Importance of between and within Subject Variability in Extracellular Vesicle Abundance and Cargo when Performing Biomarker Analyses
Abstract
1. Introduction
2. Materials and Methods
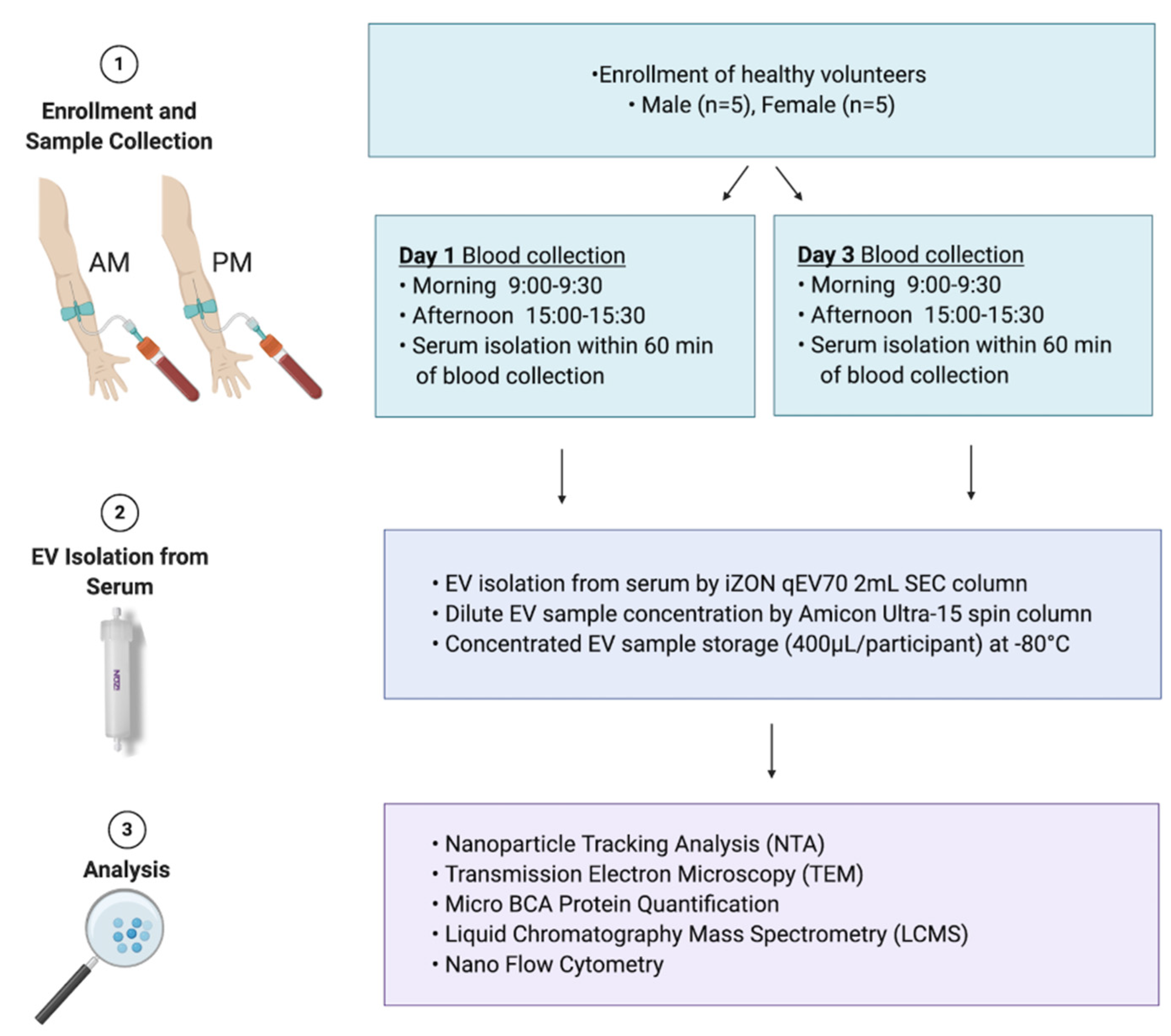
2.1. Study Cohort
2.2. Collection of Blood/Serum
2.3. Extracellular Vesicle Isolation
2.4. Human Liver Microsome Preparation
2.5. Nanoparticle Tracking Analysis
2.6. Transmission Electron Microscopy (TEM)
2.7. Micro BCA Protein Quantification
2.8. Trypsin Digest
2.9. Liquid Chromatography Mass Spectrometry (LCMS)
2.10. Nano Flow Cytometry (nFC)
2.11. Statistical Analysis
2.12. EV-TRACK
3. Results
3.1. Purity Assessment of EV Isolations from Serum
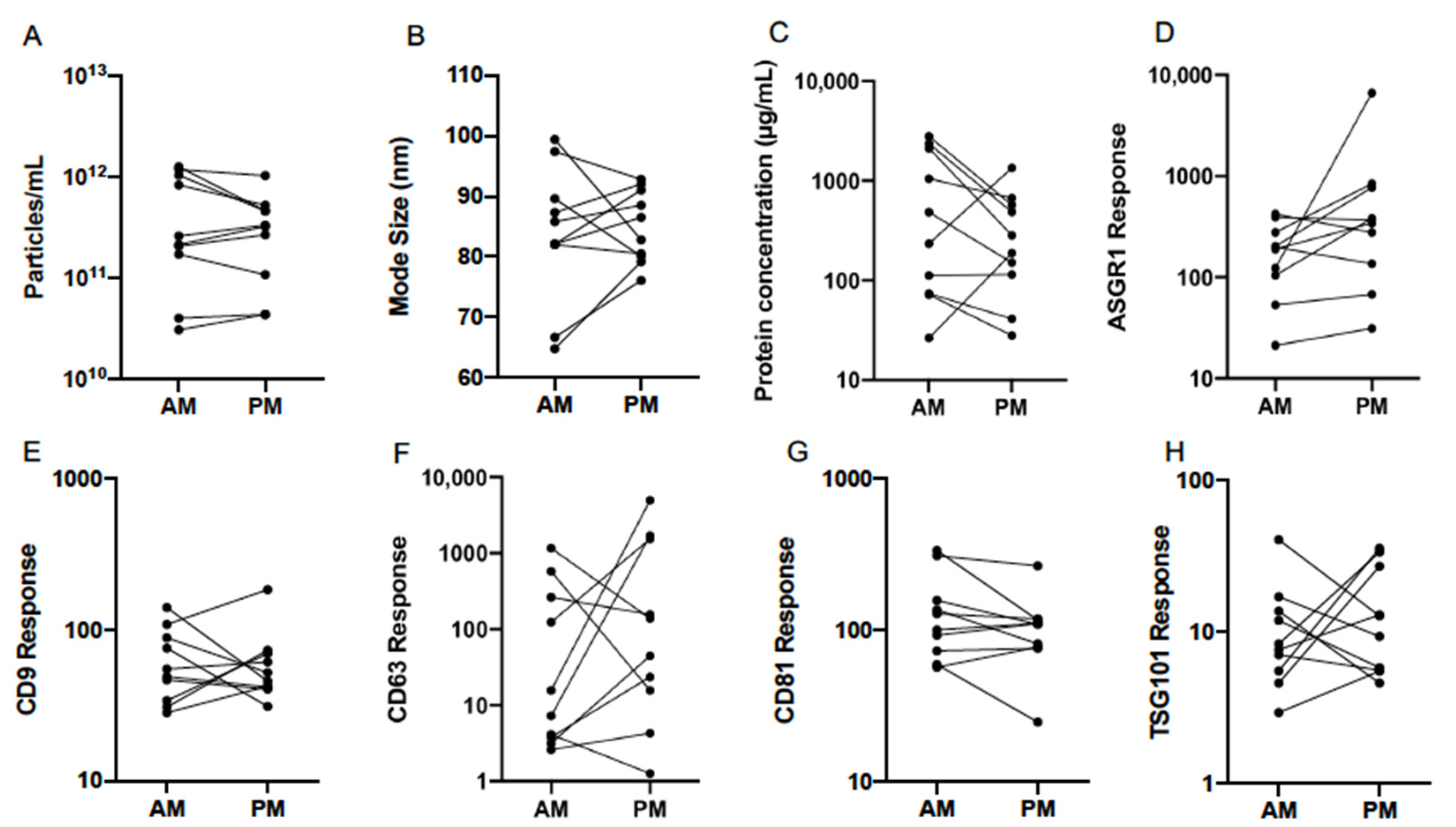
3.2. Normal Variability
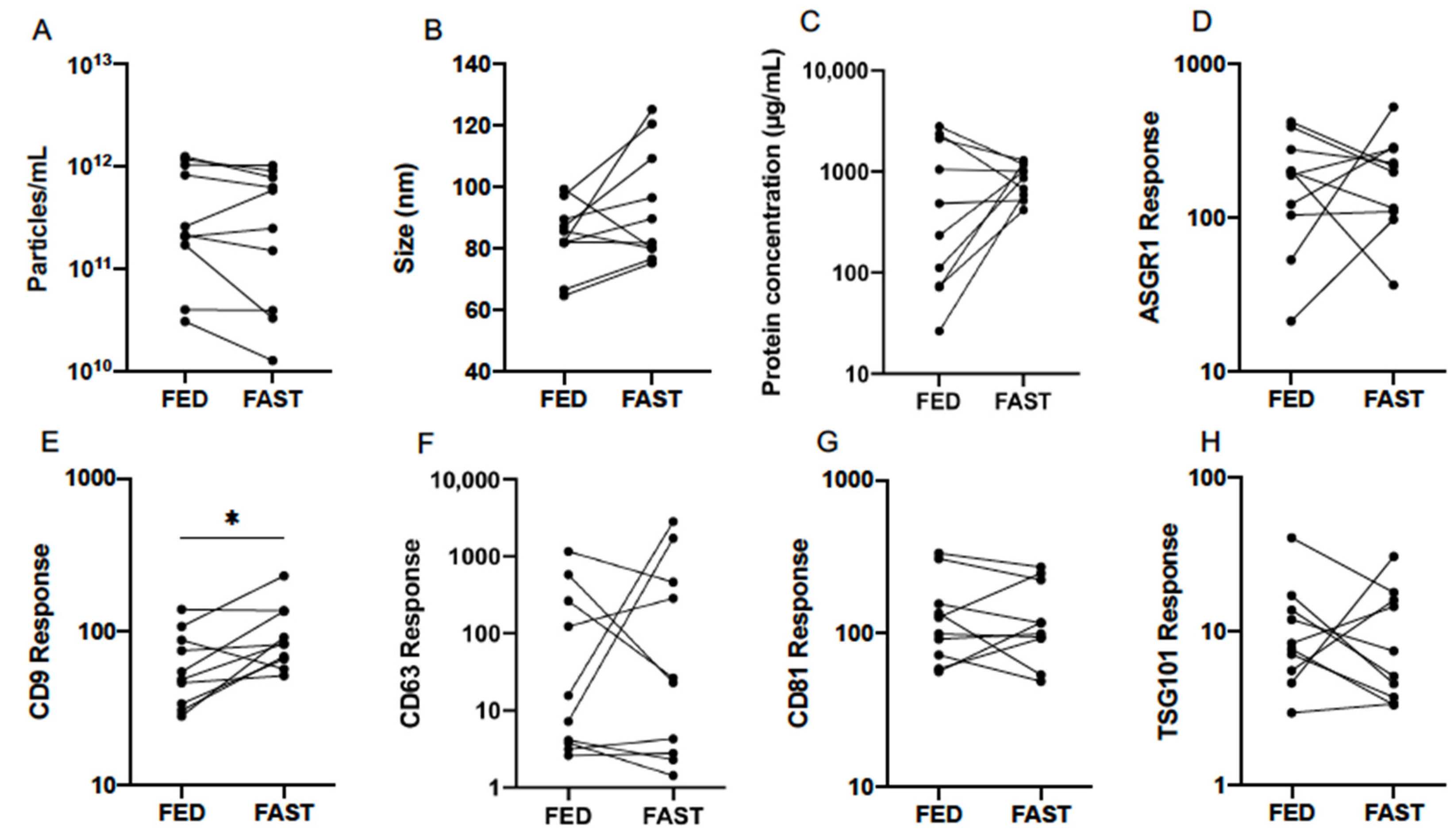
3.3. Effect of Fasting
3.4. Diurnal Variability
3.5. Effect of Sex
3.6. Single EV Analysis by Nano Flow Cytometry
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Shah, R.; Patel, T.; Freedman, J.E. Circulating Extracellular Vesicles in Human Disease. N. Engl. J. Med. 2018, 379, 958–966. [Google Scholar] [CrossRef]
- Rowland, A.; Ruanglertboon, W.; Van Dyk, M.; Wijayakumara, D.; Wood, L.S.; Meech, R.; MacKenzie, P.I.; Rodrigues, A.D.; Marshall, J.; Sorich, M.J. Plasma extracellular nanovesicle (exosome)-derived biomarkers for drug metabolism pathways: A novel approach to characterize variability in drug exposure. Br. J. Clin. Pharmacol. 2019, 85, 216–226. [Google Scholar] [CrossRef]
- Rodrigues, D.; Rowland, A. From Endogenous Compounds as Biomarkers to Plasma-Derived Nanovesicles as Liquid Biopsy; Has the Golden Age of Translational Pharmacokinetics-Absorption, Distribution, Metabolism, Excretion-Drug–Drug Interaction Science Finally Arrived? Clin. Pharmacol. Ther. 2019, 105, 1407–1420. [Google Scholar] [CrossRef]
- Newman, L.A.; Sorich, M.J.; Rowland, A. Role of Extracellular Vesicles in the Pathophysiology, Diagnosis and Tracking of Non-Alcoholic Fatty Liver Disease. J. Clin. Med. 2020, 9, 2032. [Google Scholar] [CrossRef]
- Jeppesen, D.K.; Fenix, A.M.; Franklin, J.L.; Higginbotham, J.N.; Zhang, Q.; Zimmerman, L.J.; Liebler, D.C.; Ping, J.; Liu, Q.; Evans, R.; et al. Reassessment of Exosome Composition. Cell 2019, 177, 428–445.e18. [Google Scholar] [CrossRef]
- Mir, B.; Goettsch, C. Extracellular Vesicles as Delivery Vehicles of Specific Cellular Cargo. Cells 2020, 9, 1601. [Google Scholar] [CrossRef] [PubMed]
- Kowal, J.; Arras, G.; Colombo, M.; Jouve, M.; Morath, J.P.; Primdal-Bengtson, B.; Dingli, F.; Loew, D.; Tkach, M.; Théry, C. Proteomic comparison defines novel markers to characterize heterogeneous populations of extracellular vesicle subtypes. Proc. Natl. Acad. Sci. USA 2016, 113, E968–E977. [Google Scholar] [CrossRef] [PubMed]
- Oggero, S.; Austin-Williams, S.; Norling, L.V. The Contrasting Role of Extracellular Vesicles in Vascular Inflammation and Tissue Repair. Front. Pharmacol. 2019, 10, 1479. [Google Scholar] [CrossRef] [PubMed]
- Goetzl, E.J.; Schwartz, J.B.; Mustapic, M.; Lobach, I.V.; Daneman, R.; Abner, E.L.; Jicha, G.A. Altered cargo proteins of human plasma endothelial cell–derived exosomes in atherosclerotic cerebrovascular disease. FASEB J. 2017, 31, 3689–3694. [Google Scholar] [CrossRef]
- Gotanda, K.; Hirota, T.; Saito, J.; Fukae, M.; Egashira, Y.; Izumi, N.; Deguchi, M.; Kimura, M.; Matsuki, S.; Irie, S.; et al. Circulating intestine-derived exosomal miR-328 in plasma, a possible biomarker for estimating BCRP function in the human intestines. Sci. Rep. 2016, 6, 32299. [Google Scholar] [CrossRef]
- Goetzl, E.J.; Nogueras-Ortiz, C.; Mustapic, M.; Mullins, R.J.; Abner, E.L.; Schwartz, J.B.; Kapogiannis, D. Deficient neurotrophic factors of CSPG4-type neural cell exosomes in Alzheimer disease. FASEB J. 2019, 33, 231–238. [Google Scholar] [CrossRef]
- Mustapic, M.; Eitan, E.; Werner, J.K.J.; Berkowitz, S.T.; Lazaropoulos, M.P.; Tran, J.; Goetzl, E.J.; Kapogiannis, D. Plasma Extracellular Vesicles Enriched for Neuronal Origin: A Potential Window into Brain Pathologic Processes. Front. Neurosci. 2017, 11, 278. [Google Scholar] [CrossRef]
- Monguió-Tortajada, M.; Gálvez-Montón, C.; Bayes-Genis, A.; Roura, S.; Borràs, F.E. Extracellular vesicle isolation methods: Rising impact of size-exclusion chromatography. Cell. Mol. Life Sci. 2019, 76, 2369–2382. [Google Scholar] [CrossRef] [PubMed]
- Buschmann, D.; Kirchner, B.; Hermann, S.; Märte, M.; Wurmser, C.; Brandes, F.; Kotschote, S.; Bonin, M.; Steinlein, O.K.; Pfaffl, M.W.; et al. Evaluation of serum extracellular vesicle isolation methods for profiling miRNAs by next-generation sequencing. J. Extracell. Vesicles 2018, 7, 1481321. [Google Scholar] [CrossRef] [PubMed]
- Böing, A.N.; Van Der Pol, E.; Grootemaat, A.E.; Coumans, F.A.W.; Sturk, A.; Nieuwland, R. Single-step isolation of extracellular vesicles by size-exclusion chromatography. J. Extracell. Vesicles 2014, 3, 23430. [Google Scholar] [CrossRef]
- Bowalgaha, K.; Elliot, D.J.; MacKenzie, P.I.; Knights, K.M.; Swedmark, S.; Miners, J.O. S-Naproxen and desmethylnaproxen glucuronidation by human liver microsomes and recombinant human UDP-glucuronosyltransferases (UGT): Role of UGT2B7 in the elimination of naproxen. Br. J. Clin. Pharmacol. 2005, 60, 423–433. [Google Scholar] [CrossRef] [PubMed]
- Useckaite, Z.; Mukhopadhya, A.; Moran, B.; O’Driscoll, L. Extracellular vesicles report on the MET status of their cells of origin regardless of the method used for their isolation. Sci. Rep. 2020, 10, 1–11. [Google Scholar] [CrossRef]
- Brennan, K.; Martin, K.; Fitzgerald, S.P.; O’Sullivan, J.; Wu, Y.; Blanco, A.; Richardson, C.; Mc Gee, M.M. A comparison of methods for the isolation and separation of extracellular vesicles from protein and lipid particles in human serum. Sci. Rep. 2020, 10, 1–13. [Google Scholar] [CrossRef]
- Welsh, J.A.; Van Der Pol, E.; Arkesteijn, G.J.; Bremer, M.; Brisson, A.; Coumans, F.; Dignat-George, F.; Duggan, E.; Ghiran, I.; Giebel, B.; et al. MIFlowCyt-EV: A framework for standardized reporting of extracellular vesicle flow cytometry experiments. J. Extracell. Vesicles 2020, 9, 1713526. [Google Scholar] [CrossRef]
- Van Deun, J.; EV-TRACK Consortium; Mestdagh, P.; Agostinis, P.; Akay, Ö.; Anand, S.; Anckaert, J.; Martinez, Z.A.; Baetens, T.; Beghein, E.; et al. EV-TRACK: Transparent reporting and centralizing knowledge in extracellular vesicle research. Nat. Methods 2017, 14, 228–232. [Google Scholar] [CrossRef]
- Théry, C.; Witwer, K.W.; Aikawa, E.; Alcaraz, M.J.; Anderson, J.D.; Andriantsitohaina, R.; Antoniou, A.; Arab, T.; Archer, F.; Atkin-Smith, G.K.; et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): A position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J. Extracell. Vesicles 2018, 7, 1535750. [Google Scholar] [CrossRef] [PubMed]
- Povero, D.; Yamashita, H.; Ren, W.; Subramanian, M.G.; Myers, R.P.; Eguchi, A.; Simonetto, D.A.; Goodman, Z.D.; Harrison, S.A.; Sanyal, A.J.; et al. Characterization and Proteome of Circulating Extracellular Vesicles as Potential Biomarkers for NASH. Hepatol. Commun. 2020, 4, 1263–1278. [Google Scholar] [CrossRef]
- Achour, B.; Al-Majdoub, Z.M.; Grybos-Gajniak, A.; Lea, K.; Kilford, P.; Zhang, M.; Knight, D.; Barber, J.; Schageman, J.; Rostami-Hodjegan, A. Liquid Biopsy Enables Quantification of the Abundance and Interindividual Variability of Hepatic Enzymes and Transporters. Clin. Pharmacol. Ther. 2021, 109, 222–232. [Google Scholar] [CrossRef] [PubMed]
- Greening, D.W.; Simpson, R.J. Understanding extracellular vesicle diversity–current status. Expert Rev. Proteom. 2018, 15, 887–910. [Google Scholar] [CrossRef] [PubMed]
- De Jong, O.G.; Verhaar, M.C.; Chen, Y.; Vader, P.; Gremmels, H.; Posthuma, G.; Schiffelers, R.M.; Gucek, M.; Van Balkom, B.W. Cellular stress conditions are reflected in the protein and RNA content of endothelial cell-derived exosomes. J. Extracell. Vesicles 2012, 1, 18396. [Google Scholar] [CrossRef]
- Jamaly, S.; Ramberg, C.; Olsen, R.; Latysheva, N.; Webster, P.; Sovershaev, T.; Brækkan, S.K.; Hansen, J.-B. Impact of preanalytical conditions on plasma concentration and size distribution of extracellular vesicles using Nanoparticle Tracking Analysis. Sci. Rep. 2018, 8, 1–11. [Google Scholar] [CrossRef]
- Mørk, M.; Pedersen, S.; Botha, J.; Lund, S.M.; Kristensen, S.R. Preanalytical, analytical, and biological variation of blood plasma submicron particle levels measured with nanoparticle tracking analysis and tunable resistive pulse sensing. Scand. J. Clin. Lab. Investig. 2016, 76, 349–360. [Google Scholar] [CrossRef] [PubMed]
- Witwer, K.W.; Buzás, E.I.; Bemis, L.T.; Bora, A.; Lässer, C.; Lötvall, J.; Nolte-’t Hoen, E.N.; Piper, M.G.; Sivaraman, S.; Skog, J.; et al. Standardization of sample collection, isolation and analysis methods in extracellular vesicle research. J. Extracell. Vesicles 2013, 2. [Google Scholar] [CrossRef] [PubMed]
- Tian, Y.; Gong, M.; Hu, Y.; Liu, H.; Zhang, W.; Zhang, M.; Hu, X.; Aubert, D.; Zhu, S.; Wu, L.; et al. Quality and efficiency assessment of six extracellular vesicle isolation methods by nano-flow cytometry. J. Extracell. Vesicles 2020, 9, 1697028. [Google Scholar] [CrossRef]
- Mørk, M.; Nielsen, M.H.; Bæk, R.; Jørgensen, M.M.; Pedersen, S.; Kristensen, S.R. Postprandial Increase in Blood Plasma Levels of Tissue Factor–Bearing (and Other) Microvesicles Measured by Flow Cytometry: Fact or Artifact? TH Open 2018, 2, e147–e157. [Google Scholar] [CrossRef]
- Danielson, K.M.; Estanislau, J.; Tigges, J.; Toxavidis, V.; Camacho, V.; Felton, E.J.; Khoory, J.; Kreimer, S.; Ivanov, A.R.; Mantel, P.-Y.; et al. Diurnal Variations of Circulating Extracellular Vesicles Measured by Nano Flow Cytometry. PLoS ONE 2016, 11, e0144678. [Google Scholar] [CrossRef]
- Gustafson, C.M.; Shepherd, A.J.; Miller, V.M.; Jayachandran, M. Age- and sex-specific differences in blood-borne microvesicles from apparently healthy humans. Biol. Sex Differ. 2015, 6, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Jayachandran, M.; Lugo, G.; Heiling, H.; Miller, V.M.; Rule, A.D.; Lieske, J.C. Extracellular vesicles in urine of women with but not without kidney stones manifest patterns similar to men: A case control study. Biol. Sex Differ. 2015, 6, 2. [Google Scholar] [CrossRef] [PubMed]
- Hooten, N.N.; McFarland, M.H.; Freeman, D.W.; Mode, N.A.; Ezike, N.; Zonderman, A.B.; Evans, M.K. Association of Extracellular Vesicle Protein Cargo with Race and Clinical Markers of Mortality. Sci. Rep. 2019, 9, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Denham, J.; Spencer, S.J. Emerging roles of extracellular vesicles in the intercellular communication for exercise-induced adaptations. Am. J. Physiol. Metab. 2020, 319, E320–E329. [Google Scholar] [CrossRef]
- Brahmer, A.; Neuberger, E.; Esch-Heisser, L.; Haller, N.; Jorgensen, M.M.; Baek, R.; Möbius, W.; Simon, P.; Krämer-Albers, E.-M. Platelets, endothelial cells and leukocytes contribute to the exercise-triggered release of extracellular vesicles into the circulation. J. Extracell. Vesicles 2019, 8, 1615820. [Google Scholar] [CrossRef]
- Lucchetti, D.; Battaglia, A.; Ricciardi-Tenore, C.; Colella, F.; Perelli, L.; De Maria, R.; Scambia, G.; Sgambato, A.; Fattorossi, A. Measuring Extracellular Vesicles by Conventional Flow Cytometry: Dream or Reality? Int. J. Mol. Sci. 2020, 21, 6257. [Google Scholar] [CrossRef]



| Characteristic | Healthy Females (n = 5) | Healthy Males (n = 5) |
|---|---|---|
| Age (years) Mean (Range) | 28 (22–35) | 30 (23–38) |
| Height (cm) Mean (±SD) | 166.8 (5.7) | 184.2 (8.0) |
| Weight (kg) Mean (±SD) | 55.4 (3.8) | 86.4 (5.4) |
| BMI (kg/m2) Mean (±SD) | 20.0 (2.4) | 25.5 (1.2) |
| Particle Count (Particles/mL) | Mode Size (nm) | |||
|---|---|---|---|---|
| AM | PM | AM | PM | |
| Mean | 2.82 × 1011 | 2.40 × 1011 | 83.0 | 84.7 |
| 95% CI Lower | 1.07 × 1011 | 1.12 × 1011 | 75.2 | 80.5 |
| 95% CI Upper | 7.24 × 1011 | 5.13 × 1011 | 91.8 | 89.1 |
| Minimum | 3.02 × 1010 | 4.37 × 1010 | 64.9 | 76.0 |
| Maximum | 1.26 × 1012 | 1.02 × 1012 | 99.3 | 92.9 |
| Protein Concentration (µg/mL) | ASGR1 Response | |||
| AM | PM | AM | PM | |
| Mean | 347.5 | 219.8 | 147.9 | 331.9 |
| 95% CI Lower | 104.2 | 90.8 | 76.6 | 115.9 |
| 95% CI Upper | 1161.5 | 533.3 | 286.4 | 952.8 |
| Minimum | 26.4 | 27.9 | 21.2 | 31.2 |
| Maximum | 2799.0 | 1345.9 | 421.7 | 6622.2 |
| CD9 Response | CD63 Response | |||
| AM | PM | AM | PM | |
| Mean | 57.2 | 55.9 | 28.2 | 86.9 |
| 95% CI Lower | 38.6 | 39.8 | 5.2 | 12.5 |
| 95% CI Upper | 84.5 | 79.6 | 153.5 | 605.3 |
| Minimum | 28.2 | 31.0 | 2.6 | 1.3 |
| Maximum | 140.0 | 183.7 | 1158.8 | 4954.5 |
| CD81 Response | TSG101 Response | |||
| AM | PM | AM | PM | |
| Mean | 120.0 | 94.2 | 9.11 | 11.6 |
| 95% CI Lower | 77.1 | 61.7 | −0.73 | 6.6 |
| 95% CI Upper | 187.1 | 143.9 | 15.5 | 20.4 |
| Minimum | 56.6 | 24.6 | 2.9 | 4.6 |
| Maximum | 334.2 | 264.9 | 40.9 | 35.8 |
| Particle Count (Particles/mL) | Mode Size (nm) | |||||||
|---|---|---|---|---|---|---|---|---|
| Fed | Fast | Difference | p | Fed | Fast | Difference | p | |
| Mean | 2.82 × 1011 | 2.09 × 1011 | ns | 0.193 | 83.0 | 92.0 | ns | 0.093 |
| 95% CI Lower | 1.07 × 1011 | 6.61 × 1010 | 75.2 | 80.4 | ||||
| 95% CI Upper | 7.24 × 1011 | 6.46 × 1011 | 91.8 | 105.4 | ||||
| Protein Concentration (µg/mL) | ASGR1 Response | |||||||
| Fed | Fast | Difference | p | Fed | Fast | Difference | p | |
| Mean | 347.5 | 822.2 | ns | 0.1602 | 147.9 | 168.7 | ns | 0.732 |
| 95% CI Lower | 104.2 | 619.4 | 76.6 | 99.1 | ||||
| 95% CI Upper | 1161.5 | 1094.0 | 286.4 | 287.7 | ||||
| CD9 Response | CD63 Response | |||||||
| Fed | Fast | Difference | p | Fed | Fast | Difference | p | |
| Mean | 57.2 | 90.6 | * | 0.018 | 28.2 | 41.5 | ns | 0.680 |
| 95% CI Lower | 38.6 | 65.0 | 5.2 | 5.3 | ||||
| 95% CI Upper | 84.5 | 126.2 | 153.5 | 322.9 | ||||
| CD81 Response | TSG101 Response | |||||||
| Fed | Fast | Difference | p | Fed | Fast | Difference | p | |
| Mean | 120.0 | 117.2 | ns | 0.886 | 9.1 | 7.9 | ns | 0.668 |
| 95% CI Lower | 77.1 | 76.6 | 5.4 | 4.4 | ||||
| 95% CI Upper | 187.1 | 179.5 | 15.5 | 14.1 | ||||
| Particle Count (Particles/mL) | Mode Size (nm) | |||||||
|---|---|---|---|---|---|---|---|---|
| AM | PM | Difference | p | AM | PM | Difference | p | |
| Mean | 2.29 × 1011 | 2.40 × 1011 | ns | 0.863 | 87.1 | 83.2 | ns | 0.240 |
| 95% CI Lower | 1.23 × 1011 | 1.41 × 1011 | 81.3 | 77.6 | ||||
| 95% CI Upper | 4.79 × 1011 | 3.89 × 1011 | 95.5 | 87.1 | ||||
| Protein Concentration (µg/mL) | ASGR1 Response | |||||||
| AM | PM | Difference | p | AM | PM | Difference | p | |
| Mean | 537.0 | 380.2 | ns | 0.123 | 147.9 | 331.1 | ** | 0.009 |
| 95% CI Lower | 295.1 | 223.9 | 97.7 | 169.8 | ||||
| 95% CI Upper | 955 | 660.7 | 223.9 | 645.7 | ||||
| CD9 Response | CD63 Response | |||||||
| AM | PM | Difference | p | AM | PM | Difference | p | |
| Mean | 72.4 | 52.5 | ns | 0.075 | 33.8 | 67.6 | ns | 0.239 |
| 95% CI Lower | 56.2 | 38.9 | 10.2 | 19.5 | ||||
| 95% CI Upper | 93.3 | 70.8 | 114.8 | 234.4 | ||||
| CD81 Response | TSG101 Response | |||||||
| AM | PM | Difference | p | AM | PM | Difference | p | |
| Mean | 117.5 | 72.4 | * | 0.011 | 8.5 | 11.0 | ns | 0.293 |
| 95% CI Lower | 49.0 | 12.9 | 5.9 | 7.6 | ||||
| 95% CI Upper | 331.1 | 263.0 | 12.0 | 16.2 | ||||
| Particle Count (Particles/mL) | ||||||||
|---|---|---|---|---|---|---|---|---|
| AM | PM | |||||||
| Female | Male | Difference | p | Female | Male | Difference | p | |
| Mean | 7.41 × 1010 | 7.76 × 1011 | **** | <0.0001 | 1.10 × 1011 | 5.01 × 1011 | *** | 0.0002 |
| 95% CI Lower | 3.47 × 1010 | 5.62 × 1011 | 6.03 × 1010 | 3.24 × 1011 | ||||
| 95% CI Upper | 1.58 × 1011 | 1.10 × 1012 | 1.95 × 1011 | 7.94 × 1011 | ||||
| Mode Size (nm) | ||||||||
| AM | PM | |||||||
| Female | Male | Difference | p | Female | Male | Difference | p | |
| Mean | 88.8 | 86.0 | ns | 0.610 | 83.9 | 81.3 | ns | 0.622 |
| 95% CI Lower | 77.8 | 76.5 | 79.2 | 75.8 | ||||
| 95% CI Upper | 101.4 | 96.8 | 88.8 | 87.2 | ||||
| Protein Concentration (µg/mL) | ||||||||
| AM | PM | |||||||
| Female | Male | Difference | p | Female | Male | Difference | p | |
| Mean | 330.4 | 865.0 | * | 0.037 | 289.1 | 509.3 | ns | 0.822 |
| 95% CI Lower | 154.5 | 335.0 | 97.5 | 358.1 | ||||
| 95% CI Upper | 706.3 | 2238.7 | 855.1 | 722.8 | ||||
| ASGR1 Response | ||||||||
| AM | PM | |||||||
| Female | Male | Difference | p | Female | Male | Difference | p | |
| Mean | 136.5 | 183.2 | ns | 0.801 | 248.9 | 321.4 | ns | 0.844 |
| 95% CI Lower | 70.2 | 110.4 | 94.6 | 129.1 | ||||
| 95% CI Upper | 265.5 | 304.1 | 654.6 | 799.8 | ||||
| CD9 Response | ||||||||
| AM | PM | |||||||
| Female | Male | Difference | p | Female | Male | Difference | p | |
| Mean | 70.6 | 73.5 | ns | 0.987 | 56.8 | 49.3 | ns | 0.837 |
| 95% CI Lower | 46.7 | 49.8 | 32.1 | 37.8 | ||||
| 95% CI Upper | 106.9 | 108.4 | 100.7 | 64.4 | ||||
| CD63 Response | ||||||||
| AM | PM | |||||||
| Female | Male | Difference | p | Female | Male | Difference | p | |
| Mean | 29.6 | 39.5 | ns | 0.964 | 57.9 | 78.9 | ns | 0.959 |
| 95% CI Lower | 5.1 | 5.3 | 6.5 | 15.2 | ||||
| 95% CI Upper | 171.0 | 293.1 | 515.2 | 408.3 | ||||
| CD81 Response | ||||||||
| AM | PM | |||||||
| Female | Male | Difference | p | Female | Male | Difference | p | |
| Mean | 103.50 | 135.8 | ns | 0.641 | 73.0 | 71.9 | ns | 0.999 |
| 95% CI Lower | 69.2 | 87.5 | 35.4 | 48.4 | ||||
| 95% CI Upper | 155.2 | 210.9 | 150.3 | 107.2 | ||||
| TSG101 Response | ||||||||
| AM | PM | |||||||
| Female | Male | Difference | p | Female | Male | Difference | p | |
| Mean | 8.0 | 9.0 | ns | 0.945 | 11.7 | 10.5 | ns | 0.944 |
| 95% CI Lower | 5.3 | 4.6 | 5.7 | 6.6 | ||||
| 95% CI Upper | 12.1 | 17.6 | 24.2 | 16.7 | ||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Newman, L.A.; Fahmy, A.; Sorich, M.J.; Best, O.G.; Rowland, A.; Useckaite, Z. Importance of between and within Subject Variability in Extracellular Vesicle Abundance and Cargo when Performing Biomarker Analyses. Cells 2021, 10, 485. https://doi.org/10.3390/cells10030485
Newman LA, Fahmy A, Sorich MJ, Best OG, Rowland A, Useckaite Z. Importance of between and within Subject Variability in Extracellular Vesicle Abundance and Cargo when Performing Biomarker Analyses. Cells. 2021; 10(3):485. https://doi.org/10.3390/cells10030485
Chicago/Turabian StyleNewman, Lauren A., Alia Fahmy, Michael J. Sorich, Oliver G. Best, Andrew Rowland, and Zivile Useckaite. 2021. "Importance of between and within Subject Variability in Extracellular Vesicle Abundance and Cargo when Performing Biomarker Analyses" Cells 10, no. 3: 485. https://doi.org/10.3390/cells10030485
APA StyleNewman, L. A., Fahmy, A., Sorich, M. J., Best, O. G., Rowland, A., & Useckaite, Z. (2021). Importance of between and within Subject Variability in Extracellular Vesicle Abundance and Cargo when Performing Biomarker Analyses. Cells, 10(3), 485. https://doi.org/10.3390/cells10030485








