Generation and Breeding of EGFP-Transgenic Marmoset Monkeys: Cell Chimerism and Implications for Disease Modeling
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animals and Animal Housing
2.2. Embryo Collection
2.3. Lentiviral Vector Production
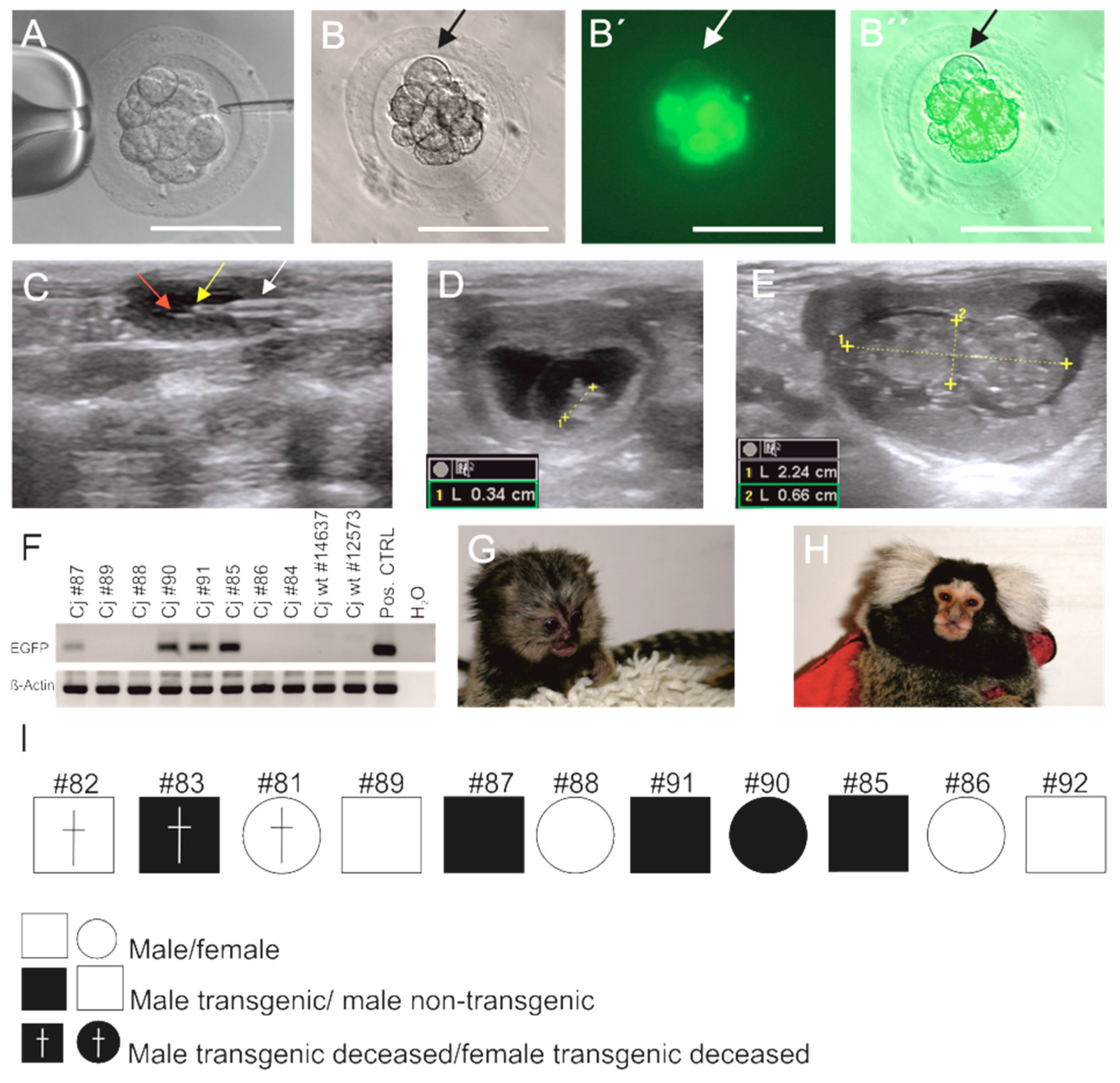
2.4. Microinjection of a Lentiviral Vector Encoding for EGFP
2.5. Embryo Transfer
2.6. Skin Biopsy, Blood, and Sperm Sampling for Expression Analysis
2.7. Cell Culture of Primary Fibroblasts
2.8. PCR Analysis of Genomic DNA
2.9. Western Blot
2.10. Flow-Cytometric Analysis
2.11. Immunohistochemistry
2.12. Genomic Southern Blot Analysis
2.13. Statistics
3. Results
3.1. Generation of EGFP-Positive Founder Animals
3.2. Sex Ratio and Normal Postnatal Development of the Transgenic Monkeys
3.3. EGFP Is Functionally Expressed in Founder Animals
3.4. Robust EGFP Transgene Expression in Organs of a Founder Animal
3.5. Germline Transmission of the Transgene by Natural Mating
3.6. Genotyping of Transgenic Offspring Reveals Consistent Cell Chimerism in Siblings
3.7. Phenotyping of Progeny
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Phillips, K.A.; Bales, K.L.; Capitanio, J.P.; Conley, A.; Czoty, P.W.; ‘t Hart, B.A.; Hopkins, W.D.; Hu, S.-L.; Miller, L.A.; Nader, M.A.; et al. Why primate models matter. Am. J. Primatol. 2014, 76, 801–827. [Google Scholar] [CrossRef] [Green Version]
- Preuss, T.M. Critique of Pure Marmoset. Brain Behav. Evol. 2019, 93, 92–107. [Google Scholar] [CrossRef] [PubMed]
- Kwiecien, T.D.; Sy, C.; Ding, Y. Rodent models of ischemic stroke lack translational relevance… are baboon models the answer? Neurol. Res. 2014, 36, 417–422. [Google Scholar] [CrossRef] [PubMed]
- Florio, M.; Huttner, W.B. Neural progenitors, neurogenesis and the evolution of the neocortex. Development 2014, 141, 2182–2194. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lemon, R.N. Descending Pathways in Motor Control. Annu. Rev. Neurosci. 2008, 31, 195–218. [Google Scholar] [CrossRef] [Green Version]
- Craig, A.D. A rat is not a monkey is not a human: Comment on Mogil. Nat. Rev. Neurosci. 2009, 10, 466. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roelfsema, P.R.; Treue, S. Basic Neuroscience Research with Nonhuman Primates: A Small but Indispensable Component of Biomedical Research. Neuron 2014, 82, 1200–1204. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feng, G.; Jensen, F.E.; Greely, H.T.; Okano, H.; Treue, S.; Roberts, A.C.; Fox, J.G.; Caddick, S.; Poo, M.-M.; Newsome, W.T.; et al. Opportunities and limitations of genetically modified nonhuman primate models for neuroscience research. Proc. Natl. Acad. Sci. USA 2020, 117, 24022–24031. [Google Scholar] [CrossRef] [PubMed]
- Hermann, B.P.; Sukhwani, M.; Hansel, M.C.; Orwig, K.E. Spermatogonial stem cells in higher primates: Are there differences from those in rodents? Reproduction 2010, 139, 479–493. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weinbauer, G.F.; Niehoff, M.; Niehaus, M.; Srivastav, S.; Fuchs, A.; Van Esch, E.; Cline, J.M. Physiology and Endocrinology of the Ovarian Cycle in Macaques. Toxicol. Pathol. 2008, 36, 7S–23S. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.E.; Long, M. New genes contribute to genetic and phenotypic novelties in human evolution. Curr. Opin. Genet. Dev. 2014, 29, 90–96. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Necsulea, A.; Kaessmann, H. Evolutionary dynamics of coding and non-coding transcriptomes. Nat. Rev. Genet. 2014, 15, 734–748. [Google Scholar] [CrossRef] [PubMed]
- Cardoso-Moreira, M.; Halbert, J.; Valloton, D.; Velten, B.; Chen, C.; Shao, Y.; Liechti, A.; Ascenção, K.; Rummel, C.; Ovchinnikova, S.; et al. Gene expression across mammalian organ development. Nature 2019, 571, 505–509. [Google Scholar] [CrossRef] [PubMed]
- Sarropoulos, I.; Marin, R.; Cardoso-Moreira, M.; Kaessmann, H. Developmental dynamics of lncRNAs across mammalian organs and species. Nature 2019, 571, 510–514. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Sharma, J.; Ke, Q.; Landman, R.; Yuan, J.; Chen, H.; Hayden, D.S.; Fisher, J.W., 3rd; Jiang, M.; Menegas, W.; et al. Atypical behaviour and connectivity in SHANK3-mutant macaques. Nature 2019, 570, 326–331. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Wan, H.; Feng, G.; Qu, J.; Wang, J.; Jing, Y.; Ren, R.; Liu, Z.; Zhang, L.; Chen, Z.; et al. SIRT6 deficiency results in developmental retardation in cynomolgus monkeys. Nature 2018, 560, 661–665. [Google Scholar] [CrossRef]
- Liu, Y.-W.; Chen, B.; Yang, X.; Fugate, J.A.; Kalucki, F.A.; Futakuchi-Tsuchida, A.; Couture, L.; Vogel, K.W.; Astley, C.A.; Baldessari, A.; et al. Human embryonic stem cell–derived cardiomyocytes restore function in infarcted hearts of non-human primates. Nat. Biotechnol. 2018, 36, 597–605. [Google Scholar] [CrossRef] [PubMed]
- Shiba, Y.; Gomibuchi, T.; Seto, T.; Wada, Y.; Ichimura, H.; Tanaka, Y.; Ogasawara, T.; Okada, K.; Shiba, N.; Sakamoto, K.; et al. Allogeneic transplantation of iPS cell-derived cardiomyocytes regenerates primate hearts. Nature 2016, 538, 388–391. [Google Scholar] [CrossRef] [PubMed]
- Kikuchi, T.; Morizane, A.; Doi, D.; Magotani, H.; Onoe, H.; Hayashi, T.; Mizuma, H.; Takara, S.; Takahashi, R.; Inoue, H.; et al. Human iPS cell-derived dopaminergic neurons function in a primate Parkinson’s disease model. Nature 2017, 548, 592–596. [Google Scholar] [CrossRef] [PubMed]
- Kriks, S.; Shim, J.-W.; Piao, J.; Ganat, Y.M.; Wakeman, D.R.; Xie, Z.; Carrillo-Reid, L.; Auyeung, G.; Antonacci, C.; Buch, A.; et al. Dopamine neurons derived from human ES cells efficiently engraft in animal models of Parkinson’s disease. Nature 2011, 480, 547–551. [Google Scholar] [CrossRef] [PubMed]
- Chong, J.J.; Yang, X.; Don, C.W.; Minami, E.; Liu, Y.W.; Weyers, J.J.; Mahoney, W.M.; Van Biber, B.; Cook, S.M.; Palpant, N.J.; et al. Human embryonic-stem-cell-derived cardiomyocytes regenerate non-human primate hearts. Nature 2014, 510, 273–277. [Google Scholar] [CrossRef] [PubMed]
- Berman, C.L.; Cannon, K.; Cui, Y.; Kornbrust, D.J.; Lagrutta, A.; Sun, S.Z.; Tepper, J.; Waldron, G.; Younis, H.S. Recommendations for Safety Pharmacology Evaluations of Oligonucleotide-Based Therapeutics. Nucleic Acid Ther. 2014, 24, 291–301. [Google Scholar] [CrossRef]
- Farooqi, I.S.; O’Rahilly, S. 20 years of leptin: Human disorders of leptin action. J. Endocrinol. 2014, 223, T63–T70. [Google Scholar] [CrossRef] [PubMed]
- Wilde, A.A.; Behr, E.R. Genetic testing for inherited cardiac disease. Nat. Rev. Cardiol. 2013, 10, 571–583. [Google Scholar] [CrossRef] [PubMed]
- Trinh, J.; Farrer, M. Advances in the genetics of Parkinson disease. Nat. Rev. Neurol. 2013, 9, 445–454. [Google Scholar] [CrossRef]
- Pouladi, M.A.; Morton, A.J.; Hayden, M.R. Choosing an animal model for the study of Huntington’s disease. Nat. Rev. Neurosci. 2013, 14, 708–721. [Google Scholar] [CrossRef]
- Bertram, L.; Lill, C.M.; Tanzi, R.E. The Genetics of Alzheimer Disease: Back to the Future. Neuron 2010, 68, 270–281. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lois, C.; Hong, E.J.; Pease, S.; Brown, E.J.; Baltimore, D. Germline Transmission and Tissue-Specific Expression of Transgenes Delivered by Lentiviral Vectors. Science 2002, 295, 868–872. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sasaki, E.; Suemizu, H.; Shimada, A.; Hanazawa, K.; Oiwa, R.; Kamioka, M.; Tomioka, I.; Sotomaru, Y.; Hirakawa, R.; Eto, T.; et al. Generation of transgenic non-human primates with germline transmission. Nature 2009, 459, 523–527. [Google Scholar] [CrossRef] [PubMed]
- Chan, A.W.; Chong, K.Y.; Martinovich, C.; Simerly, C.; Schatten, G. Transgenic Monkeys Produced by Retroviral Gene Transfer into Mature Oocytes. Science 2001, 291, 309–312. [Google Scholar] [CrossRef] [PubMed]
- Niu, Y.; Yu, Y.; Bernat, A.; Yang, S.; He, X.; Guo, X.; Chen, D.; Chen, Y.; Ji, S.; Si, W.; et al. Transgenic rhesus monkeys produced by gene transfer into early-cleavage–stage embryos using a simian immunodeficiency virus-based vector. Proc. Natl. Acad. Sci. USA 2010, 107, 17663–17667. [Google Scholar] [CrossRef] [Green Version]
- Seita, Y.; Tsukiyama, T.; Iwatani, C.; Tsuchiya, H.; Matsushita, J.; Azami, T.; Okahara, J.; Nakamura, S.; Hayashi, Y.; Hitoshi, S.; et al. Generation of transgenic cynomolgus monkeys that express green fluorescent protein throughout the whole body. Sci. Rep. 2016, 6, 24868. [Google Scholar] [CrossRef] [Green Version]
- Tomioka, I.; Nogami, N.; Nakatani, T.; Owari, K.; Fujita, N.; Motohashi, H.; Takayama, O.; Takae, K.; Nagai, Y.; Seki, K. Generation of transgenic marmosets using a tetracyclin-inducible transgene expression system as a neurodegenerative disease model. Biol. Reprod. 2017, 97, 772–780. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tomioka, I.; Ishibashi, H.; Minakawa, E.N.; Motohashi, H.H.; Takayama, O.; Saito, Y.; Popiel, H.A.; Puentes, S.; Owari, K.; Nakatani, T.; et al. Transgenic Monkey Model of the Polyglutamine Diseases Recapitulating Progressive Neurological Symptoms. eNeuro 2017, 4. [Google Scholar] [CrossRef] [Green Version]
- Niu, Y.; Guo, X.; Chen, Y.; Wang, C.E.; Gao, J.; Yang, W.; Kang, Y.; Si, W.; Wang, H.; Yang, S.-H.; et al. Early Parkinson’s disease symptoms in alpha-synuclein transgenic monkeys. Hum. Mol. Genet. 2015, 24, 2308–2317. [Google Scholar] [CrossRef] [Green Version]
- Liu, Z.; Li, X.; Zhang, J.T.; Cai, Y.J.; Cheng, T.L.; Cheng, C.; Wang, Y.; Zhang, C.C.; Nie, Y.H.; Chen, Z.F.; et al. Autism-like behaviours and germline transmission in transgenic monkeys overexpressing MeCP2. Nature 2016, 530, 98–102. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.H.; Cheng, P.H.; Banta, H.; Piotrowska-Nitsche, K.; Yang, J.J.; Cheng, E.C.; Snyder, B.; Larkin, K.; Liu, J.; Orkin, J.; et al. Towards a transgenic model of Huntington’s disease in a non-human primate. Nature 2008, 453, 921–924. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heide, M.; Haffner, C.; Murayama, A.; Kurotaki, Y.; Shinohara, H.; Okano, H.; Sasaki, E.; Huttner, W.B. Human-specific ARHGAP11B increases size and folding of primate neocortex in the fetal marmoset. Science 2020, 369, 546–550. [Google Scholar] [CrossRef] [PubMed]
- Moran, S.; Chi, T.; Prucha, M.S.; Ahn, K.S.; Connor-Stroud, F.; Jean, S.; Gould, K.; Chan, A.W. Germline transmission in transgenic Huntington’s disease monkeys. Theriogenology 2015, 84, 277–285. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, J.E.; Zhang, X.F.; Choi, S.-H.; Okahara, J.; Sasaki, E.; Silva, A.C. Generation of transgenic marmosets expressing genetically encoded calcium indicators. Sci. Rep. 2016, 6, 34931. [Google Scholar] [CrossRef] [PubMed]
- Tardif, S.; Carville, A.; Elmore, D.; Williams, L.E.; Rice, K. Reproduction and Breeding of Nonhuman Primates. In Nonhuman Primates in Biomedical Research, Volume 1: Biology and Management; Abee, C.R., Mansfield, K., Tardif, S., Morris, T., Eds.; ELSEVIER: Amsterdam, The Netherlands, 2012; pp. 197–249. [Google Scholar]
- Finstermeier, K.; Zinner, D.; Brameier, M.; Meyer, M.; Kreuz, E.; Hofreiter, M.; Roos, C. A Mitogenomic Phylogeny of Living Primates. PLoS ONE 2013, 8, e69504. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Elmore, D.; Eberle, R. Monkey B Virus (Cercopithecine herpesvirus 1). Comp. Med. 2008, 58, 11–21. [Google Scholar] [PubMed]
- Harding, R.D.; Hulme, M.J.; Lunn, S.F.; Henderson, C.; Aitken, R.J. Plasma Progesterone Levels throughout the Ovarian Cycle of the Common Marmoset (Callithrix jacchus). J. Med. Primatol. 1982, 11, 43–51. [Google Scholar] [CrossRef] [PubMed]
- Hodges, J.K.; Green, D.I.; Cottingham, P.G.; Sauer, M.J.; Edwards, C.; Lightman, S.L. Induction of luteal regression in the marmoset monkey (Callithrix jacchus) by a gonadotrophin-releasing hormone antagonist and the effects on subsequent follicular development. Reproduction 1988, 82, 743–752. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hanazawa, K.; Mueller, T.; Becker, T.; Heistermann, M.; Behr, R.; Sasaki, E. Minimally invasive transabdominal collection of preimplantation embryos from the common marmoset monkey (Callithrix jacchus). Theriogenology 2012, 78, 811–816. [Google Scholar] [CrossRef] [PubMed]
- Ishibashi, H.; Motohashi, H.H.; Kumon, M.; Yamamoto, K.; Okada, H.; Okada, T.; Seki, K. Efficient Embryo Transfer in the Common Marmoset Monkey (Callithrix jacchus) with a Reduced Transfer Volume: A Non-Surgical Approach with Cryopreserved Late-Stage Embryos. Biol. Reprod. 2013, 88, 115. [Google Scholar] [CrossRef]
- Schorpp, M.; Jäger, R.; Schellander, K.; Schenkel, J.; Wagner, E.F.; Weiher, H.; Angel, P. The Human Ubiquitin C Promoter Directs High Ubiquitous Expression of Transgenes in Mice. Nucleic Acids Res. 1996, 24, 1787–1788. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schneiders, A.; Sonksen, J.; Hodges, J.K. Penile vibratory stimulation in the marmoset monkey: A practical alternative to electro-ejaculation, yielding ejaculates of enhanced quality. J. Med. Primatol. 2004, 33, 98–104. [Google Scholar] [CrossRef] [PubMed]
- Debowski, K.; Warthemann, R.; Lentes, J.; Salinas-Riester, G.; Dressel, R.; Langenstroth, D.; Gromoll, J.; Sasaki, E.; Behr, R. Non-Viral Generation of Marmoset Monkey iPS Cells by a Six-Factor-in-One-Vector Approach. PLoS ONE 2015, 10, e0118424. [Google Scholar] [CrossRef] [PubMed]
- Aeckerle, N.; Drummer, C.; Debowski, K.; Viebahn, C.; Behr, R. Primordial germ cell development in the marmoset monkey as revealed by pluripotency factor expression: Suggestion of a novel model of embryonic germ cell translocation. Mol. Hum. Reprod. 2015, 21, 66–80. [Google Scholar] [CrossRef] [Green Version]
- Phillips, I.R. The embryology of the common marmoset (Callithrix jacchus). Adv. Anat. Embryol. Cell Biol. 1976, 52, 3–47. [Google Scholar]
- Levine, T.A.; Grunau, R.E.; McAuliffe, F.M.; Pinnamaneni, R.; Foran, A.; Alderdice, F.A. Early Childhood Neurodevelopment After Intrauterine Growth Restriction: A Systematic Review. Pediatrics 2015, 135, 126–141. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Braun, R.E.; Behringer, R.R.; Peschon, J.J.; Brinster, R.L.; Palmiter, R.D. Genetically haploid spermatids are phenotypically diploid. Nature 1989, 337, 373–376. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Benirschke, K.; Anderson, J.M.; Brownhill, L.E. Marrow Chimerism in Marmosets. Science 1962, 138, 513–515. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gengozian, N.; Batson, J.S.; Greene, C.T.; Gosslee, D.G. Hemopoietic chimerism in imported and laboratory-bred marmosets. Transplantation 1969, 8, 633–652. [Google Scholar] [CrossRef] [PubMed]
- Ross, C.N.; French, J.A.; Orti, G. Germ-line chimerism and paternal care in marmosets (Callithrix kuhlii). Proc. Natl. Acad. Sci. USA 2007, 104, 6278–6282. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sweeney, C.G.; Curran, E.; Westmoreland, S.V.; Mansfield, K.G.; Vallender, E.J. Quantitative molecular assessment of chimerism across tissues in marmosets and tamarins. BMC Genom. 2012, 13, 98. [Google Scholar] [CrossRef] [Green Version]
- Takabayashi, S.; Katoh, H. Noninvasive genotyping of common marmoset (Callithrix jacchus) by fingernail PCR. Primates 2015, 56, 235–240. [Google Scholar] [CrossRef] [PubMed]
- Haidl, G.; Duan, Y.G.; Chen, S.J.; Kohn, F.-M.; Schuppe, H.C.; Allam, J.P. The role of mast cells in male infertility. Expert Rev. Clin. Immunol. 2011, 7, 627–634. [Google Scholar] [CrossRef] [PubMed]
- Qiu, P.; Jiang, J.; Liu, Z.; Cai, Y.; Huang, T.; Wang, Y.; Liu, Q.; Nie, Y.; Liu, F.; Cheng, J.; et al. BMAL1 knockout macaque monkeys display reduced sleep and psychiatric disorders. Natl. Sci. Rev. 2019, 6, 87–100. [Google Scholar] [CrossRef] [Green Version]
- Behr, R. Potential of genetically modified nonhuman primate models for biomedicine. In Primate Biologics Research at a Crossroads; Weinbauer, G., Vogel, F., Eds.; Waxmann: Münster, Germany; New York, NY, USA, 2015; pp. 149–164. [Google Scholar]
- Sato, K.; Oiwa, R.; Kumita, W.; Henry, R.; Sakuma, T.; Ito, R.; Nozu, R.; Inoue, T.; Katano, I.; Sato, K.; et al. Generation of a Nonhuman Primate Model of Severe Combined Immunodeficiency Using Highly Efficient Genome Editing. Cell Stem Cell 2016, 19, 127–138. [Google Scholar] [CrossRef] [Green Version]
- Niu, Y.; Shen, B.; Cui, Y.; Chen, Y.; Wang, J.; Wang, L.; Kang, Y.; Zhao, X.; Si, W.; Li, W.; et al. Generation of Gene-Modified Cynomolgus Monkey via Cas9/RNA-Mediated Gene Targeting in One-Cell Embryos. Cell 2014, 156, 836–843. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, H.; Chen, Y.; Niu, Y.; Zhang, K.; Kang, Y.; Ge, W.; Liu, X.; Zhao, E.; Wang, C.; Lin, S.; et al. TALEN-Mediated Gene Mutagenesis in Rhesus and Cynomolgus Monkeys. Cell Stem Cell 2014, 14, 323–328. [Google Scholar] [CrossRef] [Green Version]
- Wan, H.; Feng, C.; Teng, F.; Yang, S.; Hu, B.; Niu, Y.; Xiang, A.P.; Fang, W.; Ji, W.; Li, W.; et al. One-step generation of p53 gene biallelic mutant Cynomolgus monkey via the CRISPR/Cas system. Cell Res. 2015, 25, 258–261. [Google Scholar] [CrossRef]
- Cui, Y.; Niu, Y.; Zhou, J.; Chen, Y.; Cheng, Y.; Li, S.; Ai, Z.; Chu, C.; Wang, H.; Zheng, B.; et al. Generation of a precise Oct4-hrGFP knockin cynomolgus monkey model via CRISPR/Cas9-assisted homologous recombination. Cell Res. 2018, 28, 383–386. [Google Scholar] [CrossRef] [PubMed]
- Park, F. Lentiviral vectors: Are they the future of animal transgenesis? Physiol. Genom. 2007, 31, 159–173. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Z.; Cai, Y.; Liao, Z.; Xu, Y.; Wang, Y.; Wang, Z.; Jiang, X.; Li, Y.; Lu, Y.; Nie, Y.; et al. Cloning of a gene-edited macaque monkey by somatic cell nuclear transfer. Natl. Sci. Rev. 2019, 6, 101–108. [Google Scholar] [CrossRef] [Green Version]
- Webb, R.; Campbell, B.K. Development of the dominant follicle: Mechanisms of selection and maintenance of oocyte quality. Soc. Reprod. Fertil. Suppl. 2007, 6, 141–163. [Google Scholar] [CrossRef] [PubMed]
- Shukla, K.K.; Mahdi, A.A.; Rajender, S. Apoptosis, spermatogenesis and male infertility. Front. Biosci (Elite Ed.) 2012, 4, 746–754. [Google Scholar] [CrossRef] [PubMed]
- Sakkas, D.; Ramalingam, M.; Garrido, N.; Barratt, C.L. Sperm selection in natural conception: What can we learn from Mother Nature to improve assisted reproduction outcomes? Hum. Reprod. Update 2015, 21, 711–726. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Laurentino, S.; Borgmann, J.; Gromoll, J. On the origin of sperm epigenetic heterogeneity. Reproduction 2016, 151, R71–R78. [Google Scholar] [CrossRef] [Green Version]
- Sousa, A.P.; Amaral, A.; Baptista, M.; Tavares, R.; Caballero Campo, P.; Caballero Peregrin, P.; Freitas, A.; Paiva, A.; Almeida-Santos, T.; Ramalho-Santos, J. Not All Sperm Are Equal: Functional Mitochondria Characterize a Subpopulation of Human Sperm with Better Fertilization Potential. PLoS ONE 2011, 6, e18112. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, A.Y.; Chughtai, A.A.; Lui, K.; Sullivan, E.A. Morbidity and mortality among very preterm singletons following fertility treatment in Australia and New Zealand, a population cohort study. BMC Pregnancy Childbirth 2017, 17, 50. [Google Scholar] [CrossRef] [Green Version]
- Belva, F.; Bonduelle, M.; Roelants, M.; Michielsen, D.; Van Steirteghem, A.; Verheyen, G.; Tournaye, H. Semen quality of young adult ICSI offspring: The first results. Hum. Reprod. 2016, 31, 2811–2820. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scherrer, U.; Rexhaj, E.; Allemann, Y.; Sartori, C.; Rimoldi, S.F. Cardiovascular dysfunction in children conceived by assisted reproductive technologies. Eur. Hear. J. 2015, 36, 1583–1589. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rexhaj, E.; Paoloni-Giacobino, A.; Rimoldi, S.F.; Fuster, D.G.; Anderegg, M.; Somm, E.; Bouillet, E.; Allemann, Y.; Sartori, C.; Scherrer, U. Mice generated by in vitro fertilization exhibit vascular dysfunction and shortened life span. J. Clin. Investig. 2013, 123, 5052–5060. [Google Scholar] [CrossRef] [Green Version]
- Wennerholm, U.B.; Henningsen, A.K.; Romundstad, L.B.; Bergh, C.; Pinborg, A.; Skjaerven, R.; Forman, J.; Gissler, M.; Nygren, K.G.; Tiitinen, A. Perinatal outcomes of children born after frozen-thawed embryo transfer: A Nordic cohort study from the CoNARTaS group. Hum. Reprod. 2013, 28, 2545–2553. [Google Scholar] [CrossRef] [Green Version]
- Kropp, J.; Di Marzo, A.; Golos, T. Assisted reproductive technologies in the common marmoset: An integral species for developing nonhuman primate models of human diseases. Biol. Reprod. 2017, 96, 277–287. [Google Scholar] [CrossRef]
- Reik, W.; Walter, J. Genomic imprinting: Parental influence on the genome. Nat. Rev. Genet. 2001, 2, 21–32. [Google Scholar] [CrossRef] [PubMed]
- Tang, W.W.; Kobayashi, T.; Irie, N.; Dietmann, S.; Surani, M.A. Specification and epigenetic programming of the human germ line. Nat. Rev. Genet. 2016, 17, 585–600. [Google Scholar] [CrossRef]
- Lukic, S.; Nicolas, J.C.; Levine, A.J. The diversity of zinc-finger genes on human chromosome 19 provides an evolutionary mechanism for defense against inherited endogenous retroviruses. Cell Death Differ. 2014, 21, 381–387. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anand, T.; Talluri, T.R.; Kumar, D.; Garrels, W.; Mukherjee, A.; Debowski, K.; Behr, R.; Kues, W.A. Differentiation of Induced Pluripotent Stem Cells to Lentoid Bodies Expressing a Lens Cell-Specific Fluorescent Reporter. PLoS ONE 2016, 11, e0157570. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garrels, W.; Talluri, T.R.; Apfelbaum, R.; Carratalá, Y.P.; Bosch, P.; Pötzsch, K.; Grueso, E.; Ivics, Z.; Kues, W.A. One-step Multiplex Transgenesis via Sleeping Beauty Transposition in Cattle. Sci. Rep. 2016, 6, 21953. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gengozian, N. Immunology and blood chimerism of the marmoset. Primates Med. 1978, 10, 173–183. [Google Scholar] [PubMed]
- Sweeney, C.; Ward, J.; Vallender, E.J. Naturally occurring, physiologically normal, primate chimeras. Chimerism 2012, 3, 43–44. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koford, C.B.; Farber, P.A.; Windle, W.F. Twins and teratisms in rhesus monkeys. Folia Primatol. 1966, 4, 221–226. [Google Scholar] [CrossRef]
- Liu, Z.; Nie, Y.H.; Zhang, C.C.; Cai, Y.J.; Wang, Y.; Lu, H.P.; Li, Y.Z.; Cheng, C.; Qiu, Z.L.; Sun, Q. Generation of macaques with sperm derived from juvenile monkey testicular xenografts. Cell Res. 2016, 26, 139–142. [Google Scholar] [CrossRef] [Green Version]
- Canfield, D.; Brignolo, L.; Peterson, P.E.; Hendrickx, A.G. Conjoined twins in a rhesus monkey (Macaca mulatta). J. Med. Primatol. 2000, 29, 427–430. [Google Scholar] [CrossRef]
- Morichika, J.; Iwatani, C.; Tsuchiya, H.; Nakamura, S.; Sankai, T.; Torii, R. Triplet Pregnancy in a Cynomolgus Monkey (Macaca fascicularis) after Double Embryo Transfer. Comp. Med. 2012, 62, 69–72. [Google Scholar] [PubMed]





| KERRYPNX | Virus Injection | Controls (Not Injected) |
|---|---|---|
| Embryos used | 113 | 23 |
| Embryos transferred | 110 | 23 |
| Number of embryo transfers (ET) | 46 | 14 |
| Embryos transferred per ET | 1–4 | 1–3 |
| Average number of embryos per ET | 2.4 | 1.6 |
| Pregnancies | 14 | 5 |
| Births | 11 | 5 |
| Birth rate (Birth per ET) | 23.9% | 35.7% |
| Animals alive | 8 | 4 |
| Number of vital transgenic animals (tg) | 4 | n.a. |
| Number of deceased transgenic animal | 1 | n.a. |
| Production rate of vital animals (tg per injected embryos) | 3.5% | n.a. |
| Transgenic | Non-Transgenic | |||
|---|---|---|---|---|
| Number of animals | 4 | 8 | ||
| Ratio ♂:♀ | 3:1 | 3:1 (6/2) | ||
| mean | range | mean | range | |
| weight at birth (g) | 39.2 | 36.6–44.7 | 35.7 | 33.0–39.0 |
| weight at 1 month | 66.2 | 60.4–76.2 | 65.3 | 44.6–80.0 |
| weight at 3 months | 171 | 149–180 | 169.4 | 145–200 |
| weight at 12 months | 359 | 312–433 | 378.5 | 288–450 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Drummer, C.; Vogt, E.-J.; Heistermann, M.; Roshani, B.; Becker, T.; Mätz-Rensing, K.; Kues, W.A.; Kügler, S.; Behr, R. Generation and Breeding of EGFP-Transgenic Marmoset Monkeys: Cell Chimerism and Implications for Disease Modeling. Cells 2021, 10, 505. https://doi.org/10.3390/cells10030505
Drummer C, Vogt E-J, Heistermann M, Roshani B, Becker T, Mätz-Rensing K, Kues WA, Kügler S, Behr R. Generation and Breeding of EGFP-Transgenic Marmoset Monkeys: Cell Chimerism and Implications for Disease Modeling. Cells. 2021; 10(3):505. https://doi.org/10.3390/cells10030505
Chicago/Turabian StyleDrummer, Charis, Edgar-John Vogt, Michael Heistermann, Berit Roshani, Tamara Becker, Kerstin Mätz-Rensing, Wilfried A. Kues, Sebastian Kügler, and Rüdiger Behr. 2021. "Generation and Breeding of EGFP-Transgenic Marmoset Monkeys: Cell Chimerism and Implications for Disease Modeling" Cells 10, no. 3: 505. https://doi.org/10.3390/cells10030505
APA StyleDrummer, C., Vogt, E.-J., Heistermann, M., Roshani, B., Becker, T., Mätz-Rensing, K., Kues, W. A., Kügler, S., & Behr, R. (2021). Generation and Breeding of EGFP-Transgenic Marmoset Monkeys: Cell Chimerism and Implications for Disease Modeling. Cells, 10(3), 505. https://doi.org/10.3390/cells10030505







