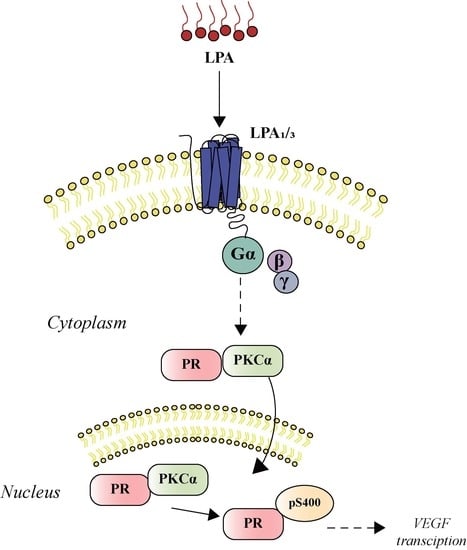
LPA1 Receptor Promotes Progesterone Receptor Phosphorylation through PKCα in Human Glioblastoma Cells
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Culture and Treatments
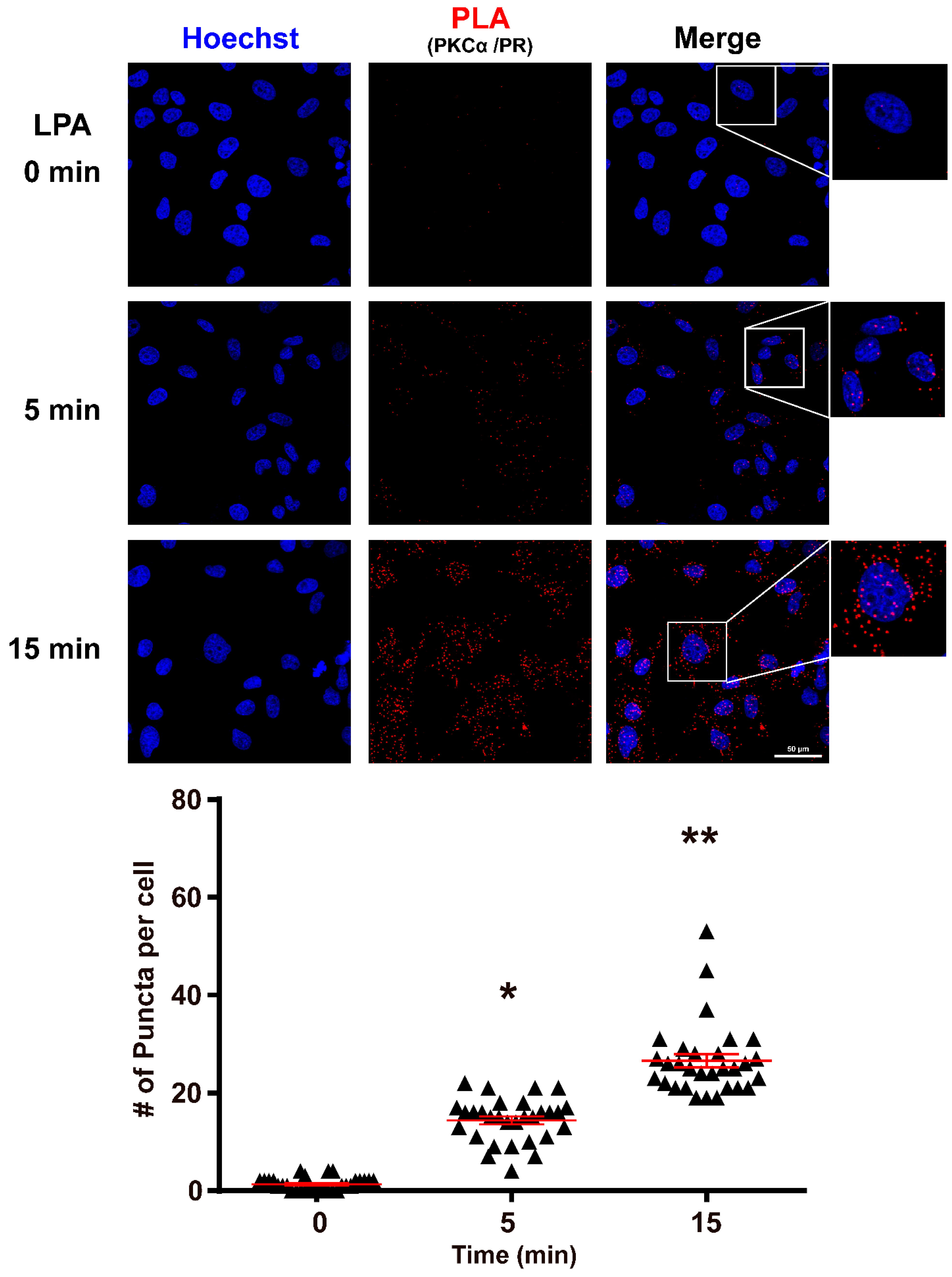
2.2. Proximity Ligation Assay
2.3. RNA Extraction and RT-qPCR
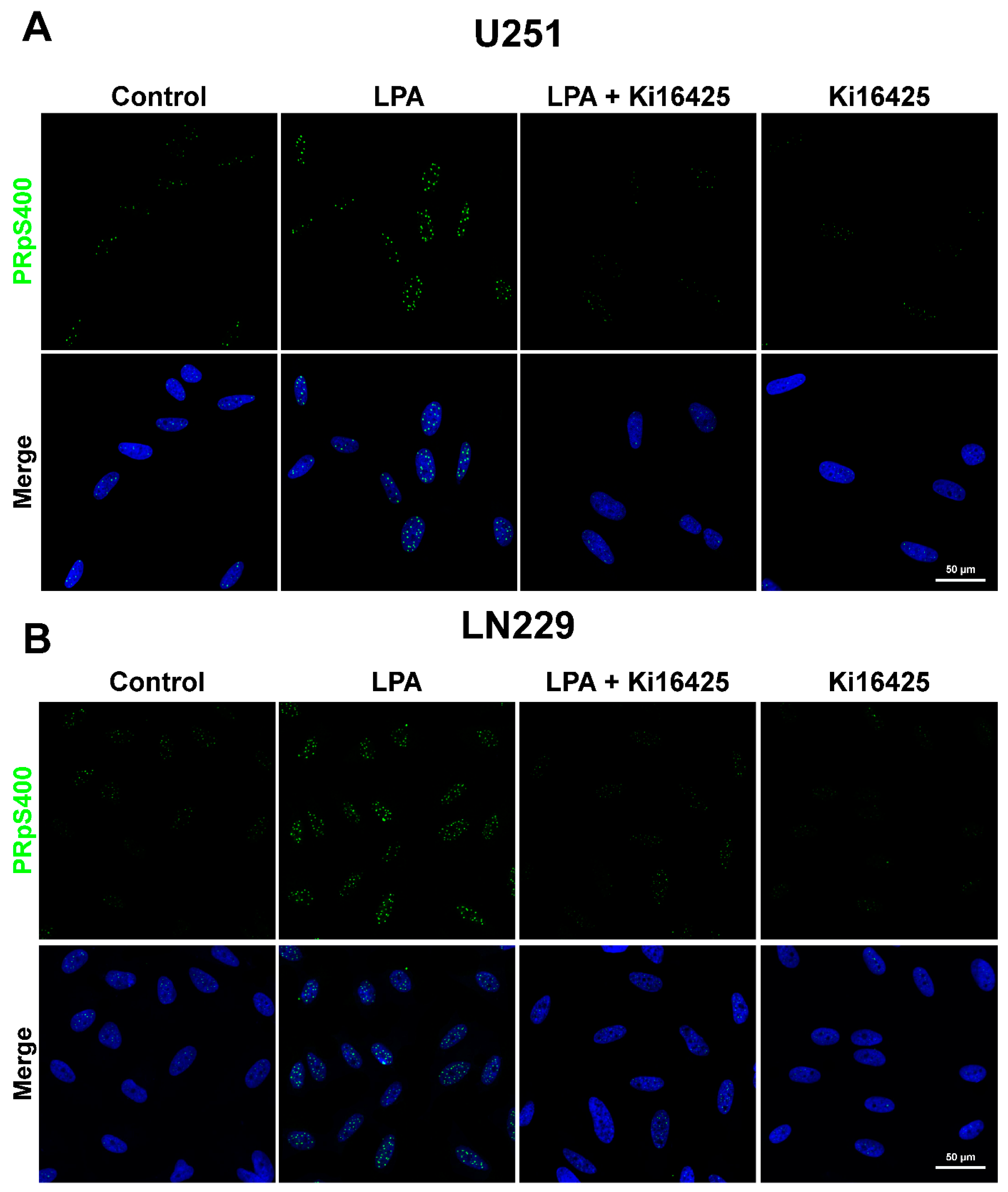
2.4. Immunofluorescence
2.5. Migration Assay
2.6. LPAR1/3 and ENPP2 Gene Expression Evaluation
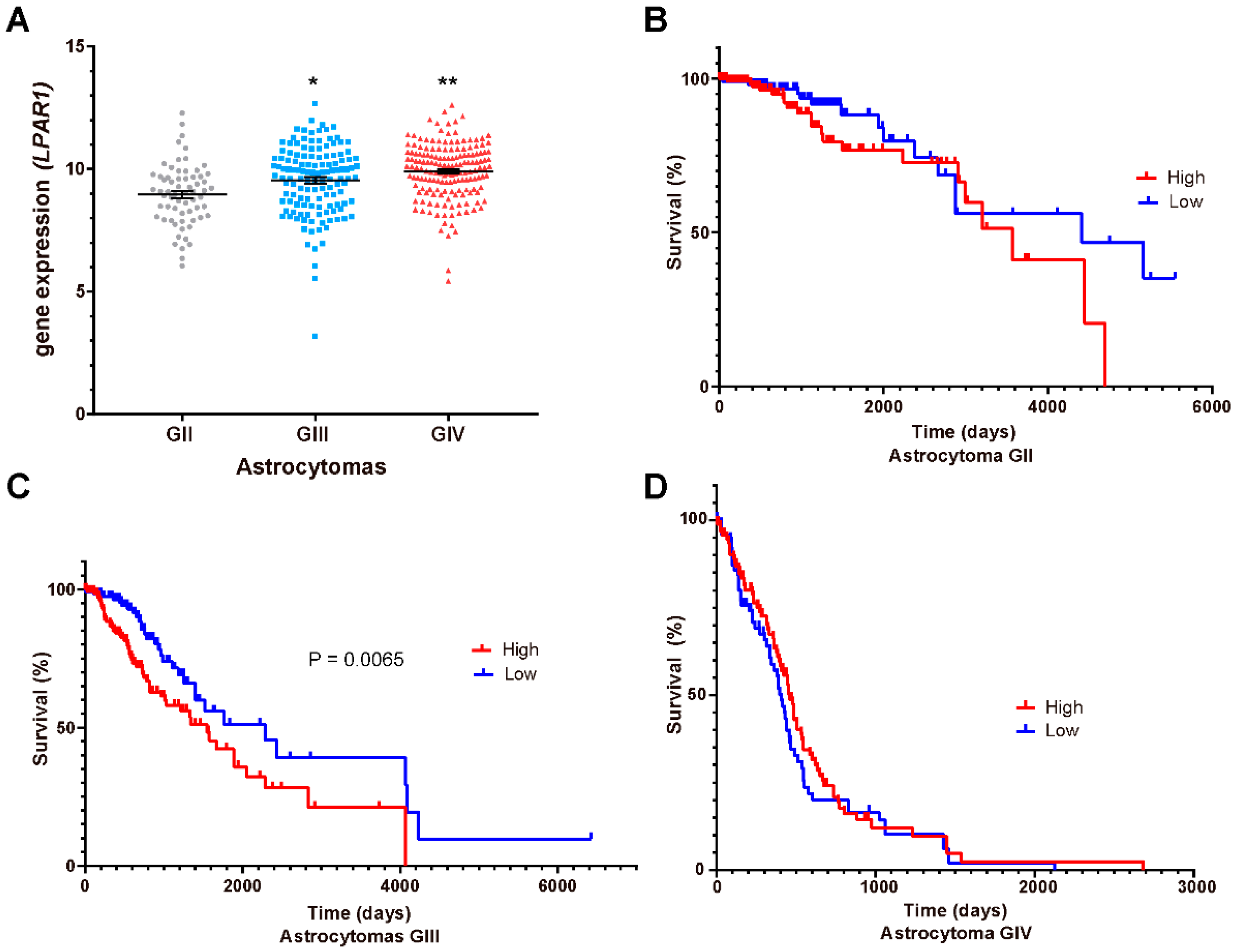
2.7. Survival Curves
2.8. Spearman Correlations
2.9. Statistical Analysis
3. Results
3.1. PKCα Interacts with PR after LPA1 Receptor Activation
3.2. PR Phosphorylated in Serine 400 (PRpS400) Has a Nuclear Localization
3.3. PR Induces the Expression of VEGF after Its Activation via the LPA1 Receptor
3.4. LPA/PR Pathway Induces Migration
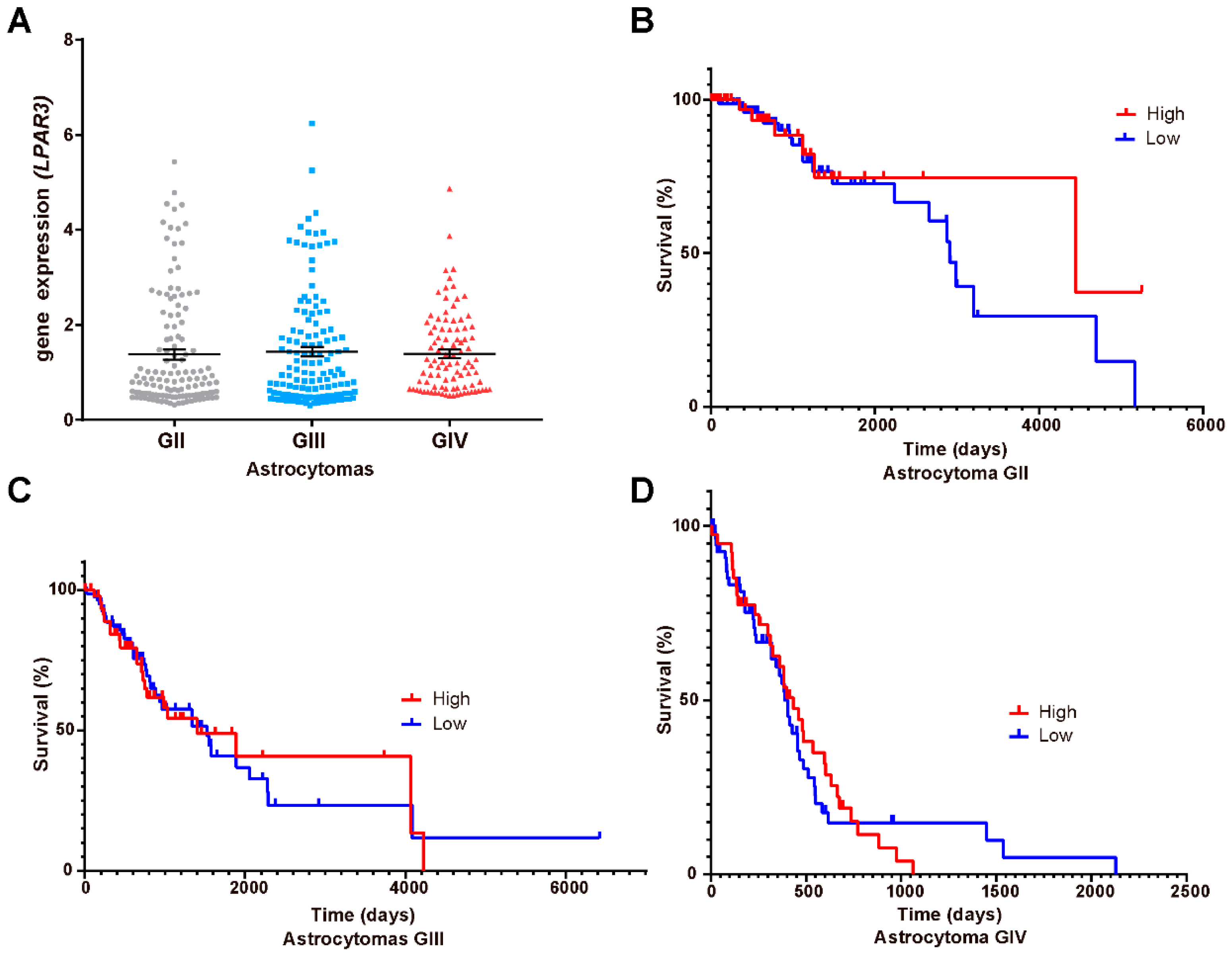
3.5. LPA1, LPA3 and ATX Expression in the Survival of GBM Patients
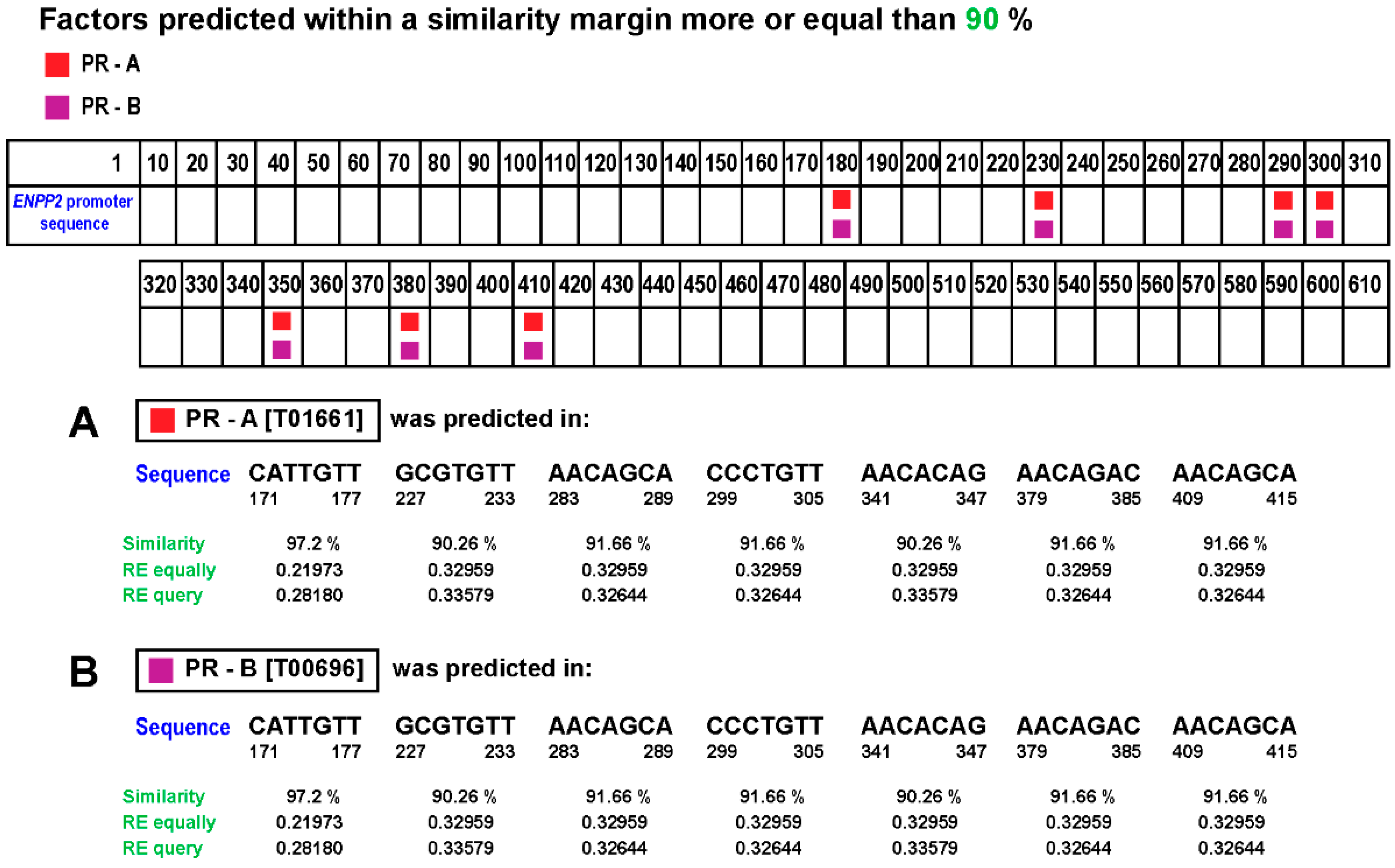
3.6. Correlation between PGR and ENPP2 Gene Expression
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Louis, D.N.; Ohgaki, H.; Wiestler, O.D.; Cavenee, W.K.; Burger, P.C.; Jouvet, A.; Scheithauer, B.W.; Kleihues, P. The 2007 WHO Classification of Tumours of the Central Nervous System. Acta Neuropathol. 2007, 114, 97–109. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ostrom, Q.T.; Gittleman, H.; Truitt, G.; Boscia, A.; Kruchko, C.; Barnholtz-Sloan, J.S. CBTRUS Statistical Report: Primary Brain and Other Central Nervous System Tumors Diagnosed in the United States in 2011–2015. Neuro. Oncol. 2018, 20, iv1–iv86. [Google Scholar] [CrossRef] [Green Version]
- Wegman-Ostrosky, T.; Reynoso-Noverón, N.; Mejía-Pérez, S.I.; Sánchez-Correa, T.E.; Alvarez-Gómez, R.M.; Vidal-Millán, S.; Cacho-Díaz, B.; Sánchez-Corona, J.; Herrera-Montalvo, L.A.; Corona-Vázquez, T. Clinical Prognostic Factors in Adults with Astrocytoma: Historic Cohort. Clin. Neurol. Neurosurg. 2016, 146, 116–122. [Google Scholar] [CrossRef] [PubMed]
- Louis, D.N.; Perry, A.; Reifenberger, G.; von Deimling, A.; Figarella-Branger, D.; Cavenee, W.K.; Ohgaki, H.; Wiestler, O.D.; Kleihues, P.; Ellison, D.W. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: A Summary. Acta. Neuropathol. 2016, 131, 803–820. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moolenar, W.H. Development of Our Current Understanding of Bioactive Lysophospholipids. Ann. N. Y. Acad. Sci. 2000, 905, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Yung, Y.C.; Stoddard, N.C.; Mirendil, H.; Chun, J. Lysophosphatidic Acid Signaling in the Nervous System. Neuron. 2015, 85, 669–682. [Google Scholar] [CrossRef] [Green Version]
- Gonzalez-Gil, I.; Zian, D.; Vazquez-Villa, H.; Ortega-Gutierrez, S.; Lopez-Rodriguez, M.L. The Status of the Lysophosphatidic Acid Receptor Type 1 (LPA1R). Medchemcomm 2015, 6, 13–23. [Google Scholar] [CrossRef] [Green Version]
- Valdés-Rives, S.A.; González-Arenas, A. Autotaxin-Lysophosphatidic Acid: From Inflammation to Cancer Development. Mediat. Inflamm. 2017, 2017. [Google Scholar] [CrossRef]
- Aoki, J.; Inoue, A.; Okudaira, S. Two Pathways for Lysophosphatidic Acid Production. Biochim. Biophys. Acta 2008, 1781, 513–518. [Google Scholar] [CrossRef]
- Perrakis, A.; Moolenaar, W.H. Autotaxin: Structure-Function and Signaling. J. Lipid Res. 2014, 55, 1010–1018. [Google Scholar] [CrossRef] [Green Version]
- Yung, Y.C.; Stoddard, N.C.; Chun, J. LPA Receptor Signaling: Pharmacology, Physiology, and Pathophysiology. J. Lipid Res. 2014, 55, 1192–1214. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, H.; Xu, X.; Gajewiak, J.; Tsukahara, R.; Fujiwara, Y.; Liu, J.; Fells, J.I.; Perygin, D.; Parrill, A.L.; Tigyi, G.; et al. Dual Activity Lysophosphatidic Acid Receptor Pan-Antagonist/Autotaxin Inhibitor Reduces Breast Cancer Cell Migration In Vitro and Causes Tumor Regression In Vivo. Cancer Res. 2009, 69, 5441–5449. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, S.-C.; Fujiwara, Y.; Liu, J.; Yue, J.; Shimizu, Y.; Norman, D.D.; Wang, Y.; Tsukahara, R.; Szabo, E.; Patil, R.; et al. Autotaxin and LPA 1 and LPA 5 Receptors Exert Disparate Functions in Tumor Cells versus the Host Tissue Microenvironment in Melanoma Invasion and Metastasis. Mol. Cancer Res. 2015, 13, 174–185. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seo, E.J.; Kwon, Y.W.; Jang, I.H.; Kim, D.K.; Lee, S.I.; Choi, E.J.; Kim, K.; Suh, D.; Lee, J.H.; Choi, K.U.; et al. Autotaxin Regulates Maintenance of Ovarian Cancer Stem Cells through Lysophosphatidic Acid-Mediated Autocrine Mechanism. Stem Cells 2016, 34, 551–564. [Google Scholar] [CrossRef] [PubMed]
- Fukushima, K.; Takahashi, K.; Yamasaki, E.; Onishi, Y.; Fukushima, N.; Honoki, K.; Tsujiuchi, T. Lysophosphatidic Acid Signaling via LPA1 and LPA3 Regulates Cellular Functions during Tumor Progression in Pancreatic Cancer Cells. Exp. Cell Res. 2017, 352, 139–145. [Google Scholar] [CrossRef]
- Koivunen, J.; Aaltonen, V.; Peltonen, J. Protein Kinase C (PKC) Family in Cancer Progression. Cancer Lett. 2006, 235, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Steinberg, S.F. Structural Basis of Protein Kinase C Isoform Function. Physiol. Rev. 2008, 88, 1341–1378. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kishi, Y.; Okudaira, S.; Tanaka, M.; Hama, K.; Shida, D.; Kitayama, J.; Yamori, T.; Aoki, J.; Fujimaki, T.; Arai, H. Autotaxin Is Overexpressed in Glioblastoma Multiforme and Contributes to Cell Motility of Glioblastoma by Converting Lysophosphatidylcholine TO Lysophosphatidic Acid. J. Biol. Chem. 2006, 281, 17492–17500. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tabuchi, S. The Autotaxin-Lysophosphatidic Acid–Lysophosphatidic Acid Receptor Cascade: Proposal of a Novel Potential Therapeutic Target for Treating Glioblastoma Multiforme. Lipids Health Dis. 2015, 14, 56. [Google Scholar] [CrossRef] [Green Version]
- Loskutov, Y.V.; Griffin, C.L.; Marinak, K.M.; Bobko, A.; Margaryan, N.V.; Geldenhuys, W.J.; Sarkaria, J.N.; Pugacheva, E.N. LPA Signaling Is Regulated through the Primary Cilium: A Novel Target in Glioblastoma. Oncogene 2018, 1. [Google Scholar] [CrossRef]
- do Carmo, A.; Balça-Silva, J.; Matias, D.; Lopes, M. PKC Signaling in Glioblastoma. Cancer Biol. Ther. 2013, 14, 287–294. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mandil, R.; Ashkenazi, E.; Blass, M.; Kronfeld, I.; Kazimirsky, G.; Rosenthal, G.; Umansky, F.; Lorenzo, P.S.; Blumberg, P.M.; Brodie, C. Protein Kinase Cα and Protein Kinase Cδ Play Opposite Roles in the Proliferation and Apoptosis of Glioma Cells. Cancer Res. 2001, 61, 4612–4619. [Google Scholar]
- Cameron, A.J.; Procyk, K.J.; Leitges, M.; Parker, P.J. PKC Alpha Protein but Not Kinase Activity Is Critical for Glioma Cell Proliferation and Survival. Int. J. Cancer 2008, 123, 769–779. [Google Scholar] [CrossRef]
- González-Arenas, A.; Peña-Ortiz, M.Á.; Hansberg-Pastor, V.; Marquina-Sánchez, B.; Baranda-Ávila, N.; Nava-Castro, K.; Cabrera-Wrooman, A.; González-Jorge, J.; Camacho-Arroyo, I. Pkcα and Pkcδ Activation Regulates Transcriptional Activity and Degradation of Progesterone Receptor in Human Astrocytoma Cells. Endocrinology 2015, 156, 1010–1022. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Valdés-Rives, S.A.; de la Fuente-Granada, M.; Velasco-Velázquez, M.A.; González-Flores, O.; González-Arenas, A. LPA1 Receptor Activation Induces PKCα Nuclear Translocation in Glioblastoma Cells. Int. J. Biochem. Cell Biol. 2019, 110, 91–102. [Google Scholar] [CrossRef] [PubMed]
- Piña-Medina, A.G.; Hansberg-Pastor, V.; González-Arenas, A.; Cerbón, M.; Camacho-Arroyo, I. Progesterone Promotes Cell Migration, Invasion and Cofilin Activation in Human Astrocytoma Cells. Steroids 2016, 105, 19–25. [Google Scholar] [CrossRef] [PubMed]
- Marquina-Sánchez, B.; González-Jorge, J.; Hansberg-Pastor, V.; Wegman-Ostrosky, T.; Baranda-Ávila, N.; Mejía-Pérez, S.; Camacho-Arroyo, I.; González-Arenas, A. The Interplay between Intracellular Progesterone Receptor and PKC Plays a Key Role in Migration and Invasion of Human Glioblastoma Cells. J. Steroid Biochem. Mol. Biol. 2016. [Google Scholar] [CrossRef] [PubMed]
- González-Agüero, G.; Gutiérrez, A.; González-Espinosa, D.; Solano, J.; Morales, R.; González-Arenas, A.; Cabrera-Muñoz, E.; Camacho-Arroyo, I. Progesterone Effects on Cell Growth of U373 and D54 Human Astrocytoma Cell Lines. Endocrine 2007, 32, 129–135. [Google Scholar] [CrossRef]
- Hernández-Hernández, O.T.; González-García, T.K.; Camacho-Arroyo, I. Progesterone Receptor and SRC-1 Participate in the Regulation of VEGF, EGFR and Cyclin D1 Expression in Human Astrocytoma Cell Lines. J. Steroid Biochem. Mol. Biol. 2012, 132, 127–134. [Google Scholar] [CrossRef]
- Schmittgen, T.D.; Livak, K.J. Analyzing Real-Time PCR Data by the Comparative CT Method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef]
- Pfaffl, M.W. A New Mathematical Model for Relative Quantification in Real-Time RT-PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef]
- Peña-Ortiz, M.Á.; Germán-Castelán, L.; González-Arenas, A. Growth Factors and Kinases in Glioblastoma Growth. Adv. Mod. Oncol. Res. 2016, 2, 248–260. [Google Scholar] [CrossRef] [Green Version]
- Cheng, S.-Y.; Chen, N.-F.; Lin, P.-Y.; Su, J.-H.; Chen, B.-H.; Kuo, H.-M.; Sung, C.-S.; Sung, P.-J.; Wen, Z.-H.; Chen, W.-F. Anti-Invasion and Antiangiogenic Effects of Stellettin B through Inhibition of the Akt/Girdin Signaling Pathway and VEGF in Glioblastoma Cells. Cancers 2019, 11, 220. [Google Scholar] [CrossRef] [Green Version]
- Fredriksson, S.; Gullberg, M.; Jarvius, J.; Olsson, C.; Pietras, K.; Gústafsdóttir, S.M.; Östman, A.; Landegren, U. Protein Detection Using Proximity-Dependent DNA Ligation Assays. Nat. Biotechnol. 2002, 20, 473–477. [Google Scholar] [CrossRef] [PubMed]
- Mueller, M.D.; Vigne, J.-L.; Pritts, E.A.; Chao, V.; Dreher, E.; Taylor, R.N. Progestins Activate Vascular Endothelial Growth Factor Gene Transcription in Endometrial Adenocarcinoma Cells. Fertil. Steril. 2003, 79, 386–392. [Google Scholar] [CrossRef]
- Tischer, E.; Mitchell, R.; Hartman, T.; Silva, M.; Gospodarowicz, D.; Fiddes, J.C.; Abraham, J.A. The Human Gene for Vascular Endothelial Growth Factor. Multiple Protein Forms Are Encoded through Alternative Exon Splicing. J. Biol. Chem. 1991, 266, 11947–11954. [Google Scholar] [CrossRef]
- Hagan, C.R.; Daniel, A.R.; Dressing, G.E.; Lange, C.A. Role of Phosphorylation in Progesterone Receptor Signaling and Specificity. Mol. Cell. Endocrinol. 2012, 357, 43–49. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hudson, L.G.; Thompson, K.L.; Xu, J.; Gill, G.N. Identification and Characterization of a Regulated Promoter Element in the Epidermal Growth Factor Receptor Gene. Proc. Nat. Acad. Sci. USA 1990, 87, 7536–7540. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lange, C.A.; Shen, T.; Horwitz, K.B. Phosphorylation of Human Progesterone Receptors at Serine-294 by Mitogen-Activated Protein Kinase Signals Their Degradation by the 26S Proteasome. Proc. Natl. Acad. Sci. USA 2000, 97, 1032–1037. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, J.; Wang, J.; Shao, J.; Gao, Y.; Xu, J.; Yu, S.; Liu, Z.; Jia, L. The Unique Pharmacological Characteristics of Mifepristone (RU486): From Terminating Pregnancy to Preventing Cancer Metastasis. Med. Res. Rev. 2014, 34, 979–1000. [Google Scholar] [CrossRef]
- Germán-Castelán, L.; Manjarrez-Marmolejo, J.; González-Arenas, A.; González-Morán, M.G.M.G.; Camacho-Arroyo, I. Progesterone Induces the Growth and Infiltration of Human Astrocytoma Cells Implanted in the Cerebral Cortex of the Rat. Biomed Res. Int. 2014, 2014. [Google Scholar] [CrossRef] [PubMed]
- Pagès, G.; Pouysségur, J. Transcriptional Regulation of the Vascular Endothelial Growth Factor Gene–a Concert of Activating Factors*. Cardiovasc. Res. 2005, 65, 564–573. [Google Scholar] [CrossRef] [PubMed]
- Lemée, J.M.; Clavreul, A.; Aubry, M.; Com, E.; De Tayrac, M.; Mosser, J.; Menei, P. Integration of Transcriptome and Proteome Profiles in Glioblastoma: Looking for the Missing Link. BMC Mol. Biol. 2018, 19, 13. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.C.; Lu, G.X.; Zhang, H.W.; Zhong, X.M.; Cong, X.L.; Xue, S.B.; Kong, R.; Li, D.; Chang, Z.Y.; Wang, X.F.; et al. Proteogenomic Characterization and Integrative Analysis of Glioblastoma Multiforme. Oncotarget 2017, 8, 97304–97312. [Google Scholar] [CrossRef] [Green Version]
- Arcos-Montoya, D.; Wegman-Ostrosky, T.; Mejía-Pérez, S.; De la Fuente-Granada, M.; Camacho-Arroyo, I.; García-Carrancá, A.; Velasco-Velazquez, M.A.; Manjarrez-Marmolejo, J.; González-Arenas, A. Progesterone Receptor Together with PKCα Expression as Prognostic Factors for Astrocytomas Malignancy. Onco. Targets. Ther. 2021. (In press) [Google Scholar]
- Maier, T.; Güell, M.; Serrano, L. Correlation of MRNA and Protein in Complex Biological Samples. FEBS Lett. 2009, 3966–3973. [Google Scholar] [CrossRef] [Green Version]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Valdés-Rives, S.A.; Arcos-Montoya, D.; de la Fuente-Granada, M.; Zamora-Sánchez, C.J.; Arias-Romero, L.E.; Villamar-Cruz, O.; Camacho-Arroyo, I.; Pérez-Tapia, S.M.; González-Arenas, A. LPA1 Receptor Promotes Progesterone Receptor Phosphorylation through PKCα in Human Glioblastoma Cells. Cells 2021, 10, 807. https://doi.org/10.3390/cells10040807
Valdés-Rives SA, Arcos-Montoya D, de la Fuente-Granada M, Zamora-Sánchez CJ, Arias-Romero LE, Villamar-Cruz O, Camacho-Arroyo I, Pérez-Tapia SM, González-Arenas A. LPA1 Receptor Promotes Progesterone Receptor Phosphorylation through PKCα in Human Glioblastoma Cells. Cells. 2021; 10(4):807. https://doi.org/10.3390/cells10040807
Chicago/Turabian StyleValdés-Rives, Silvia Anahi, Denisse Arcos-Montoya, Marisol de la Fuente-Granada, Carmen J. Zamora-Sánchez, Luis Enrique Arias-Romero, Olga Villamar-Cruz, Ignacio Camacho-Arroyo, Sonia M. Pérez-Tapia, and Aliesha González-Arenas. 2021. "LPA1 Receptor Promotes Progesterone Receptor Phosphorylation through PKCα in Human Glioblastoma Cells" Cells 10, no. 4: 807. https://doi.org/10.3390/cells10040807