A Novel Therapeutic Target, BACH1, Regulates Cancer Metabolism
Abstract
1. Introduction
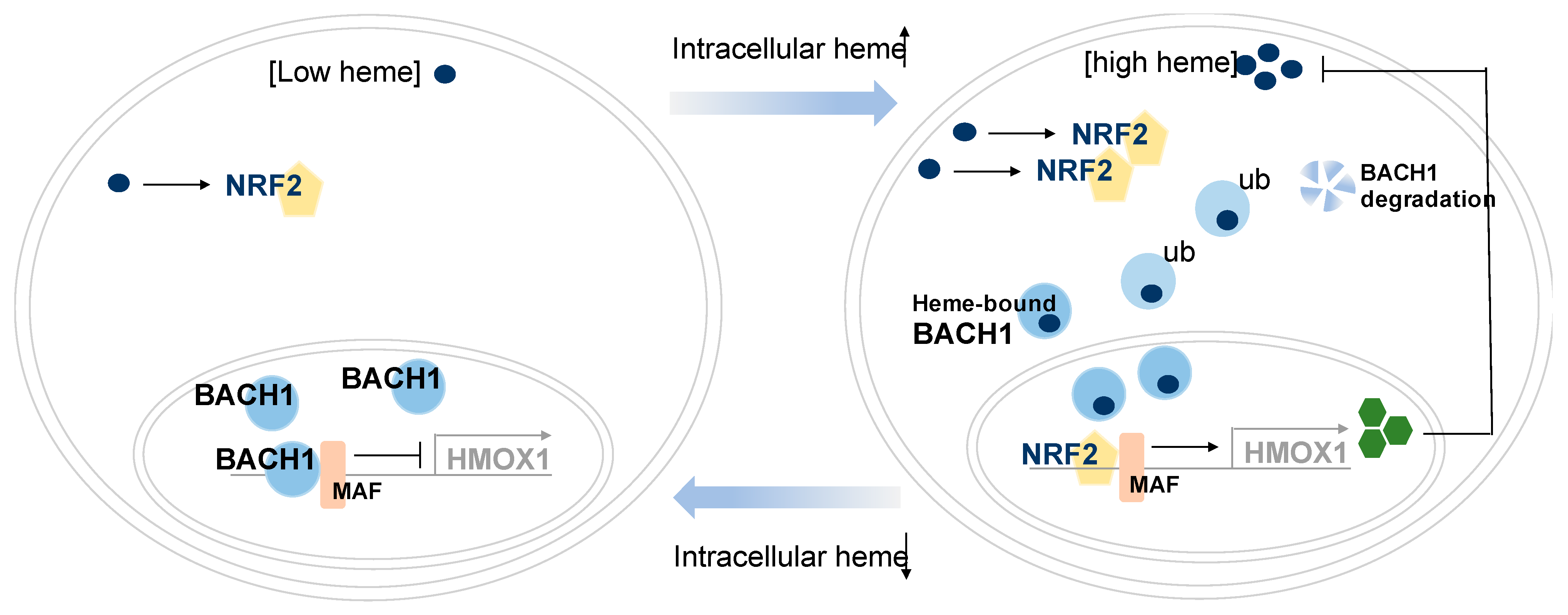
2. Heme-Binding Transcription Factor, BACH1
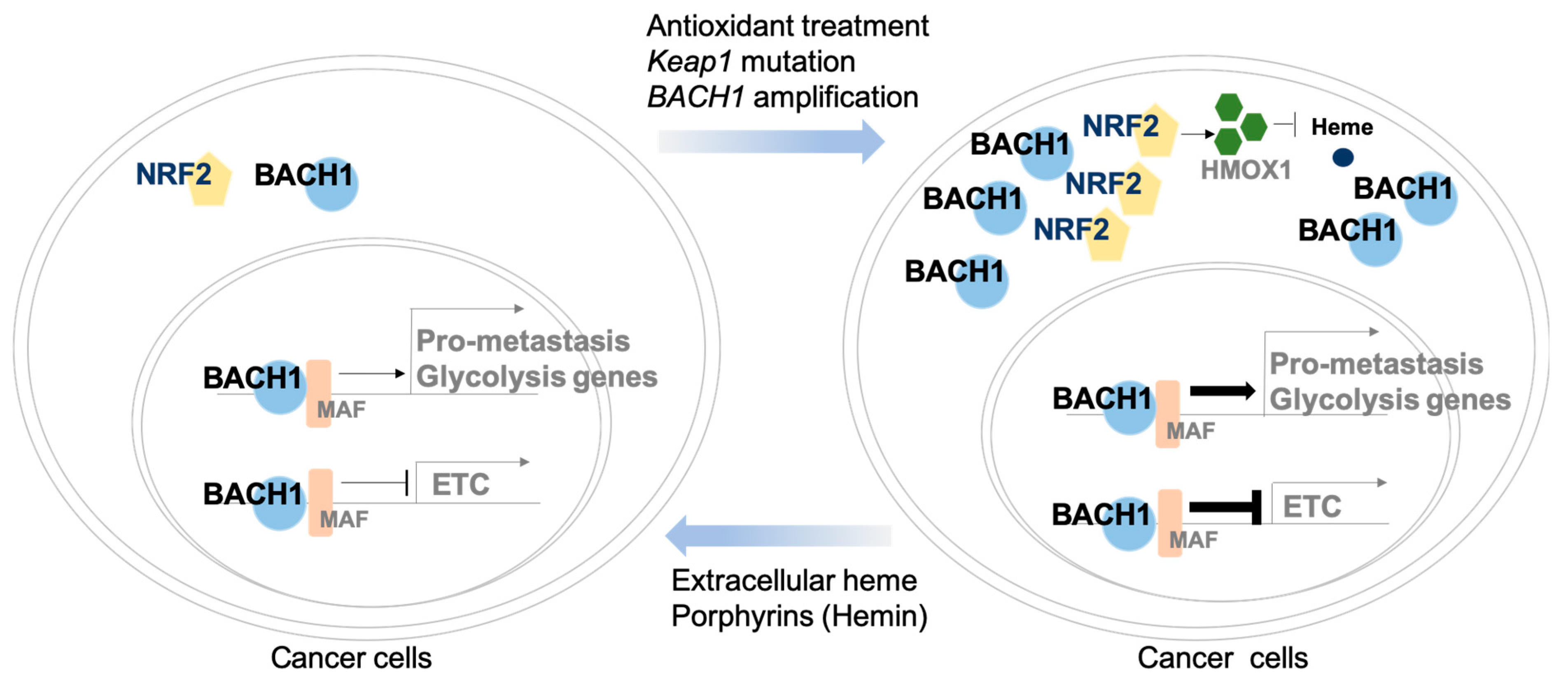
3. BACH1 Regulation in Cancer Cells
4. BACH1 Regulates Cancer Metastasis
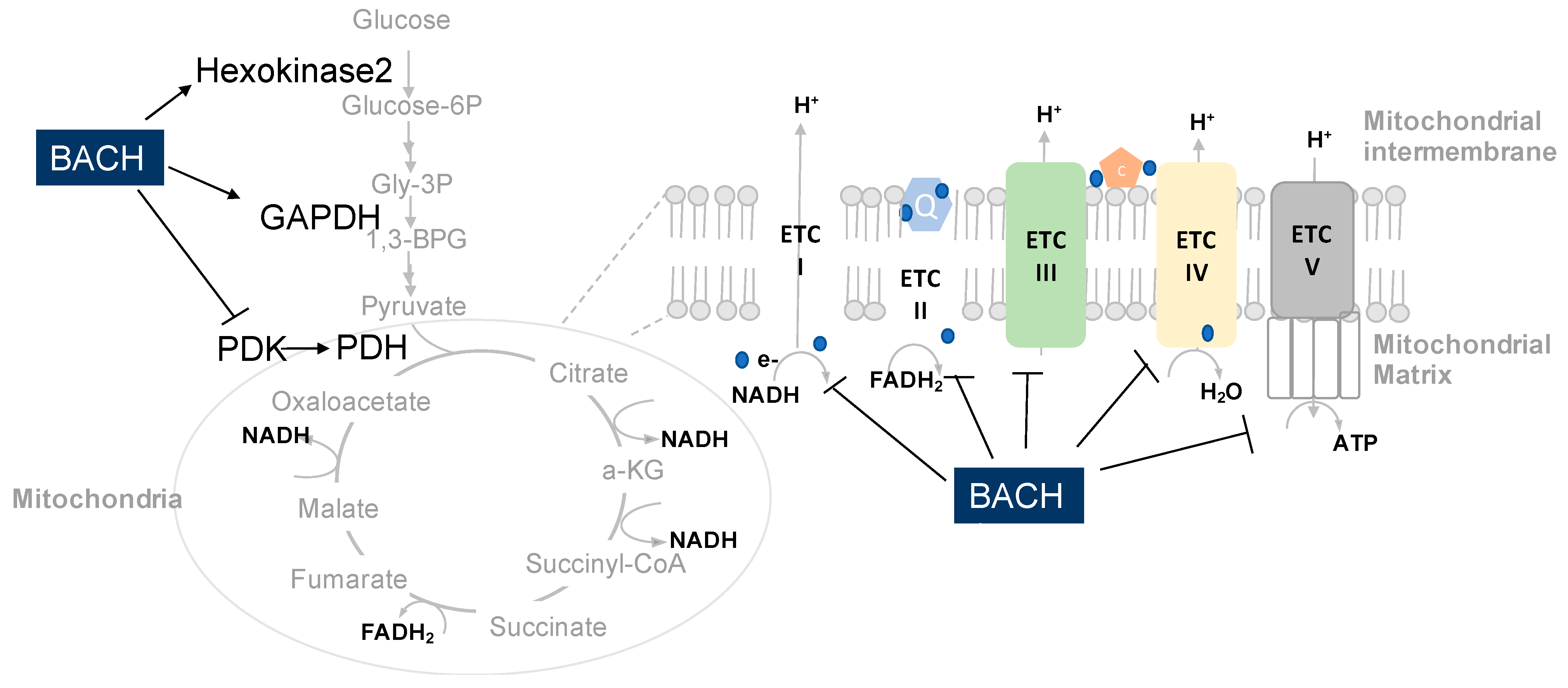
5. Function Roles of BACH1 in the Regulation of Metabolic Pathways in Cancer Cells
5.1. BACH1 Regulates Mitochondrial Metabolism of Cancer
5.2. BACH1 Regulates Redox Stress of Cancer
5.3. BACH1 Regulates Glycolysis of Cancer Cells
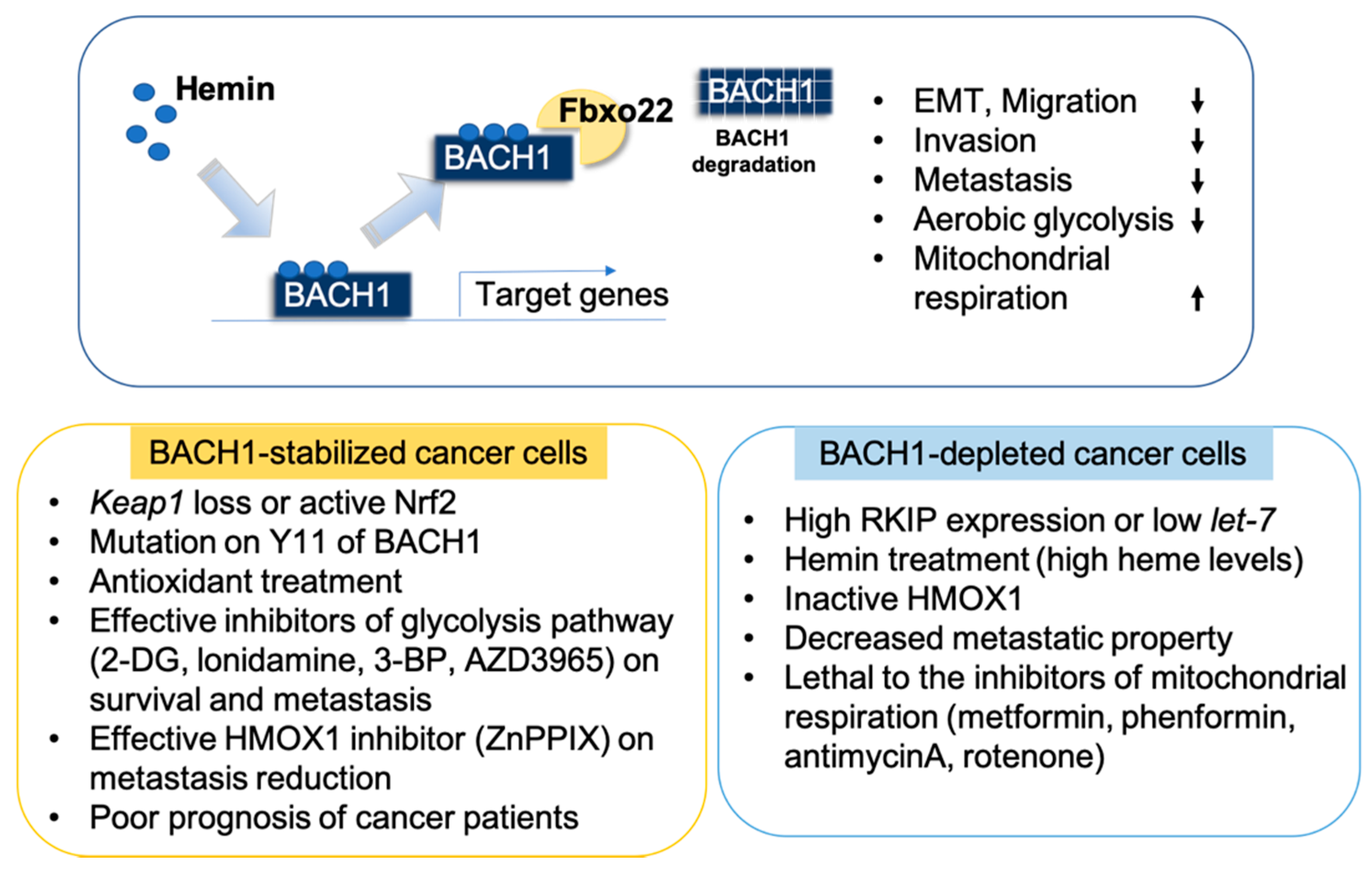
6. Pharmacological Inhibition of BACH1
7. Targeting BACH1 is Clinically Beneficial
8. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Vander Heiden, M.G.; DeBerardinis, R.J. Understanding the Intersections between Metabolism and Cancer Biology. Cell 2017, 168, 657–669. [Google Scholar] [CrossRef]
- Warburg, O. On the Origin of Cancer Cells. Science 1956, 123, 309–314. [Google Scholar] [CrossRef] [PubMed]
- Cairns, R.A.; Mak, T.W. The Current State of Cancer Metabolism. Available online: https://www.nature.com/articles/nrc.2016.100 (accessed on 21 October 2019). [CrossRef]
- DeBerardinis, R.J.; Chandel, N.S. Fundamentals of Cancer Metabolism. Sci. Adv. 2016, 2, e1600200. [Google Scholar] [CrossRef]
- Bar-Peled, L.; Sabatini, D.M. Regulation of MTORC1 by Amino Acids. Trends Cell Biol 2014, 24, 400–406. [Google Scholar] [CrossRef]
- Commisso, C.; Davidson, S.M.; Soydaner-Azeloglu, R.G.; Parker, S.J.; Kamphorst, J.J.; Hackett, S.; Grabocka, E.; Nofal, M.; Drebin, J.A.; Thompson, C.B.; et al. Macropinocytosis of Protein Is an Amino Acid Supply Route in Ras-Transformed Cells. Nature 2013, 497, 633–637. [Google Scholar] [CrossRef]
- Wolpaw, A.J.; Dang, C.V. Exploiting Metabolic Vulnerabilities of Cancer with Precision and Accuracy. Trends Cell Biol. 2018, 28, 201–212. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Lee, J.; Farquhar, K.S.; Yun, J.; Frankenberger, C.A.; Bevilacqua, E.; Yeung, K.; Kim, E.-J.; Balázsi, G.; Rosner, M.R. Network of Mutually Repressive Metastasis Regulators Can Promote Cell Heterogeneity and Metastatic Transitions. Proc. Natl. Acad. Sci. USA 2014, 111, E364–E373. [Google Scholar] [CrossRef] [PubMed]
- Fendt, S.-M.; Frezza, C.; Erez, A. Targeting Metabolic Plasticity and Flexibility Dynamics for Cancer Therapy. Cancer Discov 2020, 10, 1797–1807. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Yesilkanal, A.E.; Wynne, J.P.; Frankenberger, C.; Liu, J.; Yan, J.; Elbaz, M.; Rabe, D.C.; Rustandy, F.D.; Tiwari, P.; et al. Effective Breast Cancer Combination Therapy Targeting BACH1 and Mitochondrial Metabolism. Nature 2019, 568, 254–258. [Google Scholar] [CrossRef]
- Wiel, C.; Le Gal, K.; Ibrahim, M.X.; Jahangir, C.A.; Kashif, M.; Yao, H.; Ziegler, D.V.; Xu, X.; Ghosh, T.; Mondal, T.; et al. BACH1 Stabilization by Antioxidants Stimulates Lung Cancer Metastasis. Cell 2019, 178, 330–345.e22. [Google Scholar] [CrossRef]
- Lignitto, L.; LeBoeuf, S.E.; Homer, H.; Jiang, S.; Askenazi, M.; Karakousi, T.R.; Pass, H.I.; Bhutkar, A.J.; Tsirigos, A.; Ueberheide, B.; et al. Nrf2 Activation Promotes Lung Cancer Metastasis by Inhibiting the Degradation of Bach1. Cell 2019, 178, 316–329.e18. [Google Scholar] [CrossRef] [PubMed]
- Igarashi, K.; Kurosaki, T.; Roychoudhuri, R. BACH Transcription Factors in Innate and Adaptive Immunity. Nat. Rev. Immunol. 2017, 17, 437–450. [Google Scholar] [CrossRef] [PubMed]
- Dohi, Y.; Ikura, T.; Hoshikawa, Y.; Katoh, Y.; Ota, K.; Nakanome, A.; Muto, A.; Omura, S.; Ohta, T.; Ito, A.; et al. Bach1 Inhibits Oxidative Stress–Induced Cellular Senescence by Impeding P53 Function on Chromatin. Nat. Struct. Mol. Biol. 2008, 15, 1246–1254. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Shima, H.; Nishizawa, H.; Ikeda, M.; Brydun, A.; Matsumoto, M.; Kato, H.; Saiki, Y.; Liu, L.; Watanabe-Matsui, M.; et al. Phosphorylation of BACH1 Switches Its Function from Transcription Factor to Mitotic Chromosome Regulator and Promotes Its Interaction with HMMR. Biochem. J. 2018, 475, 981–1002. [Google Scholar] [CrossRef] [PubMed]
- Mito, S.; Ozono, R.; Oshima, T.; Yano, Y.; Watari, Y.; Yamamoto, Y.; Brydun, A.; Igarashi, K.; Yoshizumi, M. Myocardial Protection Against Pressure Overload in Mice Lacking Bach1, a Transcriptional Repressor of Heme Oxygenase-1. Hypertension 2008, 51, 1570–1577. [Google Scholar] [CrossRef] [PubMed]
- Oyake, T.; Itoh, K.; Motohashi, H.; Hayashi, N.; Hoshino, H.; Nishizawa, M.; Yamamoto, M.; Igarashi, K. Bach Proteins Belong to a Novel Family of BTB-Basic Leucine Zipper Transcription Factors That Interact with MafK and Regulate Transcription through the NF-E2 Site. Mol. Cell. Biol. 1996, 16, 6083–6095. [Google Scholar] [CrossRef]
- Zhang, X.; Guo, J.; Wei, X.; Niu, C.; Jia, M.; Li, Q.; Meng, D. Bach1: Function, Regulation, and Involvement in Disease. Oxid. Med. Cell. Longev. 2018, 2018, 1347969. [Google Scholar] [CrossRef] [PubMed]
- Reinke, A.W.; Baek, J.; Ashenberg, O.; Keating, A.E. Networks of BZIP Protein-Protein Interactions Diversified Over a Billion Years of Evolution. Science 2013, 340, 730–734. [Google Scholar] [CrossRef] [PubMed]
- Amoutzias, G.D.; Veron, A.S.; Weiner, J.; Robinson-Rechavi, M.; Bornberg-Bauer, E.; Oliver, S.G.; Robertson, D.L. One Billion Years of BZIP Transcription Factor Evolution: Conservation and Change in Dimerization and DNA-Binding Site Specificity. Mol. Biol. Evol. 2007, 24, 827–835. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Hoshino, H.; Takaku, K.; Nakajima, O.; Muto, A.; Suzuki, H.; Tashiro, S.; Takahashi, S.; Shibahara, S.; Alam, J.; et al. Hemoprotein Bach1 Regulates Enhancer Availability of Heme Oxygenase-1 Gene. EMBO J. 2002, 21, 5216–5224. [Google Scholar] [CrossRef]
- Ogawa, K.; Sun, J.; Taketani, S.; Nakajima, O.; Nishitani, C.; Sassa, S.; Hayashi, N.; Yamamoto, M.; Shibahara, S.; Fujita, H.; et al. Heme Mediates Derepression of Maf Recognition Element through Direct Binding to Transcription Repressor Bach1. EMBO J. 2001, 20, 2835–2843. [Google Scholar] [CrossRef] [PubMed]
- Zenke-Kawasaki, Y.; Dohi, Y.; Katoh, Y.; Ikura, T.; Ikura, M.; Asahara, T.; Tokunaga, F.; Iwai, K.; Igarashi, K. Heme Induces Ubiquitination and Degradation of the Transcription Factor Bach1. Mol. Cell. Biol. 2007, 27, 6962–6971. [Google Scholar] [CrossRef]
- Warnatz, H.-J.; Schmidt, D.; Manke, T.; Piccini, I.; Sultan, M.; Borodina, T.; Balzereit, D.; Wruck, W.; Soldatov, A.; Vingron, M.; et al. The BTB and CNC Homology 1 (BACH1) Target Genes Are Involved in the Oxidative Stress Response and in Control of the Cell Cycle. J. Biol Chem 2011, 286, 23521–23532. [Google Scholar] [CrossRef] [PubMed]
- Matsumoto, M.; Kondo, K.; Shiraki, T.; Brydun, A.; Funayama, R.; Nakayama, K.; Yaegashi, N.; Katagiri, H.; Igarashi, K. Genomewide Approaches for BACH1 Target Genes in Mouse Embryonic Fibroblasts Showed BACH1-Pparg Pathway in Adipogenesis. Genes Cells 2016, 21, 553–567. [Google Scholar] [CrossRef]
- Dangi-Garimella, S.; Yun, J.; Eves, E.M.; Newman, M.; Erkeland, S.J.; Hammond, S.M.; Minn, A.J.; Rosner, M.R. Raf Kinase Inhibitory Protein Suppresses a Metastasis Signalling Cascade Involving LIN28 and Let-7. EMBO J. 2009, 28, 347–358. [Google Scholar] [CrossRef]
- Yun, J.; Frankenberger, C.A.; Kuo, W.-L.; Boelens, M.C.; Eves, E.M.; Cheng, N.; Liang, H.; Li, W.-H.; Ishwaran, H.; Minn, A.J.; et al. Signalling Pathway for RKIP and Let-7 Regulates and Predicts Metastatic Breast Cancer. EMBO J. 2011, 30, 4500–4514. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Muto, A.; Hoshino, H.; Kobayashi, A.; Nishimura, S.; Yamamoto, M.; Hayashi, N.; Ito, E.; Igarashi, K. The Promoter of Mouse Transcription Repressor Bachl Is Regulated by Spl and Trans-Activated by Bachl. J. Biochem 2001, 130, 385–392. [Google Scholar] [CrossRef]
- Hou, W.; Tian, Q.; Steuerwald, N.M.; Schrum, L.W.; Bonkovsky, H.L. The Let-7 MicroRNA Enhances Heme Oxygenase-1 by Suppressing Bach1 and Attenuates Oxidant Injury in Human Hepatocytes. Biochim. Biophys. Acta 2012, 1819, 1113–1122. [Google Scholar] [CrossRef]
- Bartel, D.P. Metazoan MicroRNAs. Cell 2018, 173, 20–51. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Chen, T.; Zhong, Z.; Wang, Y.; Li, Y.; Zhao, X. MicroRNA-155 Silencing Inhibits Proliferation and Migration and Induces Apoptosis by Upregulating BACH1 in Renal Cancer Cells. Mol. Med. Rep. 2012, 5, 949–954. [Google Scholar] [CrossRef] [PubMed]
- Shirjang, S.; Mansoori, B.; Mohammadi, A.; Shajari, N.; Duijf, P.H.; Najafi, S.; Gaballu, F.A.; Nofouzi, K.; Baradaran, B. MiR-330 Regulates Colorectal Cancer Oncogenesis by Targeting BACH1. Adv. Pharm. Bull. 2020, 10, 444–451. [Google Scholar] [CrossRef] [PubMed]
- Slater, S.C.; Mansoori, B.; Mohammadi, A.; Shajari, N.; Duijf, P.H.; Najafi, S.; Gaballu, F.A.; Nofouzi, K.; Baradaran, B. MicroRNA-532-5p Regulates Pericyte Function by Targeting the Transcription Regulator BACH1 and Angiopoietin-1. Mol. Ther. 2018, 26, 2823–2837. [Google Scholar] [CrossRef] [PubMed]
- Go, H.; La, P.; Namba, F.; Ito, M.; Yang, G.; Brydun, A.; Igarashi, K.; Dennery, P.A. MiR-196a Regulates Heme Oxygenase-1 by Silencing Bach1 in the Neonatal Mouse Lung. Am. J. Physiol. -Lung Cell. Mol. Physiol. 2016, 311, L400–L411. [Google Scholar] [CrossRef] [PubMed]
- Tanioka, N.; Shimizu, H.; Takahashi, T.; Omori, E.; Kuroda, K.; Shibata, M.; Yamaoka, M.; Toda, Y.; Matsusaki, T.; Morimatsu, H. Induction of Hepatic Bach1 MRNA Expression by Carbon Tetrachloride-induced Acute Liver Injury in Rats. Biomed. Rep. 2014, 2, 359–363. [Google Scholar] [CrossRef]
- Lee, U.; Frankenberger, C.; Yun, J.; Bevilacqua, E.; Caldas, C.; Chin, S.-F.; Rueda, O.M.; Reinitz, J.; Rosner, M.R. A Prognostic Gene Signature for Metastasis-Free Survival of Triple Negative Breast Cancer Patients. PLoS ONE 2013, 8, e82125. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.; Wu, H.; Lei, R.; Chong, R.A.; Wei, Y.; Lu, X.; Tagkopoulos, I.; Kung, S.-Y.; Yang, Q.; Hu, G.; et al. Transcriptional Network Analysis Identifies BACH1 as A Master Regulator of Breast Cancer Bone Metastasis. J. Biol. Chem. 2012, jbc.M112.392332. [Google Scholar] [CrossRef]
- Sato, M.; Matsumoto, M.; Saiki, Y.; Alam, M.; Nishizawa, H.; Rokugo, M.; Brydun, A.; Yamada, S.; Kaneko, M.K.; Funayama, R.; et al. BACH1 Promotes Pancreatic Cancer Metastasis by Repressing Epithelial Genes and Enhancing Epithelial–Mesenchymal Transition. Cancer Res. 2020, 80, 1279–1292. [Google Scholar] [CrossRef]
- Fang, M.; Ou, J.; Hutchinson, L.; Green, M.R. The BRAF Oncoprotein Functions through the Transcriptional Repressor MAFG to Mediate the CpG Island Methylator Phenotype. Mol. Cell 2014, 55, 904–915. [Google Scholar] [CrossRef] [PubMed]
- Fang, M.; Hutchinson, L.; Deng, A.; Green, M.R. Common BRAF(V600E)-Directed Pathway Mediates Widespread Epigenetic Silencing in Colorectal Cancer and Melanoma. Proc. Natl. Acad. Sci. USA 2016, 113, 1250–1255. [Google Scholar] [CrossRef]
- Vasan, K.; Werner, M.; Chandel, N.S. Mitochondrial Metabolism as a Target for Cancer Therapy. Cell Metab. 2020, 32, 341–352. [Google Scholar] [CrossRef]
- Lee, P.; Chandel, N.S.; Simon, M.C. Cellular Adaptation to Hypoxia through Hypoxia Inducible Factors and Beyond. Nat. Rev. Mol. Cell Biol. 2020, 21, 268–283. [Google Scholar] [CrossRef] [PubMed]
- Gui, D.Y.; Sullivan, L.B.; Luengo, A.; Hosios, A.M.; Bush, L.N.; Gitego, N.; Davidson, S.M.; Freinkman, E.; Thomas, C.J.; Vander Heiden, M.G. Environment Dictates Dependence on Mitochondrial Complex I for NAD+ and Aspartate Production and Determines Cancer Cell Sensitivity to Metformin. Cell Metab. 2016, 24, 716–727. [Google Scholar] [CrossRef]
- Nishizawa, H.; Matsumoto, M.; Shindo, T.; Saigusa, D.; Kato, H.; Suzuki, K.; Sato, M.; Ishii, Y.; Shimokawa, H.; Igarashi, K. Ferroptosis Is Controlled by the Coordinated Transcriptional Regulation of Glutathione and Labile Iron Metabolism by the Transcription Factor BACH1. J. Biol. Chem. 2020, 295, 69–82. [Google Scholar] [CrossRef] [PubMed]
- Chiabrando, D.; Vinchi, F.; Fiorito, V.; Mercurio, S.; Tolosano, E. Heme in Pathophysiology: A Matter of Scavenging, Metabolism and Trafficking across Cell Membranes. Front. Pharmacol. 2014, 5. [Google Scholar] [CrossRef] [PubMed]
- Bissell, D.M.; Anderson, K.E.; Bonkovsky, H.L. Porphyria. N. Engl. J. Med. 2017, 377, 862–872. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, H.; Tashiro, S.; Sun, J.; Doi, H.; Satomi, S.; Igarashi, K. Cadmium Induces Nuclear Export of Bach1, a Transcriptional Repressor of Heme Oxygenase-1 Gene. J. Biol. Chem. 2003, 278, 49246–49253. [Google Scholar] [CrossRef] [PubMed]
- Hou, W.; Shan, Y.; Zheng, J.; Lambrecht, R.W.; Donohue, S.E.; Bonkovsky, H.L. Zinc Mesoporphyrin Induces Rapid and Marked Degradation of the Transcription Factor Bach1 and Up-Regulates HO-1. Biochim. Biophys. Acta 2008, 1779, 195–203. [Google Scholar] [CrossRef]
- Attucks, O.C.; Jasmer, K.J.; Hannink, M.; Kassis, J.; Zhong, Z.; Gupta, S.; Victory, S.F.; Guzel, M.; Polisetti, D.R.; Andrews, R.; et al. Induction of Heme Oxygenase I (HMOX1) by HPP-4382: A Novel Modulator of Bach1 Activity. PLoS ONE 2014, 9, e101044. [Google Scholar] [CrossRef]
- Lyssiotis, C.A.; Kimmelman, A.C. Metabolic Interactions in the Tumor Microenvironment. Trends Cell Biol. 2017, 27, 863–875. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Padilla, J.; Lee, J. A Novel Therapeutic Target, BACH1, Regulates Cancer Metabolism. Cells 2021, 10, 634. https://doi.org/10.3390/cells10030634
Padilla J, Lee J. A Novel Therapeutic Target, BACH1, Regulates Cancer Metabolism. Cells. 2021; 10(3):634. https://doi.org/10.3390/cells10030634
Chicago/Turabian StylePadilla, Joselyn, and Jiyoung Lee. 2021. "A Novel Therapeutic Target, BACH1, Regulates Cancer Metabolism" Cells 10, no. 3: 634. https://doi.org/10.3390/cells10030634
APA StylePadilla, J., & Lee, J. (2021). A Novel Therapeutic Target, BACH1, Regulates Cancer Metabolism. Cells, 10(3), 634. https://doi.org/10.3390/cells10030634





