TNFAIP8 Regulates Intestinal Epithelial Cell Differentiation and May Alter Terminal Differentiation of Secretory Progenitors
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animals
2.2. Single Cell RNA Sequencing Analysis
2.2.1. Dimensionality Reduction Using PCA and UMAP
2.2.2. Clustering and Annotation
2.2.3. Determining Cell Type Percentages
2.2.4. Learning Trajectories and Ordering Cells in Pseudotime
2.2.5. Pseudotime Binning Analysis
2.2.6. Secretory Branch Analysis
3. Results
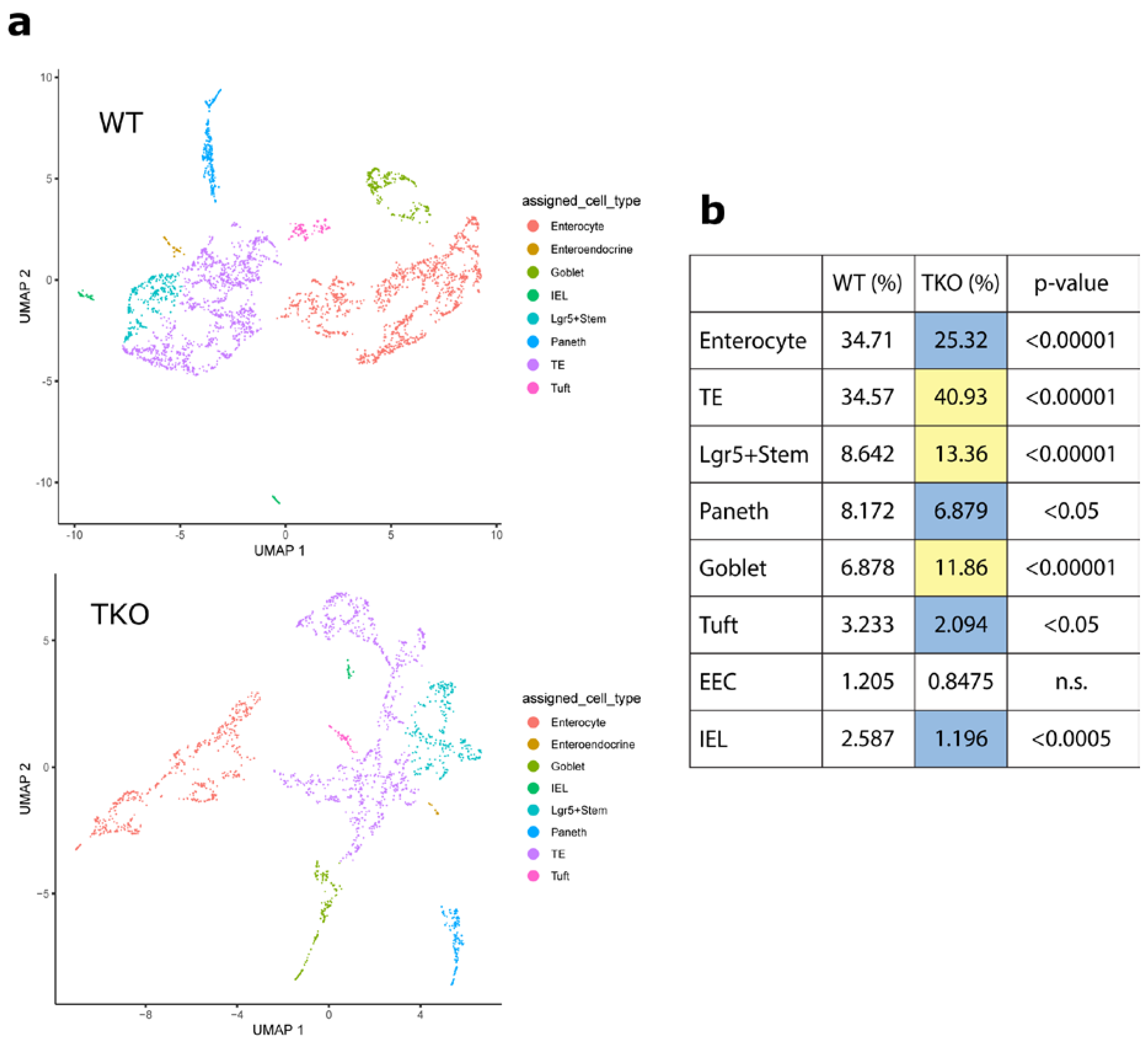
3.1. Loss of TIPE0 Alters Intestinal Differentiation
3.2. Loss of TIPE Leads to Altered Gene Expression in the Intestinal Epithelium
3.3. Nupr1, Kdm4a, and Gatad1 Are Possible Novel Regulators of Secretory Cell Fate
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- De Lau, W.; Barker, N.; Low, T.Y.; Koo, B.-K.; Li, V.S.W.; Teunissen, H.; Kujala, P.; Haegebarth, A.; Peters, P.J.; Van De Wetering, M.; et al. Lgr5 homologues associate with Wnt receptors and mediate R-spondin signalling. Nat. Cell Biol. 2011, 476, 293–297. [Google Scholar] [CrossRef] [PubMed]
- Scoville, D.H.; Sato, T.; He, X.C.; Li, L. Current View: Intestinal Stem Cells and Signaling. Gastroenterology 2008, 134, 849–864. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Bermingham, N.A.; Finegold, M.J.; Zoghbi, H.Y. Requirement of Math1 for Secretory Cell Lineage Commitment in the Mouse Intestine. Science 2001, 294, 2155–2158. [Google Scholar] [CrossRef] [PubMed]
- Shroyer, N.F.; Helmrath, M.A.; Wang, V.Y.; Antalffy, B.; Henning, S.J.; Zoghbi, H.Y. Intestine-Specific Ablation of Mouse atonal homolog 1 (Math1) Reveals a Role in Cellular Homeostasis. Gastroenterology 2007, 132, 2478–2488. [Google Scholar] [CrossRef]
- VanDussen, K.L.; Samuelson, L.C. Mouse atonal homolog 1 directs intestinal progenitors to secretory cell rather than absorptive cell fate. Dev. Biol. 2010, 346, 215–223. [Google Scholar] [CrossRef] [Green Version]
- Lee, C.S.; Perreault, N.; Brestelli, J.E.; Kaestner, K.H. Neurogenin 3 is essential for the proper specification of gastric enteroendocrine cells and the maintenance of gastric epithelial cell identity. Genes Dev. 2002, 16, 1488–1497. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jenny, M.; Uhl, C.; Roche, C.; Duluc, I.; Guillermin, V.; Guillemot, F.; Jensen, J.; Kedinger, M.; Gradwohl, G. Neurogenin3 is differentially required for endocrine cell fate specification in the intestinal and gastric epithelium. EMBO J. 2002, 21, 6338–6347. [Google Scholar] [CrossRef] [Green Version]
- López-Díaz, L.; Jain, R.N.; Keeley, T.M.; VanDussen, K.L.; Brunkan, C.S.; Gumucio, D.L.; Samuelson, L.C. Intestinal Neurogenin 3 directs differentiation of a bipotential secretory progenitor to endocrine cell rather than goblet cell fate. Dev. Biol. 2007, 309, 298–305. [Google Scholar] [CrossRef] [Green Version]
- Katz, J.P.; Perreault, N.; Goldstein, B.G.; Lee, C.S.; Labosky, P.A.; Yang, V.W.; Kaestner, K.H. The zinc-finger transcription factor Klf4 is required for terminal differentiation of goblet cells in the colon. Development 2002, 129, 2619–2628. [Google Scholar]
- Katano, T.; Bialkowska, A.B.; Yang, V.W. KLF4 Regulates Goblet Cell Differentiation in BMI1+ Reserve Intestinal Stem Cell Lineage during Homeostasis. Int. J. Stem Cells 2020, 13, 424–431. [Google Scholar] [CrossRef]
- Noah, T.K.; Kazanjian, A.; Whitsett, J.; Shroyer, N.F. SAM pointed domain ETS factor (SPDEF) regulates terminal differentiation and maturation of intestinal goblet cells. Exp. Cell Res. 2010, 316, 452–465. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shroyer, N.F.; Wallis, D.; Venken, K.J.; Bellen, H.J.; Zoghbi, H.Y. Gfi1 functions downstream of Math1 to control intestinal secretory cell subtype allocation and differentiation. Genes Dev. 2005, 19, 2412–2417. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bjerknes, M.; Khandanpour, C.; Möröy, T.; Fujiyama, T.; Hoshino, M.; Klisch, T.J.; Ding, Q.; Gan, L.; Wang, J.; Martín, M.G.; et al. Origin of the brush cell lineage in the mouse intestinal epithelium. Dev. Biol. 2012, 362, 194–218. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Spit, M.; Koo, B.-K.; Maurice, M.M. Tales from the crypt: Intestinal niche signals in tissue renewal, plasticity and cancer. Open Biol. 2018, 8, 180120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, D.; Whiteside, T.L.; Kasid, U. Identification of a Novel Tumor Necrosis Factor-α-inducible Gene, SCC-S2, Containing the Consensus Sequence of a Death Effector Domain of Fas-associated Death Domain-like Interleukin- 1β-converting Enzyme-inhibitory Protein. J. Biol. Chem. 2000, 275, 2973–2978. [Google Scholar] [CrossRef] [Green Version]
- Freundt, E.C.; Bidere, N.; Lenardo, M.J. A Different TIPE of Immune Homeostasis. Cell 2008, 133, 401–402. [Google Scholar] [CrossRef] [Green Version]
- Sun, H.; Gong, S.; Carmody, R.J.; Hilliard, A.; Li, L.; Sun, J.; Kong, L.; Xu, L.; Hilliard, B.; Hu, S.; et al. TIPE2, a Negative Regulator of Innate and Adaptive Immunity that Maintains Immune Homeostasis. Cell 2008, 133, 415–426. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.; Wang, J.; Fan, C.; Li, H.; Sun, H.; Gong, S.; Chen, Y.H.; Shi, Y. Crystal structure of TIPE2 provides insights into immune homeostasis. Nat. Struct. Mol. Biol. 2008, 16, 89–90. [Google Scholar] [CrossRef]
- Fayngerts, S.A.; Wu, J.; Oxley, C.L.; Liu, X.; Vourekas, A.; Cathopoulis, T.; Wang, Z.; Cui, J.; Liu, S.; Sun, H.; et al. TIPE3 Is the Transfer Protein of Lipid Second Messengers that Promote Cancer. Cancer Cell 2014, 26, 465–478. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Antony, P.; Baby, B.; Vijayan, R. Molecular insights into the binding of phosphoinositides to the TH domain region of TIPE proteins. J. Mol. Model. 2016, 22, 272. [Google Scholar] [CrossRef] [PubMed]
- Lin, M. Roles of TNFAIP in Cell Migration and Phosphoinositide Signaling. Ph.D. Thesis, University of Pennsylvania, Philadelphia, PA, USA, 2020. [Google Scholar]
- Goldsmith, J.R.; Spitofsky, N.; Zamani, A.; Hood, R.; Boggs, A.; Li, X.; Li, M.; Reiner, E.; Ayyaz, A.; Etwebi, Z.; et al. TNFAIP8 controls murine intestinal stem cell homeostasis and regeneration by regulating microbiome-induced Akt signaling. Nat. Commun. 2020, 11, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Porturas, T.P.; Sun, H.; Buchlis, G.; Lou, Y.; Liang, X.; Cathopoulis, T.; Fayngerts, S.; Johnson, D.S.; Wang, Z.; Chen, Y.H. Crucial Roles of TNFAIP8 Protein in Regulating Apoptosis andListeriaInfection. J. Immunol. 2015, 194, 5743–5750. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trapnell, C.; Cacchiarelli, D.; Grimsby, J.; Pokharel, P.; Li, S.; Morse, A.M.; Lennon, N.J.; Livak, K.J.; Mikkelsen, T.S.; Rinn, J.L. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat. Biotechnol. 2014, 32, 381–386. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qiu, X.; Hill, A.; Packer, J.; Lin, D.; Ma, Y.-A.; Trapnell, C. Single-cell mRNA quantification and differential analysis with Census. Nat. Methods 2017, 14, 309–315. [Google Scholar] [CrossRef] [PubMed]
- Qiu, X.; Mao, Q.; Tang, Y.; Wang, L.; Chawla, R.; Pliner, H.A.; Trapnell, C. Reversed graph embedding resolves complex single-cell trajectories. Nat. Methods 2017, 14, 979–982. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Haber, A.L.; Biton, M.; Rogel, N.; Herbst, R.H.; Shekhar, K.; Smillie, C.; Burgin, G.; Delorey, T.M.; Howitt, M.R.; Katz, Y.; et al. A single-cell survey of the small intestinal epithelium. Nat. Cell Biol. 2017, 551, 333–339. [Google Scholar] [CrossRef] [PubMed]
- Traag, V.A.; Waltman, L.; Van Eck, N.J. From Louvain to Leiden: Guaranteeing well-connected communities. Sci. Rep. 2019, 9, 1–12. [Google Scholar] [CrossRef]
- Gracz, A.D.; Samsa, L.A.; Fordham, M.J.; Trotier, D.C.; Zwarycz, B.; Lo, Y.-H.; Bao, K.; Starmer, J.; Raab, J.R.; Shroyer, N.F.; et al. Sox4 Promotes Atoh1-Independent Intestinal Secretory Differentiation Toward Tuft and Enteroendocrine Fates. Gastroenterology 2018, 155, 1508–1523.e10. [Google Scholar] [CrossRef]
- Ghaleb, A.M.; McConnell, B.B.; Kaestner, K.H.; Yang, V.W. Altered intestinal epithelial homeostasis in mice with intestine-specific deletion of the Krüppel-like factor 4 gene. Dev. Biol. 2011, 349, 310–320. [Google Scholar] [CrossRef] [Green Version]
- Hamidi, T.; Algül, H.; Cano, C.E.; Sandi, M.J.; Molejon, M.I.; Riemann, M.; Calvo, E.L.; Lomberk, G.; Dagorn, J.-C.; Weih, F.; et al. Nuclear protein 1 promotes pancreatic cancer development and protects cells from stress by inhibiting apoptosis. J. Clin. Investig. 2012, 122, 2092–2103. [Google Scholar] [CrossRef] [Green Version]
- Santofimia-Castaño, P.; Lan, W.; Bintz, J.; Gayet, O.; Carrier, A.; Lomberk, G.; Neira, J.L.; González, A.; Urrutia, R.; Soubeyran, P.; et al. Inactivation of NUPR1 promotes cell death by coupling ER-stress responses with necrosis. Sci. Rep. 2018, 8, 1–16. [Google Scholar] [CrossRef] [Green Version]
- Mead, B.E.; Ordovas-Montanes, J.; Braun, A.P.; Levy, L.E.; Bhargava, P.; Szucs, M.J.; Ammendolia, D.A.; MacMullan, M.A.; Yin, X.; Hughes, T.K.; et al. Harnessing single-cell genomics to improve the physiological fidelity of organoid-derived cell types. BMC Biol. 2018, 16, 1–24. [Google Scholar] [CrossRef] [PubMed]
- Santofimia-Castaño, P.; Rizzuti, B.; Pey, A.L.; Soubeyran, P.; Vidal, M.; Urrutia, R.; Iovanna, J.L.; Neira, J.L. Intrinsically disordered chromatin protein NUPR1 binds to the C-terminal region of Polycomb RING1B. Proc. Natl. Acad. Sci. USA 2017, 114, E6332–E6341. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chiacchiera, F.; Pasini, D. Control of adult intestinal identity by the Polycomb repressive machinery. Cell Cycle 2016, 16, 243–244. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gerbe, F.; Van Es, J.H.; Makrini, L.; Brulin, B.; Mellitzer, G.; Robine, S.; Romagnolo, B.; Shroyer, N.F.; Bourgaux, J.-F.; Pignodel, C.; et al. Distinct ATOH1 and Neurog3 requirements define tuft cells as a new secretory cell type in the intestinal epithelium. J. Cell Biol. 2011, 192, 767–780. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- VanDussen, K.L.; Carulli, A.J.; Keeley, T.M.; Patel, S.R.; Puthoff, B.J.; Magness, S.T.; Tran, I.T.; Maillard, I.; Siebel, C.; Kolterud, A.; et al. Notch signaling modulates proliferation and differentiation of intestinal crypt base columnar stem cells. Development 2011, 139, 488–497. [Google Scholar] [CrossRef] [Green Version]
- Bavetsias, V.; Lanigan, R.M.; Ruda, G.F.; Atrash, B.; McLaughlin, M.G.; Tumber, A.; Mok, N.Y.; Le Bihan, Y.-V.; Dempster, S.; Boxall, K.J.; et al. 8-Substituted Pyrido[3,4-d]pyrimidin-4(3H)-one Derivatives As Potent, Cell Permeable, KDM4 (JMJD2) and KDM5 (JARID1) Histone Lysine Demethylase Inhibitors. J. Med. Chem. 2016, 59, 1388–1409. [Google Scholar] [CrossRef]
- Cardamone, M.D.; Tanasa, B.; Chan, M.; Cederquist, C.T.; Andricovich, J.; Rosenfeld, M.G.; Perissi, V. GPS2/KDM4A Pioneering Activity Regulates Promoter-Specific Recruitment of PPARγ. Cell Rep. 2014, 8, 163–176. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Verrier, L.; Escaffit, F.; Chailleux, C.; Trouche, D.; Vandromme, M. A New Isoform of the Histone Demethylase JMJD2A/KDM4A Is Required for Skeletal Muscle Differentiation. PLoS Genet. 2011, 7, e1001390. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.; Banerjee, A.; Herring, C.A.; Attalla, J.; Hu, R.; Xu, Y.; Shao, Q.; Simmons, A.J.; Dadi, P.K.; Wang, S.; et al. Neurog3-Independent Methylation Is the Earliest Detectable Mark Distinguishing Pancreatic Progenitor Identity. Dev. Cell 2019, 48, 49–63.e7. [Google Scholar] [CrossRef]
- Wu, L.; Wary, K.K.; Revskoy, S.; Gao, X.; Tsang, K.; Komarova, Y.A.; Rehman, J.; Malik, A.B. Histone Demethylases KDM4A and KDM4C Regulate Differentiation of Embryonic Stem Cells to Endothelial Cells. Stem Cell Rep. 2015, 5, 10–21. [Google Scholar] [CrossRef] [Green Version]
- Theis, J.L.; Sharpe, K.M.; Matsumoto, M.E.; Chai, H.S.; Nair, A.A.; Theis, J.D.; De Andrade, M.; Wieben, E.D.; Michels, V.V.; Olson, T.M. Homozygosity Mapping and Exome Sequencing Reveal GATAD1 Mutation in Autosomal Recessive Dilated Cardiomyopathy. Circ. Cardiovasc. Genet. 2011, 4, 585–594. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Knight, H.C. Alternative Non-canonical Translation Initiation Codons are Used to Synthesize Novel Isoforms. Ph.D. Thesis, University of Southampton, Southampton, UK, April 2017. [Google Scholar]
- Zhao, Y.; Zheng, D.; Cvekl, A. Profiling of chromatin accessibility and identification of general cis-regulatory mechanisms that control two ocular lens differentiation pathways. Epigenetics Chromatin 2019, 12, 27. [Google Scholar] [CrossRef] [PubMed]
- Su, X.; Shi, Y.; Zou, X.; Lu, Z.-N.; Xie, G.; Yang, J.Y.H.; Wu, C.-C.; Cui, X.-F.; He, K.-Y.; Luo, Q.; et al. Single-cell RNA-Seq analysis reveals dynamic trajectories during mouse liver development. BMC Genom. 2017, 18, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Sun, W.; Zhang, Y.; Wong, K.C.; Liu, K.; Yang, Y.; Wu, B.; Tong, J.H.; Chan, A.W.; Chan, H.L.; Yu, J. Increased expression of GATA zinc finger domain containing 1 through gene amplification promotes liver cancer by directly inducing phosphatase of regenerating liver 3. Hepatology 2017, 67, 2302–2319. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hood, R.; Chen, Y.H.; Goldsmith, J.R. TNFAIP8 Regulates Intestinal Epithelial Cell Differentiation and May Alter Terminal Differentiation of Secretory Progenitors. Cells 2021, 10, 871. https://doi.org/10.3390/cells10040871
Hood R, Chen YH, Goldsmith JR. TNFAIP8 Regulates Intestinal Epithelial Cell Differentiation and May Alter Terminal Differentiation of Secretory Progenitors. Cells. 2021; 10(4):871. https://doi.org/10.3390/cells10040871
Chicago/Turabian StyleHood, Ryan, Youhai H. Chen, and Jason R. Goldsmith. 2021. "TNFAIP8 Regulates Intestinal Epithelial Cell Differentiation and May Alter Terminal Differentiation of Secretory Progenitors" Cells 10, no. 4: 871. https://doi.org/10.3390/cells10040871
APA StyleHood, R., Chen, Y. H., & Goldsmith, J. R. (2021). TNFAIP8 Regulates Intestinal Epithelial Cell Differentiation and May Alter Terminal Differentiation of Secretory Progenitors. Cells, 10(4), 871. https://doi.org/10.3390/cells10040871





