A Genome-Wide Screen in Saccharomyces cerevisiae Reveals a Critical Role for Oxidative Phosphorylation in Cellular Tolerance to Lithium Hexafluorophosphate
Abstract
:1. Introduction
2. Materials and Methods
2.1. Growth Curve Measurement
2.2. Inhibition Zone Experiment
2.3. Genome-Wide LiPF6 Screen
2.4. Functional Enrichment and Interaction Network Analysis
2.5. Complementation Strain Construction and Spot Tests
2.6. Mitochondrial Morphology Observation
2.7. ROS Measurement
2.8. Western Blot
2.9. Measurement of Mitochondrial ATP Synthesis
3. Results
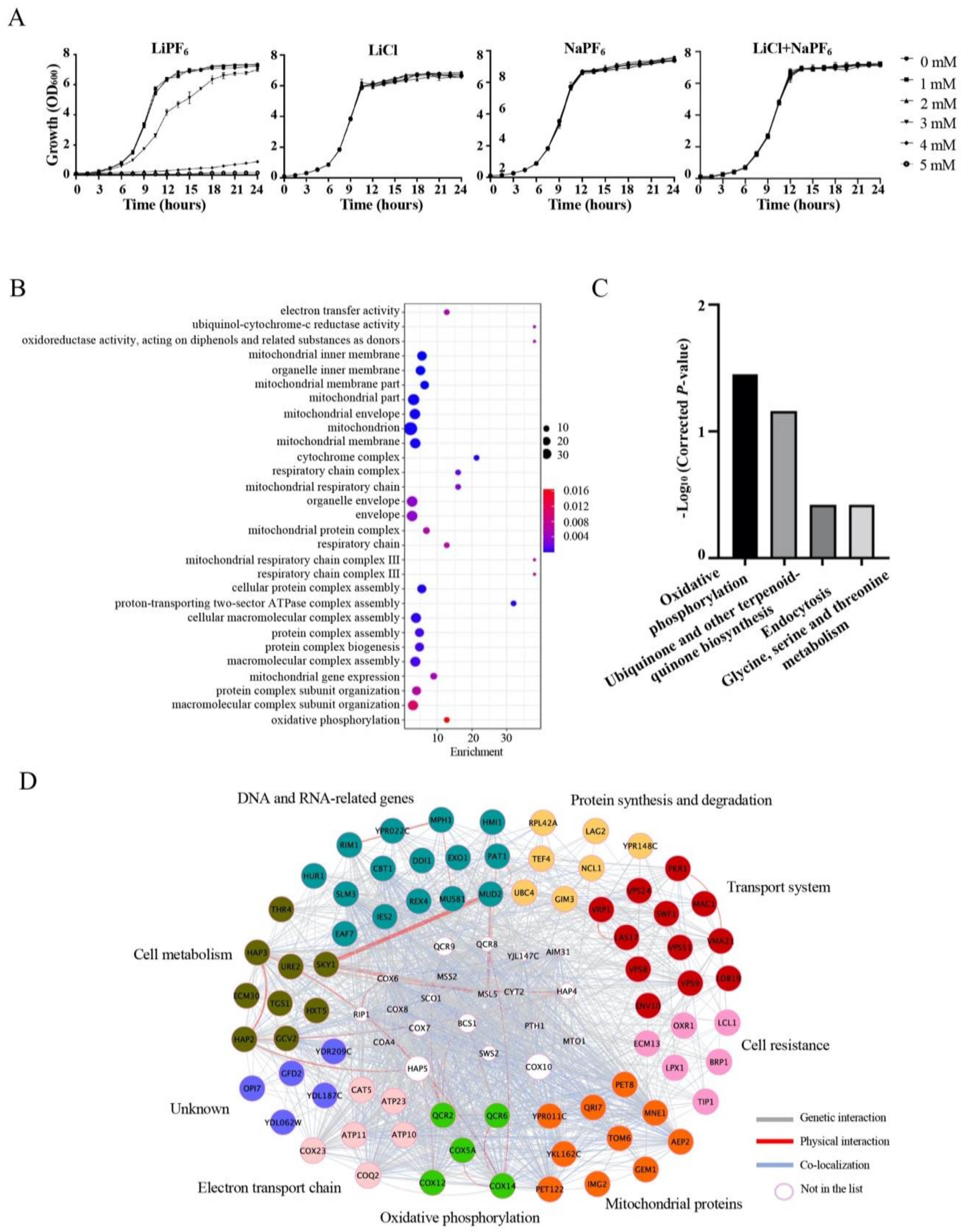
3.1. Growth in LiPF6-, LiCl-, and NaPF6-Supplemented Medium
3.2. A Genome-Wide Screen Identifies Deletion Strains with Increased Sensitivity to LiPF6
3.3. Oxidative Phosphorylation-Related Genes Are Required for Tolerance to LiPF6
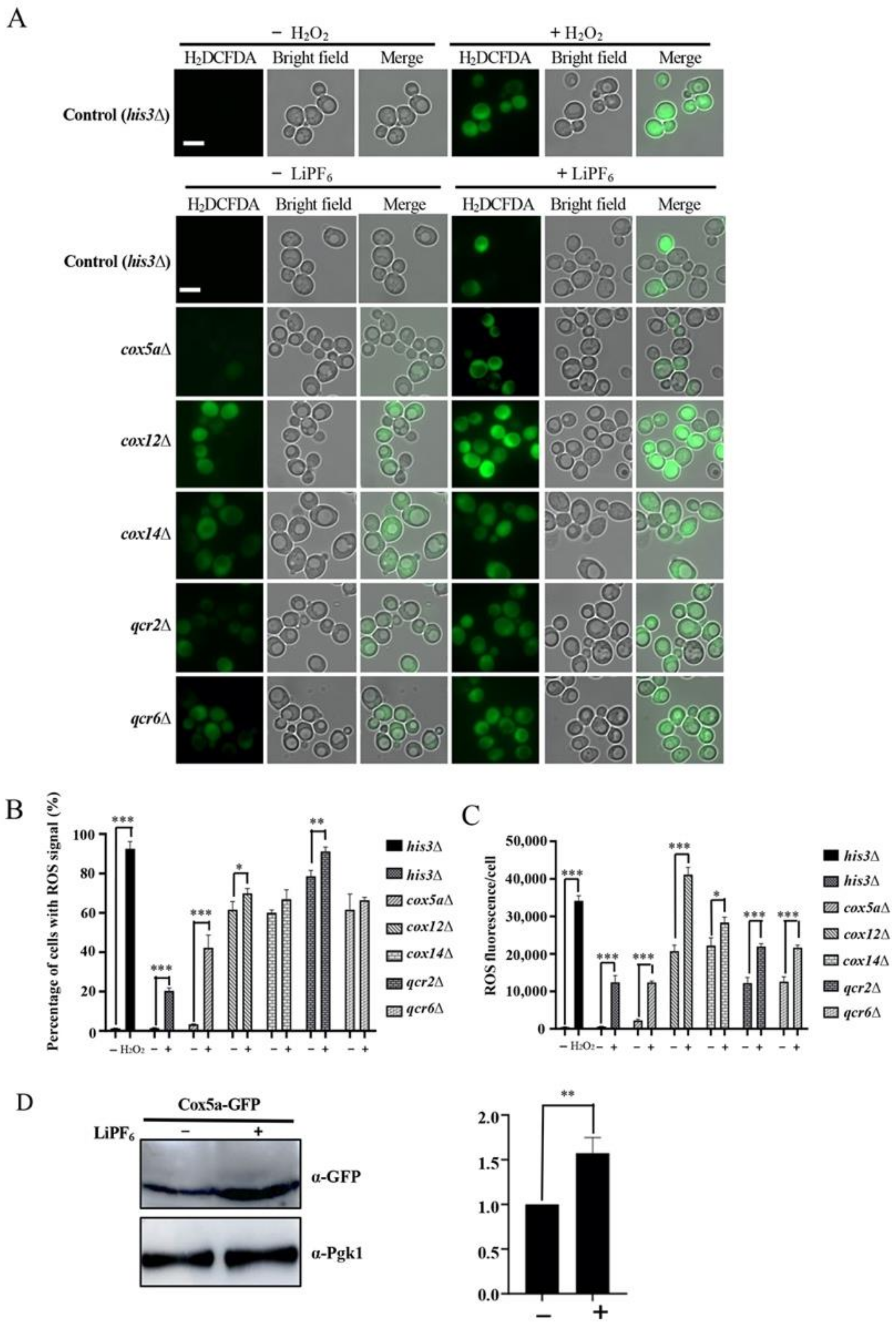
3.4. High Concentration of LiPF6 Alters Mitochondrial Morphology, Induces ROS Accumulation, and Reduces ATP Levels
3.5. Oxidative Phosphorylation-Related Genes Are Required for Counteraction of LiPF6-Induced ROS
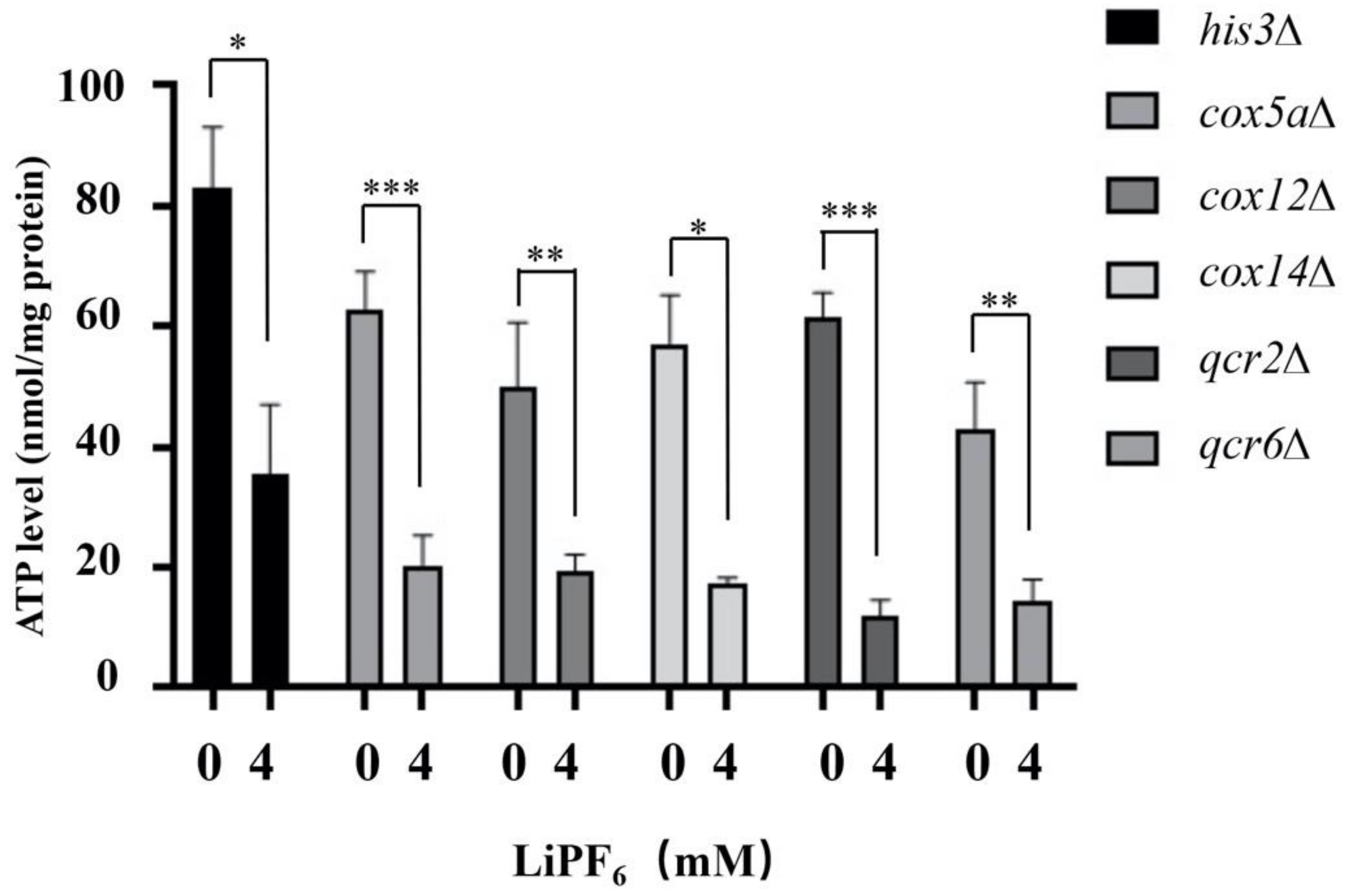
3.6. Deletion of Oxidative Phosphorylation-Related Genes Alters ATP Synthesis Abilities under High Concentration of LiPF6 Treatment
3.7. Oxidative Phosphorylation-Related Mutants Were Specifically Hypersensitive to LiPF6
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Li, J.; He, X.; Zeng, X. Designing and examining e-waste recycling process: Methodology and case studies. Environ. Technol. 2017, 38, 652–660. [Google Scholar] [CrossRef]
- He, Y.; Yuan, X.; Zhang, G.; Wang, H.; Zhang, T.; Xie, W.; Li, L. A critical review of current technologies for the liberation of electrode materials from foils in the recycling process of spent lithium-ion batteries. Sci. Total Environ. 2020, 142382. [Google Scholar] [CrossRef]
- Sun, X.; Hao, H.; Zhao, F.; Liu, Z. Global lithium flow 1994-2015: Implications for improving resource efficiency and security. Environ. Sci. Technol. 2018, 52, 2827–2834. [Google Scholar] [CrossRef] [PubMed]
- Sironval, V.; Reylandt, L.; Chaurand, P.; Ibouraadaten, S.; Palmai-Pallag, M.; Yakoub, Y.; Ucakar, B.; Rose, J.; Poleunis, C.; Vanbever, R.; et al. Respiratory hazard of Li-ion battery components: Elective toxicity of lithium cobalt oxide (LiCoO2) particles in a mouse bioassay. Arch. Toxicol. 2018, 92, 1673–1684. [Google Scholar] [CrossRef] [PubMed]
- Aral, H.; Vecchio-Sadus, A. Toxicity of lithium to humans and the environment--a literature review. Ecotoxicol. Environ. Saf. 2008, 70, 349–356. [Google Scholar] [CrossRef] [PubMed]
- Becker, R.W.; Tyobeka, E.M. Lithium enhances the proliferation of HL-60 promyelocytic leukemia cells. Leuk. Res. 1990, 14, 879–884. [Google Scholar] [CrossRef]
- Kalinowska, M.; Hawrylak-Nowak, B.; Szymanska, M. The influence of two lithium forms on the growth, L-ascorbic acid content and lithium accumulation in lettuce plants. Biol. Trace Elem. Res. 2013, 152, 251–257. [Google Scholar] [CrossRef] [Green Version]
- Castillo-Quan, J.I.; Li, L.; Kinghorn, K.J.; Ivanov, D.K.; Tain, L.S.; Slack, C.; Kerr, F.; Nespital, T.; Thornton, J.; Hardy, J.; et al. Lithium promotes longevity through GSK3/NRF2-dependent hormesis. Cell Rep. 2016, 15, 638–650. [Google Scholar] [CrossRef] [Green Version]
- Leonard, A.; Hantson, P.; Gerber, G.B. Mutagenicity, carcinogenicity and teratogenicity of lithium compounds. Mutat. Res. 1995, 339, 131–137. [Google Scholar] [CrossRef]
- Naranjo, M.A.; Romero, C.; Belles, J.M.; Montesinos, C.; Vicente, O.; Serrano, R. Lithium treatment induces a hypersensitive-like response in tobacco. Planta 2003, 217, 417–424. [Google Scholar] [CrossRef]
- Shahzad, B.; Mughal, M.N.; Tanveer, M.; Gupta, D.; Abbas, G. Is lithium biologically an important or toxic element to living organisms? An overview. Environ. Sci. Pollut. Res. Int. 2017, 24, 103–115. [Google Scholar] [CrossRef]
- Hawrylak-Nowak, B.; Kalinowska, M.; Szymanska, M. A study on selected physiological parameters of plants grown under lithium supplementation. Biol. Trace Elem. Res. 2012, 149, 425–430. [Google Scholar] [CrossRef] [Green Version]
- Tandon, A.; Dhawan, D.K.; Nagpaul, J.P. Effect of lithium on hepatic lipid peroxidation and antioxidative enzymes under different dietary protein regimens. J. Appl. Toxicol. 1998, 18, 187–190. [Google Scholar] [CrossRef]
- Eskandari, M.R.; Fard, J.K.; Hosseini, M.J.; Pourahmad, J. Glutathione mediated reductive activation and mitochondrial dysfunction play key roles in lithium induced oxidative stress and cytotoxicity in liver. Biometals 2012, 25, 863–873. [Google Scholar] [CrossRef] [PubMed]
- Lenox, R.H.; McNamara, R.K.; Papke, R.L.; Manji, H.K. Neurobiology of lithium: An update. J. Clin. Psychiatry 1998, 59 (Suppl. S6), 37–47. [Google Scholar]
- Holstein-Rathlou, N.H. Lithium transport across biological membranes. Kidney Int. Suppl. 1990, 28, S4–S9. [Google Scholar] [PubMed]
- Birch, N.J. Possible mechanism for biological action of lithium. Nature 1976, 264, 681. [Google Scholar] [CrossRef] [PubMed]
- Nagy, T.; Frank, D.; Katai, E.; Yahiro, R.K.; Poor, V.S.; Montsko, G.; Zrinyi, Z.; Kovacs, G.L.; Miseta, A. Lithium induces ER stress and N-glycan modification in galactose-grown Jurkat cells. PLoS ONE 2013, 8, e70410. [Google Scholar] [CrossRef] [Green Version]
- Biczak, R.; Telesinski, A.; Pawlowska, B. Oxidative stress in spring barley and common radish exposed to quaternary ammonium salts with hexafluorophosphate anion. Plant Physiol. Biochem. 2016, 107, 248–256. [Google Scholar] [CrossRef]
- Wei, Q.; Deng, H.; Cui, H.; Fang, J.; Zuo, Z.; Deng, J.; Li, Y.; Wang, X.; Zhao, L. A mini review of fluoride-induced apoptotic pathways. Environ. Sci. Pollut. Res. Int. 2018, 25, 33926–33935. [Google Scholar] [CrossRef] [PubMed]
- Dec, K.; Lukomska, A.; Maciejewska, D.; Jakubczyk, K.; Baranowska-Bosiacka, I.; Chlubek, D.; Wasik, A.; Gutowska, I. The influence of fluorine on the disturbances of homeostasis in the central nervous system. Biol. Trace Elem. Res. 2017, 177, 224–234. [Google Scholar] [CrossRef] [Green Version]
- Zhu, F.; Li, Q.; Zhang, F.; Sun, X.; Cai, G.; Zhang, W.; Chen, X. Chronic lithium treatment diminishes the female advantage in lifespan in Drosophila melanogaster. Clin. Exp. Pharmacol. Physiol. 2015, 42, 617–621. [Google Scholar] [CrossRef]
- Bachmann, R.F.; Wang, Y.; Yuan, P.; Zhou, R.; Li, X.; Alesci, S.; Du, J.; Manji, H.K. Common effects of lithium and valproate on mitochondrial functions: Protection against methamphetamine-induced mitochondrial damage. Int. J. Neuropsychopharmacol. 2009, 12, 805–822. [Google Scholar] [CrossRef] [Green Version]
- Kleineidam, A.; Vavassori, S.; Wang, K.; Schweizer, L.M.; Griac, P.; Schweizer, M. Valproic acid- and lithium-sensitivity in prs mutants of Saccharomyces cerevisiae. Biochem. Soc. Trans. 2009, 37, 1115–1120. [Google Scholar] [CrossRef] [Green Version]
- Hellauer, K.; Lesage, G.; Sdicu, A.M.; Turcotte, B. Large-scale analysis of genes that alter sensitivity to the anticancer drug tirapazamine in Saccharomyces cerevisiae. Mol. Pharmacol. 2005, 68, 1365–1375. [Google Scholar] [CrossRef] [Green Version]
- Barberis, A.; Gunde, T.; Berset, C.; Audetat, S.; Luthi, U. Yeast as a screening tool. Drug Discov. Today Technol. 2005, 2, 187–192. [Google Scholar] [CrossRef]
- Norcliffe, J.L.; Alvarez-Ruiz, E.; Martin-Plaza, J.J.; Steel, P.G.; Denny, P.W. The utility of yeast as a tool for cell-based, target-directed high-throughput screening. Parasitology 2014, 141, 8–16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zimmermann, A.; Hofer, S.; Pendl, T.; Kainz, K.; Madeo, F.; Carmona-Gutierrez, D. Yeast as a tool to identify anti-aging compounds. FEMS Yeast Res. 2018, 18. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Lin, W.; Ma, X.; Lu, Q.; Ma, X.; Bian, G.; Jiang, L. The protein kinase Hal5p is the high-copy suppressor of lithium-sensitive mutations of genes involved in the sporulation and meiosis as well as the ergosterol biosynthesis in Saccharomyces cerevisiae. Genomics 2010, 95, 290–298. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- van Leeuwen, J.; Pons, C.; Mellor, J.C.; Yamaguchi, T.N.; Friesen, H.; Koschwanez, J.; Ušaj, M.M.; Pechlaner, M.; Takar, M.; Ušaj, M.; et al. Exploring genetic suppression interactions on a global scale. Science 2016, 354, aag0839. [Google Scholar] [CrossRef] [Green Version]
- Tong, A.H.; Evangelista, M.; Parsons, A.B.; Xu, H.; Bader, G.D.; Page, N.; Robinson, M.; Raghibizadeh, S.; Hogue, C.W.; Bussey, H.; et al. Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science 2001, 294, 2364–2368. [Google Scholar] [CrossRef] [PubMed]
- Tong, A.H.; Lesage, G.; Bader, G.D.; Ding, H.; Xu, H.; Xin, X.; Young, J.; Berriz, G.F.; Brost, R.L.; Chang, M.; et al. Global mapping of the yeast genetic interaction network. Science 2004, 303, 808–813. [Google Scholar] [CrossRef] [Green Version]
- Wagih, O.; Usaj, M.; Baryshnikova, A.; VanderSluis, B.; Kuzmin, E.; Costanzo, M.; Myers, C.L.; Andrews, B.J.; Boone, C.M.; Parts, L. SGAtools: One-stop analysis and visualization of array-based genetic interaction screens. Nucleic Acids Res. 2013, 41, W591–W596. [Google Scholar] [CrossRef] [Green Version]
- Babazadeh, R.; Ahmadpour, D.; Jia, S.; Hao, X.; Widlund, P.; Schneider, K.; Eisele, F.; Edo, L.D.; Smits, G.J.; Liu, B.; et al. Syntaxin 5 is required for the formation and clearance of protein inclusions during proteostatic stress. Cell. Rep. 2019, 28, 2096–2110.e2098. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hong, E.L.; Balakrishnan, R.; Dong, Q.; Christie, K.R.; Park, J.; Binkley, G.; Costanzo, M.C.; Dwight, S.S.; Engel, S.R.; Fisk, D.G.; et al. Gene Ontology annotations at SGD: New data sources and annotation methods. Nucleic Acids Res. 2008, 36, D577–D581. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xie, C.; Mao, X.; Huang, J.; Ding, Y.; Wu, J.; Dong, S.; Kong, L.; Gao, G.; Li, C.Y.; Wei, L. KOBAS 2.0: A web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 2011, 39, W316–W322. [Google Scholar] [CrossRef] [Green Version]
- Zuberi, K.; Franz, M.; Rodriguez, H.; Montojo, J.; Lopes, C.T.; Bader, G.D.; Morris, Q. GeneMANIA prediction server 2013 update. Nucleic Acids Res. 2013, 41, W115–W122. [Google Scholar] [CrossRef] [PubMed]
- Ho, C.H.; Magtanong, L.; Barker, S.L.; Gresham, D.; Nishimura, S.; Natarajan, P.; Koh, J.L.Y.; Porter, J.; Gray, C.A.; Andersen, R.J.; et al. A molecular barcoded yeast ORF library enables mode-of-action analysis of bioactive compounds. Nat. Biotechnol. 2009, 27, 369–377. [Google Scholar] [CrossRef] [Green Version]
- Huh, W.K.; Falvo, J.V.; Gerke, L.C.; Carroll, A.S.; Howson, R.W.; Weissman, J.S.; O’Shea, E.K. Global analysis of protein localization in budding yeast. Nature 2003, 425, 686–691. [Google Scholar] [CrossRef]
- Peng, X.; Li, F.; Li, S.; Zhu, Y. Expression of a mitochondrial gene orfH79 from the CMS-HongLian rice inhibits Saccharomyces cerevisiae growth and causes excessive ROS accumulation and decrease in ATP. Biotechnol. Lett. 2009, 31, 409–414. [Google Scholar] [CrossRef]
- Fletcher, E.; Gao, K.; Mercurio, K.; Ali, M.; Baetz, K. Yeast chemogenomic screen identifies distinct metabolic pathways required to tolerate exposure to phenolic fermentation inhibitors ferulic acid, 4-hydroxybenzoic acid and coniferyl aldehyde. Metab. Eng. 2019, 52, 98–109. [Google Scholar] [CrossRef]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef]
- Ye, X.; Morikawa, K.; Ho, S.-H.; Araki, M.; Nishida, K.; Hasunuma, T.; Hara, K.Y.; Kondo, A. Evaluation of genes involved in oxidative phosphorylation in yeast by developing a simple and rapid method to measure mitochondrial ATP synthetic activity. Microb. Cell Fact. 2015, 14, e56. [Google Scholar] [CrossRef] [Green Version]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Wilson, D.F. Oxidative phosphorylation: Regulation and role in cellular and tissue metabolism. J. Physiol. 2017, 595, 7023–7038. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schafer, J.; Dawitz, H.; Ott, M.; Adelroth, P.; Brzezinski, P. Structural and functional heterogeneity of cytochrome c oxidase in S. cerevisiae. Biochim. Biophys. Acta Bioenerg. 2018, 1859, 699–704. [Google Scholar] [CrossRef]
- Wikstrom, M.; Sharma, V. Proton pumping by cytochrome c oxidase—A 40 year anniversary. Biochim. Biophys. Acta Bioenerg. 2018, 1859, 692–698. [Google Scholar] [CrossRef] [PubMed]
- Timon-Gomez, A.; Nyvltova, E.; Abriata, L.A.; Vila, A.J.; Hosler, J.; Barrientos, A. Mitochondrial cytochrome c oxidase biogenesis: Recent developments. Semin. Cell Dev. Biol. 2018, 76, 163–178. [Google Scholar] [CrossRef] [PubMed]
- Fontanesi, F.; Clemente, P.; Barrientos, A. Cox25 teams up with Mss51, Ssc1, and Cox14 to regulate mitochondrial cytochrome c oxidase subunit 1 expression and assembly in Saccharomyces cerevisiae. J. Biol. Chem. 2011, 286, 555–566. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mick, D.U.; Vukotic, M.; Piechura, H.; Meyer, H.E.; Warscheid, B.; Deckers, M.; Rehling, P. Coa3 and Cox14 are essential for negative feedback regulation of COX1 translation in mitochondria. J. Cell Biol. 2010, 191, 141–154. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chandel, N.S. Evolution of mitochondria as signaling organelles. Cell Metab. 2015, 22, 204–206. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shadel, G.S.; Horvath, T.L. Mitochondrial ROS signaling in organismal homeostasis. Cell 2015, 163, 560–569. [Google Scholar] [CrossRef] [Green Version]
- Labbe, K.; Murley, A.; Nunnari, J. Determinants and functions of mitochondrial behavior. Annu. Rev. Cell. Dev. Biol. 2014, 30, 357–391. [Google Scholar] [CrossRef]
- Kwon, Y.Y.; Choi, K.M.; Cho, C.; Lee, C.K. Mitochondrial efficiency-dependent viability of Saccharomyces cerevisiae mutants carrying individual electron transport chain component deletions. Mol. Cells 2015, 38, 1054–1063. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zorov, D.B.; Juhaszova, M.; Sollott, S.J. Mitochondrial reactive oxygen species (ROS) and ROS-induced ROS release. Physiol. Rev. 2014, 94, 909–950. [Google Scholar] [CrossRef] [Green Version]
- Laporte, D.; Gouleme, L.; Jimenez, L.; Khemiri, I.; Sagot, I. Mitochondria reorganization upon proliferation arrest predicts individual yeast cell fate. Elife 2018, 7, e35685. [Google Scholar] [CrossRef]
- Klaunig, J.E.; Wang, Z.; Pu, X.; Zhou, S. Oxidative stress and oxidative damage in chemical carcinogenesis. Toxicol. Appl. Pharmacol. 2011, 254, 86–99. [Google Scholar] [CrossRef]
- James, J.; Fiji, N.; Roy, D.; Andrew, M.G.D.; Shihabudeen, M.S.; Chattopadhyay, D.; Thirumurugan, K. A rapid method to assess reactive oxygen species in yeast using H2DCF-DA. Anal. Methods 2015, 7, 8572–8575. [Google Scholar] [CrossRef]
- Markgraf, D.F.; Ahnert, F.; Arlt, H.; Mari, M.; Peplowska, K.; Epp, N.; Griffith, J.; Reggiori, F.; Ungermann, C. The CORVET subunit Vps8 cooperates with the Rab5 homolog Vps21 to induce clustering of late endosomal compartments. Mol. Biol. Cell. 2009, 20, 5276–5289. [Google Scholar] [CrossRef] [Green Version]
- Robinson, J.S.; Klionsky, D.J.; Banta, L.M.; Emr, S.D. Protein sorting in Saccharomyces cerevisiae: Isolation of mutants defective in the delivery and processing of multiple vacuolar hydrolases. Mol. Cell. Biol. 1988, 8, 4936–4948. [Google Scholar] [CrossRef]
- Davies, B.A.; Topp, J.D.; Sfeir, A.J.; Katzmann, D.J.; Carney, D.S.; Tall, G.G.; Friedberg, A.S.; Deng, L.; Chen, Z.; Horazdovsky, B.F. Vps9p CUE domain ubiquitin binding is required for efficient endocytic protein traffic. J. Biol. Chem. 2003, 278, 19826–19833. [Google Scholar] [CrossRef] [Green Version]
- Rothman, J.H.; Howald, I.; Stevens, T.H. Characterization of genes required for protein sorting and vacuolar function in the yeast Saccharomyces cerevisiae. EMBO J. 1989, 8, 2057–2065. [Google Scholar] [CrossRef] [PubMed]
- Reggiori, F.; Wang, C.W.; Stromhaug, P.E.; Shintani, T.; Klionsky, D.J. Vps51 is part of the yeast Vps fifty-three tethering complex essential for retrograde traffic from the early endosome and Cvt vesicle completion. J. Biol. Chem. 2003, 278, 5009–5020. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Conibear, E.; Cleck, J.N.; Stevens, T.H. Vps51p mediates the association of the GARP (Vps52/53/54) complex with the late Golgi t-SNARE Tlg1p. Mol. Biol. Cell. 2003, 14, 1610–1623. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ott, M.; Gogvadze, V.; Orrenius, S.; Zhivotovsky, B. Mitochondria, oxidative stress and cell death. Apoptosis 2007, 12, 913–922. [Google Scholar] [CrossRef] [PubMed]
- Valko, M.; Morris, H.; Cronin, M.T. Metals, toxicity and oxidative stress. Curr. Med. Chem. 2005, 12, 1161–1208. [Google Scholar] [CrossRef] [Green Version]
- Valko, M.; Leibfritz, D.; Moncol, J.; Cronin, M.T.; Mazur, M.; Telser, J. Free radicals and antioxidants in normal physiological functions and human disease. Int. J. Biochem. Cell Biol. 2007, 39, 44–84. [Google Scholar] [CrossRef]
- Salimi, A.; Gholamifar, E.; Naserzadeh, P.; Hosseini, M.J.; Pourahmad, J. Toxicity of lithium on isolated heart mitochondria and cardiomyocyte: A justification for its cardiotoxic adverse effect. J. Biochem. Mol. Toxicol. 2017, 31. [Google Scholar] [CrossRef]
- Hroudova, J.; Fisar, Z. Activities of respiratory chain complexes and citrate synthase influenced by pharmacologically different antidepressants and mood stabilizers. Neuro. Endocrinol. Lett. 2010, 31, 336–342. [Google Scholar]
- Luptak, M.; Hroudova, J. Important role of mitochondria and the effect of mood stabilizers on mitochondrial function. Physiol. Res. 2019, 68, S3–S15. [Google Scholar] [CrossRef] [PubMed]
- Zuo, H.; Chen, L.; Kong, M.; Qiu, L.; Lu, P.; Wu, P.; Yang, Y.; Chen, K. Toxic effects of fluoride on organisms. Life Sci. 2018, 198, 18–24. [Google Scholar] [CrossRef] [PubMed]
- Srinivasan, S.; Avadhani, N.G. Cytochrome c oxidase dysfunction in oxidative stress. Free Radic. Biol. Med. 2012, 53, 1252–1263. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rich, P.R.; Marechal, A. The mitochondrial respiratory chain. Essays Biochem. 2010, 47, 1–23. [Google Scholar] [CrossRef] [PubMed] [Green Version]



| ORF | Gene | Score 1 a | p-Value 1 a | Score 2 a | p-Value 2 a | Score 3 a | p-Value 3 a | Location b |
|---|---|---|---|---|---|---|---|---|
| Oxidative Phosphorylation | ||||||||
| YNL052W | COX5A | −0.3751 | 0.00006 | −0.2880 | 0.00027 | −0.3322 | 0.00011 | mitochondrion |
| YLR038C | COX12 | −0.3392 | 0.00002 | −0.7994 | 0.00339 | −0.7440 | 0.01456 | cytoplasm |
| YML129C | COX14 | −0.5864 | 0.00055 | −0.7346 | 0.00388 | −0.3517 | 0.00018 | mitochondrion |
| YPR191W | QCR2 | −0.5095 | 0.00365 | −0.7160 | 0.00026 | −0.3503 | 0.00001 | mitochondrion |
| YFR033C | QCR6 | −0.2124 | 0.00002 | −0.5472 | 0.00001 | −0.2844 | 0.00006 | cytoplasm |
| Electron Transport Chain | ||||||||
| YNL315C | ATP11 | −0.4236 | 0.00001 | −0.4884 | 0.00003 | −0.2834 | 0.00001 | mitochondrion |
| YNR020C | ATP23 | −0.4009 | 0.00000 | −0.4283 | 0.00013 | −0.3016 | 0.00001 | - |
| YLR393W | ATP10 | −0.3061 | 0.00000 | −0.3744 | 0.00107 | −0.4161 | 0.00003 | mitochondrion |
| YOR125C | CAT5 | −0.6673 | 0.00062 | −0.6531 | 0.00018 | −0.4810 | 0.00038 | - |
| YNR041C | COQ2 | −0.5151 | 0.00196 | −0.6484 | 0.00043 | −0.4307 | 0.00005 | mitochondrion |
| YHR116W | COX23 | −0.4963 | 0.00000 | −0.6415 | 0.00326 | −0.4725 | 0.00035 | cytoplasm |
| Mitochondrial Proteins | ||||||||
| YMR282C | AEP2 | −0.6256 | 0.00030 | −0.6545 | 0.00012 | −0.4035 | 0.00046 | mitochondrion |
| YNL003C | PET8 | −0.6297 | 0.00277 | −0.6204 | 0.01065 | −0.4815 | 0.00005 | mitochondrion |
| YAL048C | GEM1 | −0.5227 | 0.00222 | −0.4628 | 0.00008 | −0.3097 | 0.00008 | - |
| YPR011C | - | −0.3696 | 0.00006 | −0.3274 | 0.00013 | −0.2720 | 0.00212 | mitochondrion |
| YOR045W | TOM6 | −0.2975 | 0.00012 | −0.2318 | 0.00004 | −0.3106 | 0.00001 | mitochondrion |
| YKL162C | - | −0.4414 | 0.00001 | −0.3662 | 0.00005 | −0.2634 | 0.00001 | mitochondrion |
| YOR350C | MNE1 | −0.2384 | 0.00003 | −0.6479 | 0.00014 | −0.2888 | 0.00001 | mitochondrion |
| YDL104C | QRI7 | −0.6281 | 0.00128 | −0.6104 | 0.00025 | −0.3629 | 0.00001 | mitochondrion |
| YER153C | PET122 | −0.2664 | 0.00032 | −0.6257 | 0.00064 | −0.3802 | 0.00001 | mitochondrion |
| YCR071C | IMG2 | −0.5618 | 0.00349 | −0.5690 | 0.00003 | −0.3813 | 0.00003 | mitochondrion |
| DNA and RNA-Related Genes | ||||||||
| YCR028C-A | RIM1 | −0.5714 | 0.00325 | −0.5815 | 0.00024 | −0.2990 | 0.00003 | mitochondrion |
| YKL208W | CBT1 | −0.4650 | 0.00064 | −0.8063 | 0.00017 | −0.4649 | 0.00002 | mitochondrion |
| YOL080C | REX4 | −0.5478 | 0.00000 | −0.5925 | 0.00001 | −0.4610 | 0.00001 | nucleolus, nucleus |
| YDL033C | SLM3 | −0.4263 | 0.00014 | −0.5773 | 0.00007 | −0.4009 | 0.00002 | mitochondrion |
| YKL074C | MUD2 | −0.3940 | 0.00001 | −0.2244 | 0.00011 | −0.2730 | 0.00001 | cytoplasm, nucleus |
| YOR033C | EXO1 | −0.6815 | 0.00162 | −0.5806 | 0.00015 | −0.2738 | 0.00001 | nucleus |
| YNL215W | IES2 | −0.4173 | 0.00006 | −0.3761 | 0.00000 | −0.2594 | 0.00003 | nucleus |
| YDR386W | MUS81 | −0.2506 | 0.00000 | −0.3685 | 0.00003 | −0.2387 | 0.00001 | - |
| YIR002C | MPH1 | −0.3117 | 0.00000 | −0.2264 | 0.00001 | −0.5003 | 0.00021 | cytoplasm, nucleus |
| YOL095C | HMI1 | −0.6735 | 0.00339 | −0.5763 | 0.00010 | −0.2354 | 0.00002 | - |
| YPR022C | - | −0.3192 | 0.00020 | −0.3845 | 0.00000 | −0.4943 | 0.00003 | cytoplasm, nucleus |
| YNL136W | EAF7 | −0.4389 | 0.00004 | −0.2674 | 0.00001 | −0.2094 | 0.00001 | nucleus |
| YER143W | DDI1 | −0.3362 | 0.00000 | −0.2392 | 0.00001 | −0.2629 | 0.00002 | cytoplasm |
| YCR077C | PAT1 | −0.3727 | 0.00008 | −0.3601 | 0.00001 | −0.3147 | 0.00005 | cytoplasm |
| YGL168W | HUR1 | −0.3131 | 0.00001 | −0.5268 | 0.00000 | −0.4062 | 0.00001 | - |
| Transport System | ||||||||
| YAL002W | VPS8 | −0.6784 | 0.00002 | −0.6423 | 0.00002 | −0.4657 | 0.00001 | endosome |
| YML097C | VPS9 | −0.6179 | 0.00001 | −0.5143 | 0.00003 | −0.5346 | 0.00008 | cytoplasm |
| YKR020W | VPS51 | −0.4877 | 0.00012 | −0.4249 | 0.00002 | −0.3440 | 0.00004 | punctate composite |
| YKL041W | VPS24 | −0.4652 | 0.00000 | −0.4097 | 0.00026 | −0.2860 | 0.00006 | punctate composite, endosome |
| YOR322C | LDB19 | −0.6997 | 0.00006 | −0.4052 | 0.00000 | −0.2972 | 0.00003 | cytoplasm, late Golgi |
| YLR065C | ENV10 | −0.2237 | 0.00003 | −0.2707 | 0.00001 | −0.3896 | 0.00009 | ambiguous |
| YMR021C | MAC1 | −0.5325 | 0.00027 | −0.5749 | 0.00014 | −0.5273 | 0.00029 | cytoplasm, nucleus |
| YGR105W | VMA21 | −0.5096 | 0.00006 | −0.4375 | 0.00002 | −0.3984 | 0.00002 | vacuole |
| YDR126W | SWF1 | −0.4926 | 0.00004 | −0.2602 | 0.00000 | −0.3249 | 0.00002 | - |
| YMR123W | PKR1 | −0.2993 | 0.00000 | −0.2286 | 0.00002 | −0.2083 | 0.00005 | ER |
| YOR181W | LAS17 | −0.2113 | 0.00001 | −0.2026 | 0.00002 | −0.2052 | 0.00002 | actin |
| YLR337C | VRP1 | −0.5067 | 0.00006 | −0.3804 | 0.00000 | −0.6244 | 0.00002 | punctate composite, actin |
| Cell Metabolism | ||||||||
| YMR189W | GCV2 | −0.2085 | 0.00000 | −0.4420 | 0.00030 | −0.5500 | 0.00002 | mitochondrion |
| YGL237C | HAP2 | −0.5162 | 0.00004 | −0.2985 | 0.00000 | −0.3660 | 0.00000 | nucleus |
| YCR053W | THR4 | −0.2961 | 0.00004 | −0.3573 | 0.00001 | −0.3767 | 0.00016 | cytoplasm, nucleus |
| YPL157W | TGS1 | −0.4255 | 0.00001 | −0.2466 | 0.00001 | −0.2310 | 0.00002 | nucleolus |
| YMR216C | SKY1 | −0.6509 | 0.00035 | −0.5810 | 0.00003 | −0.5751 | 0.00002 | cytoplasm |
| YLR436C | ECM30 | −0.2724 | 0.00002 | −0.4312 | 0.00000 | −0.4328 | 0.00002 | cytoplasm |
| YHR096C | HXT5 | −0.2206 | 0.00004 | −0.2280 | 0.00002 | −0.2911 | 0.00003 | - |
| YBL021C | HAP3 | −0.3353 | 0.00082 | −0.2217 | 0.00000 | −0.3064 | 0.00000 | cytoplasm, nucleus |
| YNL229C | URE2 | −0.2639 | 0.00012 | −0.3145 | 0.00001 | −0.2208 | 0.00000 | cytoplasm |
| Protein Synthesis and Degradation | ||||||||
| YNL162W | RPL42A | −0.2885 | 0.00000 | −0.2105 | 0.00000 | −0.2294 | 0.00000 | cytoplasm |
| YBL024W | NCL1 | −0.5915 | 0.00002 | −0.5668 | 0.00001 | −0.4087 | 0.00007 | nucleus |
| YPR148C | - | −0.4126 | 0.00003 | −0.3759 | 0.00002 | −0.4687 | 0.00003 | punctate composite |
| YKL081W | TEF4 | −0.3932 | 0.00001 | −0.3216 | 0.00003 | −0.3571 | 0.00000 | cytoplasm |
| YBR082C | UBC4 | −0.2783 | 0.00002 | −0.3556 | 0.00001 | −0.3065 | 0.00000 | cytoplasm, nucleus |
| YOL025W | LAG2 | −0.3460 | 0.00001 | −0.2428 | 0.00000 | −0.3409 | 0.00001 | - |
| YNL153C | GIM3 | −0.2539 | 0.00004 | −0.2300 | 0.00002 | −0.4280 | 0.00005 | cytoplasm |
| Cell Resistance | ||||||||
| YOR084W | LPX1 | −0.4131 | 0.00000 | −0.3710 | 0.00029 | −0.2318 | 0.00011 | ambiguous |
| YPL196W | OXR1 | −0.3809 | 0.00001 | −0.2965 | 0.00000 | −0.2073 | 0.00000 | - |
| YBL043W | ECM13 | −0.4461 | 0.00002 | −0.4689 | 0.00000 | −0.3406 | 0.00001 | - |
| YPL056C | LCL1 | −0.3480 | 0.00005 | −0.3583 | 0.00004 | −0.3946 | 0.00002 | - |
| YBR067C | TIP1 | −0.2071 | 0.00006 | −0.2896 | 0.00003 | −0.2285 | 0.00000 | ER |
| YGL007W | BRP1 | −0.7219 | 0.00031 | −0.7044 | 0.00002 | −0.6213 | 0.00005 | - |
| Unknown | ||||||||
| YDL062W | −0.4626 | 0.00485 | −0.5224 | 0.00048 | −0.3412 | 0.00002 | - | |
| YCL036W | GFD2 | −0.3546 | 0.00005 | −0.4342 | 0.00023 | −0.3447 | 0.00037 | - |
| YDR360W | OPI7 | −0.4144 | 0.00001 | −0.3920 | 0.00002 | −0.3042 | 0.00000 | - |
| YDR209C | - | −0.3699 | 0.00002 | −0.3455 | 0.00001 | −0.3306 | 0.00000 | - |
| YDL187C | - | −0.4446 | 0.00048 | −0.3041 | 0.00013 | −0.4404 | 0.00030 | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jin, X.; Zhang, J.; An, T.; Zhao, H.; Fu, W.; Li, D.; Liu, S.; Cao, X.; Liu, B. A Genome-Wide Screen in Saccharomyces cerevisiae Reveals a Critical Role for Oxidative Phosphorylation in Cellular Tolerance to Lithium Hexafluorophosphate. Cells 2021, 10, 888. https://doi.org/10.3390/cells10040888
Jin X, Zhang J, An T, Zhao H, Fu W, Li D, Liu S, Cao X, Liu B. A Genome-Wide Screen in Saccharomyces cerevisiae Reveals a Critical Role for Oxidative Phosphorylation in Cellular Tolerance to Lithium Hexafluorophosphate. Cells. 2021; 10(4):888. https://doi.org/10.3390/cells10040888
Chicago/Turabian StyleJin, Xuejiao, Jie Zhang, Tingting An, Huihui Zhao, Wenhao Fu, Danqi Li, Shenkui Liu, Xiuling Cao, and Beidong Liu. 2021. "A Genome-Wide Screen in Saccharomyces cerevisiae Reveals a Critical Role for Oxidative Phosphorylation in Cellular Tolerance to Lithium Hexafluorophosphate" Cells 10, no. 4: 888. https://doi.org/10.3390/cells10040888
APA StyleJin, X., Zhang, J., An, T., Zhao, H., Fu, W., Li, D., Liu, S., Cao, X., & Liu, B. (2021). A Genome-Wide Screen in Saccharomyces cerevisiae Reveals a Critical Role for Oxidative Phosphorylation in Cellular Tolerance to Lithium Hexafluorophosphate. Cells, 10(4), 888. https://doi.org/10.3390/cells10040888






