, and Doris Hendig. 2021. "Statins as a Therapeutic Approach for the Treatment of Pseudoxanthoma Elasticum Patients: Evaluation of the Spectrum Efficacy of Atorvastatin In Vitro" Cells 10, no. 2: 442. https://doi.org/10.3390/cells10020442
APA StyleTiemann, J., Lindenkamp, C., Plümers, R., Faust, I., Knabbe, C., & Hendig, D. (2021). Statins as a Therapeutic Approach for the Treatment of Pseudoxanthoma Elasticum Patients: Evaluation of the Spectrum Efficacy of Atorvastatin In Vitro. Cells, 10(2), 442. https://doi.org/10.3390/cells10020442






