Initiation and Pathogenesis of Severe Asthma with Fungal Sensitization
Abstract
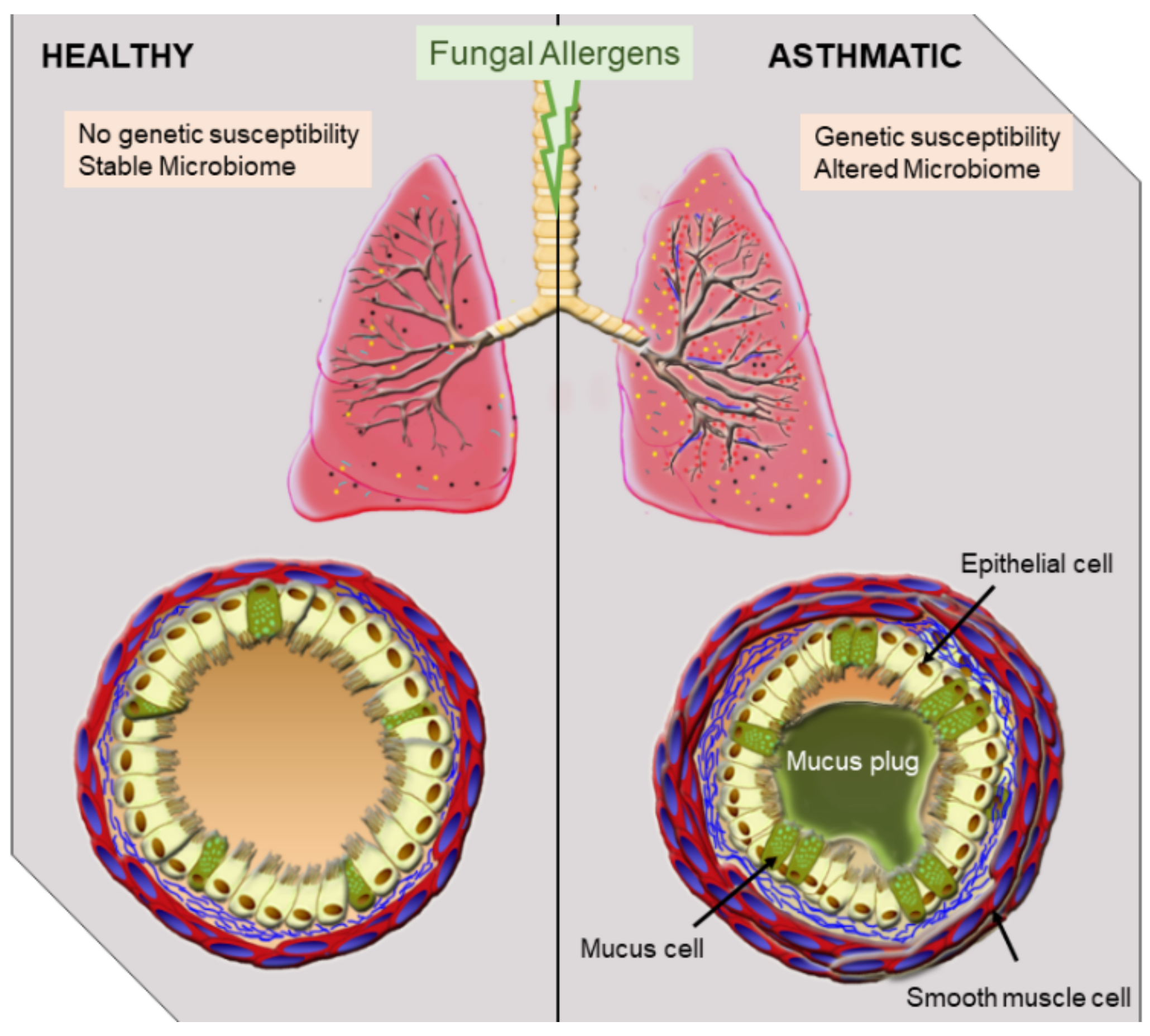
1. Introduction
2. Overview of Fungal Allergen-Mediated Immune Responses Leading to SAFS
3. Animal Models of Severe Asthma with Fungal Sensitization
4. Mechanisms of SAFS Induction at the Respiratory Barrier
5. T Cell Response to Fungal Allergen Exposure in the Airways
6. Eosinophils in SAFS
7. B Cells in SAFS
8. Commensals and Allergic Asthma
9. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sakula, A. Sir John Floyer’s A Treatise of the Asthma (1698). Thorax 1984, 39, 248–254. [Google Scholar] [CrossRef] [PubMed]
- Braman, S.S. The global burden of asthma. Chest 2006, 130, 4S–12S. [Google Scholar] [CrossRef] [PubMed]
- Nurmagambetov, T.; Kuwahara, R.; Garbe, P. The Economic Burden of Asthma in the United States, 2008–2013. Ann. Am. Thorac. Soc. 2018, 15, 348–356. [Google Scholar] [CrossRef] [PubMed]
- US Department of Health and Human Services. National Heart, Lung and Blood Institute: Expert Panel Report 3: Guidelines for the Diagnosis and Management of Asthma (EPR-3 2007); NIH Item No. 08-4051; US Department of Health and Human Services: Washington, DC, USA, 2007.
- Lötvall, J.; Akdis, C.A.; Bacharier, L.B.; Bjermer, L.; Casale, T.B.; Custovic, A.; Lemanske, R.F., Jr.; Wardlaw, A.J.; Wenzel, S.E.; Greenberger, P.A. Asthma endotypes: A new approach to classification of disease entities within the asthma syndrome. J. Allergy Clin. Immunol. 2011, 127, 355–360. [Google Scholar] [CrossRef]
- Romanet-Manent, S.; Charpin, D.; Magnan, A.; Lanteaume, A.; Vervloet, D.; Group, E.C. Allergic vs nonallergic asthma: What makes the difference? Allergy 2002, 57, 607–613. [Google Scholar] [CrossRef]
- Munthe-Kaas, M.C.; Carlsen, K.H.; Håland, G.; Devulapalli, C.S.; Gervin, K.; Egeland, T.; Carlsen, K.L.; Undlien, D. Immunology, C. T cell–specific T-box transcription factor haplotype is associated with allergic asthma in children. J. Allergy Clin. Immunol. 2008, 121, 51–56. [Google Scholar] [CrossRef]
- Nguyen, K.D.; Vanichsarn, C.; Nadeau, K.C. Impaired IL-10–dependent Induction of Tolerogenic Dendritic Cells by CD4+ CD25hiCD127lo/− Natural Regulatory T Cells in Human Allergic Asthma. Am. J. Respir. Crit. Care Med. 2009, 180, 823–833. [Google Scholar] [CrossRef]
- Sears, M.R.; Greene, J.M.; Willan, A.R.; Wiecek, E.M.; Taylor, D.R.; Flannery, E.M.; Cowan, J.O.; Herbison, G.P.; Silva, P.A.; Poulton, R. A longitudinal, population-based, cohort study of childhood asthma followed to adulthood. N. Engl. J. Med. 2003, 349, 1414–1422. [Google Scholar] [CrossRef]
- DeVries, A.; Wlasiuk, G.; Miller, S.J.; Bosco, A.; Stern, D.A.; Nicodemus-Johnson, J.; Jones, A.C.; Rothers, J.; Lohman, I.C.; Wright, A.L.; et al. Neonatal epigenetic predictors of childhood asthma map to immunoregulatory and pro-inflammatory pathways. Am. J. Respir. Crit. Care Med. 2015, 191, A3524. [Google Scholar]
- Lockett, G.A.; Soto-Ramírez, N.; Ray, M.A.; Everson, T.M.; Xu, C.J.; Patil, V.K.; Terry, W.; Kaushal, A.; Rezwan, F.I.; Ewart, S.L.J.A. Association of season of birth with DNA methylation and allergic disease. Allergy 2016, 71, 1314–1324. [Google Scholar] [CrossRef]
- DeVries, A.; Vercelli, D. Early predictors of asthma and allergy in children: The role of epigenetics. Curr. Opin. Allergy Clin. Immunol. 2015, 15, 435. [Google Scholar] [CrossRef]
- Nelson, H.S. The importance of allergens in the development of asthma and the persistence of symptoms. J. Allergy Clin. Immunol. 2000, 105, S628–S632. [Google Scholar] [CrossRef]
- Frohlich-Nowoisky, J.; Pickersgill, D.A.; Despres, V.R.; Poschl, U. High diversity of fungi in air particulate matter. Proc. Natl. Acad. Sci. USA 2009, 106, 12814–12819. [Google Scholar] [CrossRef]
- Womble, S.E.; Burton, L.E.; Kolb, L.; Girman, J.R.; Carpenter, M.; McCarthy, J.F. Prevalence and concentrations of culturable airborne fungal spores in 86 office buildings from the Building Assessment Survey and Evaluation (BASE) study. In Proceedings of the 8th Inernational Conference on Indoor Air and Climate, Ediburgh, Scotland, 8–13 August 1999; Volume 1, pp. 261–266. [Google Scholar]
- Bozek, A.; Pyrkosz, K. Immunotherapy of mold allergy: A review. Hum. Vaccin. Immunother. 2017, 13, 2397–2401. [Google Scholar] [CrossRef]
- Samarasinghe, A.E.; Woolard, S.N.; Boyd, K.L.; Hoselton, S.A.; Schuh, J.M.; McCullers, J.A. The immune profile associated with acute allergic asthma accelerates clearance of influenza virus. Immunol. Cell. Biol. 2014, 92, 449–459. [Google Scholar] [CrossRef]
- Leino, M.S.; Loxham, M.; Blume, C.; Swindle, E.J.; Jayasekera, N.P.; Dennison, P.W.; Shamji, B.W.; Edwards, M.J.; Holgate, S.T.; Howarth, P.H.; et al. Barrier disrupting effects of alternaria alternata extract on bronchial epithelium from asthmatic donors. PLoS ONE 2013, 8, e71278. [Google Scholar] [CrossRef]
- Tomee, J.F.; Wierenga, A.T.; Hiemstra, P.S.; Kauffman, H.K. Proteases from Aspergillus fumigatus induce release of proinflammatory cytokines and cell detachment in airway epithelial cell lines. J. Infect. Dis. 1997, 176, 300–303. [Google Scholar] [CrossRef]
- Kogan, T.V.; Jadoun, J.; Mittelman, L.; Hirschberg, K.; Osherov, N. Involvement of secreted Aspergillus fumigatus proteases in disruption of the actin fiber cytoskeleton and loss of focal adhesion sites in infected A549 lung pneumocytes. J. Infect. Dis. 2004, 189, 1965–1973. [Google Scholar] [CrossRef]
- Tai, H.Y.; Tam, M.F.; Chou, H.; Peng, H.J.; Su, S.N.; Perng, D.W.; Shen, H.D. Pen ch 13 allergen induces secretion of mediators and degradation of occludin protein of human lung epithelial cells. Allergy 2006, 61, 382–388. [Google Scholar] [CrossRef]
- Chen, J.C.; Chuang, J.G.; Su, Y.Y.; Chiang, B.L.; Lin, Y.S.; Chow, L.P. The protease allergen Pen c 13 induces allergic airway inflammation and changes in epithelial barrier integrity and function in a murine model. J. Biol. Chem. 2011, 286, 26667–26679. [Google Scholar] [CrossRef]
- Redes, J.L.; Basu, T.; Ram-Mohan, S.; Ghosh, C.C.; Chan, E.C.; Sek, A.C.; Zhao, M.; Krishnan, R.; Rosenberg, H.F.; Druey, K.M. Aspergillus fumigatus-Secreted Alkaline Protease 1 Mediates Airways Hyperresponsiveness in Severe Asthma. Immunohorizons 2019, 3, 368–377. [Google Scholar] [CrossRef] [PubMed]
- Balenga, N.A.; Klichinsky, M.; Xie, Z.; Chan, E.C.; Zhao, M.; Jude, J.; Laviolette, M.; Panettieri, R.A., Jr.; Druey, K.M. A fungal protease allergen provokes airway hyper-responsiveness in asthma. Nat. Commun. 2015, 6, 6763. [Google Scholar] [CrossRef] [PubMed]
- Hurst, S.D.; Muchamuel, T.; Gorman, D.M.; Gilbert, J.M.; Clifford, T.; Kwan, S.; Menon, S.; Seymour, B.; Jackson, C.; Kung, T.T.; et al. New IL-17 family members promote Th1 or Th2 responses in the lung: In vivo function of the novel cytokine IL-25. J. Immunol. 2002, 169, 443–453. [Google Scholar] [CrossRef] [PubMed]
- Hogaboam, C.M.; Blease, K.; Mehrad, B.; Steinhauser, M.L.; Standiford, T.J.; Kunkel, S.L.; Lukacs, N.W. Chronic airway hyperreactivity, goblet cell hyperplasia, and peribronchial fibrosis during allergic airway disease induced by Aspergillus fumigatus. Am. J. Pathol. 2000, 156, 723–732. [Google Scholar] [CrossRef]
- Vincent, M.; Percier, P.; De Prins, S.; Huygen, K.; Potemberg, G.; Muraille, E.; Romano, M.; Michel, O.; Denis, O. Investigation of inflammatory and allergic responses to common mold species: Results from in vitro experiments, from a mouse model of asthma, and from a group of asthmatic patients. Indoor Air 2017, 27, 933–945. [Google Scholar] [CrossRef]
- Toki, S.; Goleniewska, K.; Zhang, J.; Zhou, W.; Newcomb, D.C.; Zhou, B.; Kita, H.; Boyd, K.L.; Peebles, R.S., Jr. TSLP and IL-33 reciprocally promote each other’s lung protein expression and ILC2 receptor expression to enhance innate type-2 airway inflammation. Allergy 2020, 75, 1606–1617. [Google Scholar] [CrossRef]
- Lv, J.; Yu, Q.; Lv, J.; Di, C.; Lin, X.; Su, W.; Wu, M.; Xia, Z. Airway epithelial TSLP production of TLR2 drives type 2 immunity in allergic airway inflammation. Eur. J. Immunol. 2018, 48, 1838–1850. [Google Scholar] [CrossRef]
- Kim, Y.H.; Lee, S.H. TGF-beta/SMAD4 mediated UCP2 downregulation contributes to Aspergillus protease-induced inflammation in primary bronchial epithelial cells. Redox Biol. 2018, 18, 104–113. [Google Scholar] [CrossRef]
- Kobayashi, T.; Iijima, K.; Dent, A.L.; Kita, H. Follicular helper T cells mediate IgE antibody response to airborne allergens. J. Allergy Clin. Immunol. 2017, 139, 300–313.e7. [Google Scholar] [CrossRef]
- Moreira, A.P.; Cavassani, K.A.; Ismailoglu, U.B.; Hullinger, R.; Dunleavy, M.P.; Knight, D.A.; Kunkel, S.L.; Uematsu, S.; Akira, S.; Hogaboam, C.M. The protective role of TLR6 in a mouse model of asthma is mediated by IL-23 and IL-17A. J. Clin. Investig. 2011, 121, 4420–4432. [Google Scholar] [CrossRef]
- Fei, M.; Bhatia, S.; Oriss, T.B.; Yarlagadda, M.; Khare, A.; Akira, S.; Saijo, S.; Iwakura, Y.; Fallert Junecko, B.A.; Reinhart, T.A.; et al. TNF-alpha from inflammatory dendritic cells (DCs) regulates lung IL-17A/IL-5 levels and neutrophilia versus eosinophilia during persistent fungal infection. Proc. Natl. Acad. Sci. USA 2011, 108, 5360–5365. [Google Scholar] [CrossRef]
- Ramirez-Ortiz, Z.G.; Means, T.K. The role of dendritic cells in the innate recognition of pathogenic fungi (A. fumigatus, C. neoformans and C. albicans). Virulence 2012, 3, 635–646. [Google Scholar] [CrossRef]
- Hoselton, S.A.; Samarasinghe, A.E.; Seydel, J.M.; Schuh, J.M. An inhalation model of airway allergic response to inhalation of environmental Aspergillus fumigatus conidia in sensitized BALB/c mice. Med. Mycol. 2010, 48, 1056–1065. [Google Scholar] [CrossRef]
- Ghosh, S.; Hoselton, S.A.; Schuh, J.M. mu-chain-deficient mice possess B-1 cells and produce IgG and IgE, but not IgA, following systemic sensitization and inhalational challenge in a fungal asthma model. J. Immunol. 2012, 189, 1322–1329. [Google Scholar] [CrossRef]
- Doorley, L.A.; LeMessurier, K.S.; Iverson, A.R.; Palipane, M.; Samarasinghe, A.E. Humoral immune responses during asthma and influenza co-morbidity in mice. Immunobiology 2017, 222, 1064–1073. [Google Scholar] [CrossRef]
- Guerra, E.S.; Lee, C.K.; Specht, C.A.; Yadav, B.; Huang, H.; Akalin, A.; Huh, J.R.; Mueller, C.; Levitz, S.M. Central Role of IL-23 and IL-17 Producing Eosinophils as Immunomodulatory Effector Cells in Acute Pulmonary Aspergillosis and Allergic Asthma. PLoS Pathog. 2017, 13, e1006175. [Google Scholar] [CrossRef]
- Muniz, V.S.; Silva, J.C.; Braga, Y.A.V.; Melo, R.C.N.; Ueki, S.; Takeda, M.; Hebisawa, A.; Asano, K.; Figueiredo, R.T.; Neves, J.S. Eosinophils release extracellular DNA traps in response to Aspergillus fumigatus. J. Allergy Clin. Immunol. 2018, 141, 571–585.e7. [Google Scholar] [CrossRef]
- Hawksworth, D.L.; Lucking, R. Fungal Diversity Revisited: 2.2 to 3.8 Million Species. Microbiol. Spectr. 2017, 5. [Google Scholar] [CrossRef]
- Kohler, J.R.; Casadevall, A.; Perfect, J. The spectrum of fungi that infects humans. Cold Spring Harb. Perspect. Med. 2014, 5, a019273. [Google Scholar] [CrossRef]
- Templeton, S.P.; Rivera, A.; Hube, B.; Jacobsen, I.D. Editorial: Immunity to Human Fungal Pathogens: Mechanisms of Host Recognition, Protection, Pathology, and Fungal Interference. Front. Immunol. 2018, 9, 2337. [Google Scholar] [CrossRef]
- Kuek, L.E.; Lee, R.J. First contact: The role of respiratory cilia in host-pathogen interactions in the airways. Am. J. Physiol. Lung Cell. Mol. Physiol. 2020, 319, L603–L619. [Google Scholar] [CrossRef] [PubMed]
- Silva, P.M.; Goncalves, S.; Santos, N.C. Defensins: Antifungal lessons from eukaryotes. Front. Microbiol. 2014, 5, 97. [Google Scholar] [CrossRef] [PubMed]
- Salazar, F.; Ghaemmaghami, A.M. Allergen recognition by innate immune cells: Critical role of dendritic and epithelial cells. Front. Immunol. 2013, 4, 356. [Google Scholar] [CrossRef] [PubMed]
- Gandhi, V.D.; Vliagoftis, H. Airway epithelium interactions with aeroallergens: Role of secreted cytokines and chemokines in innate immunity. Front. Immunol. 2015, 6, 147. [Google Scholar] [CrossRef] [PubMed]
- Bernton, H.S. Asthma due to a mold—Aspergillus fumigatus. J. Am. Med. Assoc. 1930, 95, 189–191. [Google Scholar] [CrossRef]
- Brown, G.T. Hypersensitiveness to fungi. J. Allergy 1936, 7, 455–470. [Google Scholar] [CrossRef]
- Hopkins, J.G.; Benham, R.W.; Kesten, B.M. Asthma due to a fungus—Alternaria. J. Am. Med. Assoc. 1930, 94, 6–10. [Google Scholar] [CrossRef]
- Black, P.N.; Udy, A.A.; Brodie, S.M. Sensitivity to fungal allergens is a risk factor for life-threatening asthma. Allergy 2000, 55, 501–504. [Google Scholar] [CrossRef]
- Targonski, P.V.; Persky, V.W.; Ramekrishnan, V. Effect of environmental molds on risk of death from asthma during the pollen season. J. Allergy Clin. Immunol. 1995, 95, 955–961. [Google Scholar] [CrossRef]
- Zureik, M.; Neukirch, C.; Leynaert, B.; Liard, R.; Bousquet, J.; Neukirch, F. Sensitisation to airborne moulds and severity of asthma: Cross sectional study from European Community respiratory health survey. BMJ 2002, 325, 411–414. [Google Scholar] [CrossRef]
- Agarwal, R.; Gupta, D. Severe asthma and fungi: Current evidence. Med. Mycol. 2011, 49, S150–S157. [Google Scholar] [CrossRef]
- Denning, D.W.; Pashley, C.; Hartl, D.; Wardlaw, A.; Godet, C.; Del Giacco, S.; Delhaes, L.; Sergejeva, S. Fungal allergy in asthma-state of the art and research needs. Clin. Transl. Allergy 2014, 4, 14. [Google Scholar] [CrossRef]
- Denning, D.W.; O’Driscoll, B.R.; Hogaboam, C.M.; Bowyer, P.; Niven, R.M. The link between fungi and severe asthma: A summary of the evidence. Eur. Respir. J. 2006, 27, 615–626. [Google Scholar] [CrossRef]
- Croisant, S. Epidemiology of asthma: Prevalence and burden of disease. In Heterogeneity in Asthma; Humana Press: Boston, MA, USA, 2014; pp. 17–29. [Google Scholar]
- Masaki, K.; Fukunaga, K.; Matsusaka, M.; Kabata, H.; Tanosaki, T.; Mochimaru, T.; Kamatani, T.; Ohtsuka, K.; Baba, R.; Ueda, S.; et al. Characteristics of severe asthma with fungal sensitization. Ann. Allergy Asthma Immunol. 2017, 119, 253–257. [Google Scholar] [CrossRef]
- Swirski, F.K.; Sajic, D.; Robbins, C.S.; Gajewska, B.U.; Jordana, M.; Stampfli, M.R. Chronic exposure to innocuous antigen in sensitized mice leads to suppressed airway eosinophilia that is reversed by granulocyte macrophage colony-stimulating factor. J. Immunol. 2002, 169, 3499–3506. [Google Scholar] [CrossRef]
- Van Hove, C.L.; Maes, T.; Joos, G.F.; Tournoy, K.G. Prolonged inhaled allergen exposure can induce persistent tolerance. Am. J. Respir. Cell Mol. Biol. 2007, 36, 573–584. [Google Scholar] [CrossRef]
- Kumar, R.K.; Herbert, C.; Foster, P.S. Mouse models of acute exacerbations of allergic asthma. Respirology 2016, 21, 842–849. [Google Scholar] [CrossRef] [PubMed]
- Beezhold, D.H.; Green, B.J.; Blachere, F.M.; Schmechel, D.; Weissman, D.N.; Velickoff, D.; Hogan, M.B.; Wilson, N.W. Prevalence of allergic sensitization to indoor fungi in West Virginia. Allergy Asthma Proc. 2008, 29, 29–34. [Google Scholar] [CrossRef]
- Knutsen, A.P.; Bush, R.K.; Demain, J.G.; Denning, D.W.; Dixit, A.; Fairs, A.; Greenberger, P.A.; Kariuki, B.; Kita, H.; Kurup, V.P.; et al. Fungi and allergic lower respiratory tract diseases. J. Allergy Clin. Immunol. 2012, 129, 280–291; quiz 292–283. [Google Scholar] [CrossRef]
- Samarasinghe, A.E.; Hoselton, S.A.; Schuh, J.M. A comparison between intratracheal and inhalation delivery of Aspergillus fumigatus conidia in the development of fungal allergic asthma in C57BL/6 mice. Fungal Biol. 2011, 115, 21–29. [Google Scholar] [CrossRef]
- Havaux, X.; Zeine, A.; Dits, A.; Denis, O. A new mouse model of lung allergy induced by the spores of Alternaria alternata and Cladosporium herbarum molds. Clin. Exp. Immunol. 2005, 139, 179–188. [Google Scholar] [CrossRef] [PubMed]
- Salo, P.M.; Arbes, S.J., Jr.; Sever, M.; Jaramillo, R.; Cohn, R.D.; London, S.J.; Zeldin, D.C. Exposure to Alternaria alternata in US homes is associated with asthma symptoms. J. Allergy Clin. Immunol. 2006, 118, 892–898. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, T.; Iijima, K.; Radhakrishnan, S.; Mehta, V.; Vassallo, R.; Lawrence, C.B.; Cyong, J.C.; Pease, L.R.; Oguchi, K.; Kita, H. Asthma-related environmental fungus, Alternaria, activates dendritic cells and produces potent Th2 adjuvant activity. J. Immunol. 2009, 182, 2502–2510. [Google Scholar] [CrossRef]
- Snelgrove, R.J.; Gregory, L.G.; Peiro, T.; Akthar, S.; Campbell, G.A.; Walker, S.A.; Lloyd, C.M. Alternaria-derived serine protease activity drives IL-33-mediated asthma exacerbations. J. Allergy Clin. Immunol. 2014, 134, 583–592.e6. [Google Scholar] [CrossRef]
- Green, B.J.; Tovey, E.R.; Sercombe, J.K.; Blachere, F.M.; Beezhold, D.H.; Schmechel, D. Airborne fungal fragments and allergenicity. Med. Mycol. 2006, 44, S245–S255. [Google Scholar] [CrossRef]
- Samarasinghe, A.E.; Hoselton, S.A.; Schuh, J.M. The absence of VPAC2 leads to aberrant antibody production in Aspergillus fumigatus sensitized and challenged mice. Peptides 2011, 32, 131–137. [Google Scholar] [CrossRef]
- Watanabe, H.; Numata, K.; Ito, T.; Takagi, K.; Matsukawa, A. Innate immune response in Th1- and Th2-dominant mouse strains. Shock 2004, 22, 460–466. [Google Scholar] [CrossRef]
- Atochina, E.N.; Beers, M.F.; Tomer, Y.; Scanlon, S.T.; Russo, S.J.; Panettieri, R.A., Jr.; Haczku, A. Attenuated allergic airway hyperresponsiveness in C57BL/6 mice is associated with enhanced surfactant protein (SP)-D production following allergic sensitization. Respir. Res. 2003, 4, 15. [Google Scholar] [CrossRef]
- Pandey, S.; Hoselton, S.A.; Schuh, J.M. The impact of Aspergillus fumigatus viability and sensitization to its allergens on the murine allergic asthma phenotype. Biomed Res. Int. 2013, 2013, 619614. [Google Scholar] [CrossRef]
- Buskirk, A.D.; Green, B.J.; Lemons, A.R.; Nayak, A.P.; Goldsmith, W.T.; Kashon, M.L.; Anderson, S.E.; Hettick, J.M.; Templeton, S.P.; Germolec, D.R.; et al. A murine inhalation model to characterize pulmonary exposure to dry Aspergillus fumigatus conidia. PLoS ONE 2014, 9, e109855. [Google Scholar] [CrossRef]
- Roy, R.M.; Klein, B.S. Fungal glycan interactions with epithelial cells in allergic airway disease. Curr. Opin. Microbiol. 2013, 16, 404–408. [Google Scholar] [CrossRef]
- Kwon-Chung, K.J.; Sugui, J.A. Aspergillus fumigatus--what makes the species a ubiquitous human fungal pathogen? PLoS Pathog. 2013, 9, e1003743. [Google Scholar] [CrossRef]
- Brown, G.D. Dectin-1: A signalling non-TLR pattern-recognition receptor. Nat. Rev. Immunol. 2006, 6, 33–43. [Google Scholar] [CrossRef]
- Taylor, P.R.; Tsoni, S.V.; Willment, J.A.; Dennehy, K.M.; Rosas, M.; Findon, H.; Haynes, K.; Steele, C.; Botto, M.; Gordon, S.; et al. Dectin-1 is required for beta-glucan recognition and control of fungal infection. Nat. Immunol. 2007, 8, 31–38. [Google Scholar] [CrossRef]
- Speakman, E.A.; Dambuza, I.M.; Salazar, F.; Brown, G.D. T Cell Antifungal Immunity and the Role of C-Type Lectin Receptors. Trends Immunol. 2020, 41, 61–76. [Google Scholar] [CrossRef]
- Chung, Y.J.; Copeland, L.B.; Doerfler, D.L.; Ward, M.D. The relative allergenicity of Stachybotrys chartarum compared to house dust mite extracts in a mouse model. Inhal. Toxicol. 2010, 22, 460–468. [Google Scholar] [CrossRef]
- Da Silva, C.A.; Hartl, D.; Liu, W.; Lee, C.G.; Elias, J.A. TLR-2 and IL-17A in chitin-induced macrophage activation and acute inflammation. J. Immunol. 2008, 181, 4279–4286. [Google Scholar] [CrossRef]
- Roy, R.M.; Wuthrich, M.; Klein, B.S. Chitin elicits CCL2 from airway epithelial cells and induces CCR2-dependent innate allergic inflammation in the lung. J. Immunol. 2012, 189, 2545–2552. [Google Scholar] [CrossRef]
- Levy, M.; Kolodziejczyk, A.A.; Thaiss, C.A.; Elinav, E. Dysbiosis and the immune system. Nat. Rev. Immunol. 2017, 17, 219–232. [Google Scholar] [CrossRef]
- Van Dyken, S.J.; Mohapatra, A.; Nussbaum, J.C.; Molofsky, A.B.; Thornton, E.E.; Ziegler, S.F.; McKenzie, A.N.; Krummel, M.F.; Liang, H.E.; Locksley, R.M. Chitin activates parallel immune modules that direct distinct inflammatory responses via innate lymphoid type 2 and gammadelta T cells. Immunity 2014, 40, 414–424. [Google Scholar] [CrossRef]
- Hammad, H.; Lambrecht, B.N. Barrier Epithelial Cells and the Control of Type 2 Immunity. Immunity 2015, 43, 29–40. [Google Scholar] [CrossRef] [PubMed]
- Andreakos, E.; Papadopoulos, N.G. IL-25: The Missing Link between Allergy, Viral Infection, and Asthma? Sci. Transl. Med. 2014, 6, 256fs238. [Google Scholar] [CrossRef] [PubMed]
- Stolarski, B.; Kurowska-Stolarska, M.; Kewin, P.; Xu, D.; Liew, F.Y. IL-33 Exacerbates Eosinophil-Mediated Airway Inflammation. J. Immunol. 2010, 185, 3472–3480. [Google Scholar] [CrossRef] [PubMed]
- Hristova, M.; Habibovic, A.; Veith, C.; Janssen-Heininger, Y.M.; Dixon, A.E.; Geiszt, M.; van der Vliet, A. Airway epithelial dual oxidase 1 mediates allergen-induced IL-33 secretion and activation of type 2 immune responses. J. Allergy Clin. Immunol. 2016, 137, 1545–1556.e11. [Google Scholar] [CrossRef] [PubMed]
- Suzukawa, M.; Koketsu, R.; Iikura, M.; Nakae, S.; Matsumoto, K.; Nagase, H.; Saito, H.; Matsushima, K.; Ohta, K.; Yamamoto, K.; et al. Interleukin-33 enhances adhesion, CD11b expression and survival in human eosinophils. Lab. Investig. 2008, 88, 1245–1253. [Google Scholar] [CrossRef]
- Holmes, D.A.; Yeh, J.H.; Yan, D.; Xu, M.; Chan, A.C. Dusp5 negatively regulates IL-33-mediated eosinophil survival and function. EMBO J. 2015, 34, 218–235. [Google Scholar] [CrossRef]
- Daines, M.; Zhu, L.; Pereira, R.; Zhou, X.; Bondy, C.; Pryor, B.M.; Zhou, J.; Chen, Y. Alternaria induces airway epithelial cytokine expression independent of protease-activated receptor. Respirology 2020, 25, 502–510. [Google Scholar] [CrossRef]
- LeMessurier, K.S.; Rooney, R.; Ghoneim, H.E.; Liu, B.; Li, K.; Smallwood, H.S.; Samarasinghe, A.E. Influenza A virus directly modulates mouse eosinophil responses. J. Leukoc. Biol. 2020, 108, 151–168. [Google Scholar] [CrossRef]
- Artis, D.; Wang, M.L.; Keilbaugh, S.A.; He, W.; Brenes, M.; Swain, G.P.; Knight, P.A.; Donaldson, D.D.; Lazar, M.A.; Miller, H.R.; et al. RELMbeta/FIZZ2 is a goblet cell-specific immune-effector molecule in the gastrointestinal tract. Proc. Natl. Acad. Sci. USA 2004, 101, 13596–13600. [Google Scholar] [CrossRef]
- Mishra, A.; Wang, M.; Schlotman, J.; Nikolaidis, N.M.; DeBrosse, C.W.; Karow, M.L.; Rothenberg, M.E. Resistin-like molecule-beta is an allergen-induced cytokine with inflammatory and remodeling activity in the murine lung. Am. J. Physiol. Lung Cell. Mol. Physiol. 2007, 293, L305–L313. [Google Scholar] [CrossRef]
- LeMessurier, K.S.; Palipane, M.; Tiwary, M.; Gavin, B.; Samarasinghe, A.E. Chronic features of allergic asthma are enhanced in the absence of resistin-like molecule-beta. Sci. Rep. 2018, 8, 7061. [Google Scholar] [CrossRef]
- Brown, G.D.; Denning, D.W.; Levitz, S.M. Tackling human fungal infections. Science 2012, 336, 647. [Google Scholar] [CrossRef]
- Paris, S.; Boisvieux-Ulrich, E.; Crestani, B.; Houcine, O.; Taramelli, D.; Lombardi, L.; Latge, J.P. Internalization of Aspergillus fumigatus conidia by epithelial and endothelial cells. Infect. Immun. 1997, 65, 1510–1514. [Google Scholar] [CrossRef]
- Hamelmann, E.; Schwarze, J.; Takeda, K.; Oshiba, A.; Larsen, G.L.; Irvin, C.G.; Gelfand, E.W. Noninvasive measurement of airway responsiveness in allergic mice using barometric plethysmography. Am. J. Respir. Crit. Care Med. 1997, 156, 766–775. [Google Scholar] [CrossRef]
- Sweerus, K.; Lachowicz-Scroggins, M.; Gordon, E.; LaFemina, M.; Huang, X.; Parikh, M.; Kanegai, C.; Fahy, J.V.; Frank, J.A. Claudin-18 deficiency is associated with airway epithelial barrier dysfunction and asthma. J. Allergy Clin. Immunol. 2017, 139, 72–81.e1. [Google Scholar] [CrossRef]
- Zaidman, N.A.; O’Grady, K.E.; Patil, N.; Milavetz, F.; Maniak, P.J.; Kita, H.; O’Grady, S.M. Airway epithelial anion secretion and barrier function following exposure to fungal aeroallergens: Role of oxidative stress. Am. J. Physiol. Cell. Physiol. 2017, 313, C68–C79. [Google Scholar] [CrossRef]
- Martin, N.T.; Martin, M.U. Interleukin 33 is a guardian of barriers and a local alarmin. Nat. Immunol. 2016, 17, 122–131. [Google Scholar] [CrossRef]
- Piehler, D.; Eschke, M.; Schulze, B.; Protschka, M.; Muller, U.; Grahnert, A.; Richter, T.; Heyen, L.; Kohler, G.; Brombacher, F.; et al. The IL-33 receptor (ST2) regulates early IL-13 production in fungus-induced allergic airway inflammation. Mucosal Immunol. 2016, 9, 937–949. [Google Scholar] [CrossRef]
- Namvar, S.; Warn, P.; Farnell, E.; Bromley, M.; Fraczek, M.; Bowyer, P.; Herrick, S. Aspergillus fumigatus proteases, Asp f 5 and Asp f 13, are essential for airway inflammation and remodelling in a murine inhalation model. Clin. Exp. Allergy 2015, 45, 982–993. [Google Scholar] [CrossRef]
- Ghosh, S.; Hoselton, S.A.; Wanjara, S.B.; Carlson, J.; McCarthy, J.B.; Dorsam, G.P.; Schuh, J.M. Hyaluronan stimulates ex vivo B lymphocyte chemotaxis and cytokine production in a murine model of fungal allergic asthma. Immunobiology 2015, 220, 899–909. [Google Scholar] [CrossRef]
- Drake, L.Y.; Iijima, K.; Hara, K.; Kobayashi, T.; Kephart, G.M.; Kita, H. B cells play key roles in th2-type airway immune responses in mice exposed to natural airborne allergens. PLoS ONE 2015, 10, e0121660. [Google Scholar] [CrossRef]
- Albacker, L.A.; Chaudhary, V.; Chang, Y.J.; Kim, H.Y.; Chuang, Y.T.; Pichavant, M.; DeKruyff, R.H.; Savage, P.B.; Umetsu, D.T. Invariant natural killer T cells recognize a fungal glycosphingolipid that can induce airway hyperreactivity. Nat. Med. 2013, 19, 1297–1304. [Google Scholar] [CrossRef] [PubMed]
- Voehringer, D. Basophils in allergic immune responses. Curr. Opin. Immunol. 2011, 23, 789–793. [Google Scholar] [CrossRef] [PubMed]
- Halim, T.Y.; Steer, C.A.; Matha, L.; Gold, M.J.; Martinez-Gonzalez, I.; McNagny, K.M.; McKenzie, A.N.; Takei, F. Group 2 innate lymphoid cells are critical for the initiation of adaptive T helper 2 cell-mediated allergic lung inflammation. Immunity 2014, 40, 425–435. [Google Scholar] [CrossRef] [PubMed]
- Barnes, P.J. The cytokine network in asthma and chronic obstructive pulmonary disease. J. Clin. Investig. 2008, 118, 3546–3556. [Google Scholar] [CrossRef] [PubMed]
- Wills-Karp, M.; Finkelman, F.D. Untangling the complex web of IL-4- and IL-13-mediated signaling pathways. Sci. Signal. 2008, 1, pe55. [Google Scholar] [CrossRef] [PubMed]
- Kuperman, D.A.; Huang, X.; Koth, L.L.; Chang, G.H.; Dolganov, G.M.; Zhu, Z.; Elias, J.A.; Sheppard, D.; Erle, D.J. Direct effects of interleukin-13 on epithelial cells cause airway hyperreactivity and mucus overproduction in asthma. Nat. Med. 2002, 8, 885–889. [Google Scholar] [CrossRef] [PubMed]
- Mishina, K.; Shinkai, M.; Shimokawaji, T.; Nagashima, A.; Hashimoto, Y.; Inoue, Y.; Inayama, Y.; Rubin, B.K.; Ishigatsubo, Y.; Kaneko, T. HO-1 inhibits IL-13-induced goblet cell hyperplasia associated with CLCA1 suppression in normal human bronchial epithelial cells. Int. Immunopharmacol. 2015, 29, 448–453. [Google Scholar] [CrossRef]
- Blanchard, C.; Rothenberg, M.E. Biology of the eosinophil. Adv. Immunol. 2009, 101, 81–121. [Google Scholar] [CrossRef]
- Yu, S.; Kim, H.Y.; Chang, Y.J.; DeKruyff, R.H.; Umetsu, D.T. Innate lymphoid cells and asthma. J. Allergy Clin. Immunol. 2014, 133, 943–950; quiz 951. [Google Scholar] [CrossRef]
- Vazquez-Tello, A.; Halwani, R.; Li, R.; Nadigel, J.; Bar-Or, A.; Mazer, B.D.; Eidelman, D.H.; Al-Muhsen, S.; Hamid, Q. IL-17A and IL-17F expression in B lymphocytes. Int. Arch. Allergy Immunol. 2012, 157, 406–416. [Google Scholar] [CrossRef]
- Taylor, P.R.; Roy, S.; Leal, S.M., Jr.; Sun, Y.; Howell, S.J.; Cobb, B.A.; Li, X.; Pearlman, E. Activation of neutrophils by autocrine IL-17A-IL-17RC interactions during fungal infection is regulated by IL-6, IL-23, RORgammat and dectin-2. Nat. Immunol. 2014, 15, 143–151. [Google Scholar] [CrossRef]
- Murdoch, J.R.; Lloyd, C.M. Resolution of allergic airway inflammation and airway hyperreactivity is mediated by IL-17-producing {gamma}{delta}T cells. Am. J. Respir. Crit. Care Med. 2010, 182, 464–476. [Google Scholar] [CrossRef]
- Ghosh, S.; Hoselton, S.A.; Asbach, S.V.; Steffan, B.N.; Wanjara, S.B.; Dorsam, G.P.; Schuh, J.M. B lymphocytes regulate airway granulocytic inflammation and cytokine production in a murine model of fungal allergic asthma. Cell. Mol. Immunol. 2015, 12, 202–212. [Google Scholar] [CrossRef]
- Sutmuller, R.P.; den Brok, M.H.; Kramer, M.; Bennink, E.J.; Toonen, L.W.; Kullberg, B.J.; Joosten, L.A.; Akira, S.; Netea, M.G.; Adema, G.J. Toll-like receptor 2 controls expansion and function of regulatory T cells. J. Clin. Investig. 2006, 116, 485–494. [Google Scholar] [CrossRef]
- Van de Veerdonk, F.L.; Netea, M.G. T-cell Subsets and Antifungal Host Defenses. Curr. Fungal Infect. Rep. 2010, 4, 238–243. [Google Scholar] [CrossRef]
- Kurup, V.P.; Raju, R.; Manickam, P. Profile of gene expression in a murine model of allergic bronchopulmonary aspergillosis. Infect. Immun. 2005, 73, 4381–4384. [Google Scholar] [CrossRef]
- Lee, J.J.; Jacobsen, E.A.; McGarry, M.P.; Schleimer, R.P.; Lee, N.A. Eosinophils in health and disease: The LIAR hypothesis. Clin. Exp. Allergy 2010, 40, 563–575. [Google Scholar] [CrossRef]
- Conroy, D.M.; Williams, T.J. Eotaxin and the attraction of eosinophils to the asthmatic lung. Respir. Res. 2001, 2, 150–156. [Google Scholar] [CrossRef] [PubMed]
- Robinson, D.S.; Damia, R.; Zeibecoglou, K.; Molet, S.; North, J.; Yamada, T.; Kay, A.B.; Hamid, Q. CD34(+)/interleukin-5Ralpha messenger RNA+ cells in the bronchial mucosa in asthma: Potential airway eosinophil progenitors. Am. J. Respir. Cell. Mol. Biol. 1999, 20, 9–13. [Google Scholar] [CrossRef]
- Menzies-Gow, A.; Flood-Page, P.; Sehmi, R.; Burman, J.; Hamid, Q.; Robinson, D.S.; Kay, A.B.; Denburg, J. Anti-IL-5 (mepolizumab) therapy induces bone marrow eosinophil maturational arrest and decreases eosinophil progenitors in the bronchial mucosa of atopic asthmatics. J. Allergy Clin. Immunol. 2003, 111, 714–719. [Google Scholar] [CrossRef] [PubMed]
- Dhariwal, J.; Hearn, A.P.; Kavanagh, J.E.; d’Ancona, G.; Green, L.; Fernandes, M.; Thomson, L.; Roxas, C.; Kent, B.D.; Nanzer, A.M.; et al. Real world effectiveness of anti-IL5/5R therapy in severe atopic eosinophilic asthma with fungal sensitisation. J. Allergy Clin. Immunol. Pract 2021. [Google Scholar] [CrossRef]
- Yoon, J.; Ponikau, J.U.; Lawrence, C.B.; Kita, H. Innate antifungal immunity of human eosinophils mediated by a beta 2 integrin, CD11b. J. Immunol. 2008, 181, 2907–2915. [Google Scholar] [CrossRef] [PubMed]
- Inoue, Y.; Matsuwaki, Y.; Shin, S.H.; Ponikau, J.U.; Kita, H. Nonpathogenic, environmental fungi induce activation and degranulation of human eosinophils. J. Immunol. 2005, 175, 5439–5447. [Google Scholar] [CrossRef]
- Lilly, L.M.; Scopel, M.; Nelson, M.P.; Burg, A.R.; Dunaway, C.W.; Steele, C. Eosinophil deficiency compromises lung defense against Aspergillus fumigatus. Infect. Immun. 2014, 82, 1315–1325. [Google Scholar] [CrossRef]
- Schmid-Grendelmeier, P.; Altznauer, F.; Fischer, B.; Bizer, C.; Straumann, A.; Menz, G.; Blaser, K.; Wuthrich, B.; Simon, H.U. Eosinophils express functional IL-13 in eosinophilic inflammatory diseases. J. Immunol. 2002, 169, 1021–1027. [Google Scholar] [CrossRef]
- Gibson, P.G. Inflammatory phenotypes in adult asthma: Clinical applications. Clin. Respir. J. 2009, 3, 198–206. [Google Scholar] [CrossRef]
- Kay, A.B. Mediators of hypersensitivity and inflammatory cells in the pathogenesis of bronchial asthma. Eur. J. Respir. Dis. 1983, 129, 1–44. [Google Scholar]
- Elovic, A.E.; Ohyama, H.; Sauty, A.; McBride, J.; Tsuji, T.; Nagai, M.; Weller, P.F.; Wong, D.T. IL-4-dependent regulation of TGF-α and TGF-β1 expression in human eosinophils. J. Immunol. 1998, 160, 6121–6127. [Google Scholar]
- Fine, A.; Goldstein, R.J. Regulation of type I collagen mRNA translation by TGF-beta. Reg. Immunol. 1993, 5, 218–224. [Google Scholar]
- Wicks, J.; Haitchi, H.M.; Holgate, S.T.; Davies, D.E.; Powell, R.M. Enhanced upregulation of smooth muscle related transcripts by TGFβ2 in asthmatic (myo) fibroblasts. Thorax 2006, 61, 313–319. [Google Scholar] [CrossRef]
- Flood-Page, P.; Menzies-Gow, A.; Phipps, S.; Ying, S.; Wangoo, A.; Ludwig, M.; Barnes, N.; Robinson, D.; Kay, A.J. Anti-IL-5 treatment (mepolizumab) reduces deposition of extracellular matrix proteins in the bronchial subepithelial basement membrane of mild atopic asthmatics: Evidence for a role for eosinophils in airways remodeling. J. Clin. Investig. 2003, 112, 1029–1036. [Google Scholar] [CrossRef]
- Boxall, C.; Holgate, S.; Davies, D.J. The contribution of transforming growth factor-β and epidermal growth factor signalling to airway remodelling in chronic asthma. Eur. Respir. J. 2006, 27, 208–229. [Google Scholar] [CrossRef]
- Liu, T.; Jin, H.; Ullenbruch, M.; Hu, B.; Hashimoto, N.; Moore, B.; McKenzie, A.; Lukacs, N.W.; Phan, S.H. Regulation of found in inflammatory zone 1 expression in bleomycin-induced lung fibrosis: Role of IL-4/IL-13 and mediation via STAT-6. J. Immunol. 2004, 173, 3425–3431. [Google Scholar] [CrossRef]
- Fang, C.; Meng, Q.; Wu, H.; Eid, G.; Zhang, G.; Zhang, X.; Yang, S.; Huang, K.; Lee, T.H.; Corrigan, C.J.; et al. Resistin-like molecule-beta is a human airway remodelling mediator. Eur. Respir. J. 2012, 39, 458–466. [Google Scholar] [CrossRef]
- Figueiredo, R.T.; Neves, J.S. Eosinophils in fungal diseases: An overview. J. Leukoc. Biol. 2018, 104, 49–60. [Google Scholar] [CrossRef] [PubMed]
- Melo, R.C.; Liu, L.; Xenakis, J.J.; Spencer, L.A. Eosinophil-derived cytokines in health and disease: Unraveling novel mechanisms of selective secretion. Allergy 2013, 68, 274–284. [Google Scholar] [CrossRef]
- Jacobsen, E.A.; Helmers, R.A.; Lee, J.J.; Lee, N.A. The expanding role(s) of eosinophils in health and disease. Blood 2012, 120, 3882–3890. [Google Scholar] [CrossRef]
- Percopo, C.M.; Dyer, K.D.; Ochkur, S.I.; Luo, J.L.; Fischer, E.R.; Lee, J.J.; Lee, N.A.; Domachowske, J.B.; Rosenberg, H.F. Activated mouse eosinophils protect against lethal respiratory virus infection. Blood 2014, 123, 743–752. [Google Scholar] [CrossRef]
- Samarasinghe, A.E.; Melo, R.C.; Duan, S.; LeMessurier, K.S.; Liedmann, S.; Surman, S.L.; Lee, J.J.; Hurwitz, J.L.; Thomas, P.G.; McCullers, J.A. Eosinophils Promote Antiviral Immunity in Mice Infected with Influenza A Virus. J. Immunol. 2017, 198, 3214–3226. [Google Scholar] [CrossRef]
- LeMessurier, K.S.; Samarasinghe, A.E. Eosinophils: Nemeses of Pulmonary Pathogens? Curr. Allergy Asthma Rep. 2019, 19, 36. [Google Scholar] [CrossRef] [PubMed]
- Agache, I.; Beltran, J.; Akdis, C.; Akdis, M.; Canelo-Aybar, C.; Canonica, G.W.; Casale, T.; Chivato, T.; Corren, J.; Del Giacco, S.; et al. Efficacy and safety of treatment with biologicals (benralizumab, dupilumab, mepolizumab, omalizumab and reslizumab) for severe eosinophilic asthma. A systematic review for the EAACI Guidelines—Recommendations on the use of biologicals in severe asthma. Allergy 2020, 75, 1023–1042. [Google Scholar] [CrossRef] [PubMed]
- Lindell, D.M.; Berlin, A.A.; Schaller, M.A.; Lukacs, N.W. B cell antigen presentation promotes Th2 responses and immunopathology during chronic allergic lung disease. PLoS ONE 2008, 3, e3129. [Google Scholar] [CrossRef] [PubMed]
- Bice, D.E.; Gray, R.H.; Evans, M.J.; Muggenburg, B.A. Identification of plasma cells in lung alveoli and interstitial tissues after localized lung immunization. J. Leukoc. Biol. 1987, 41, 1–7. [Google Scholar] [CrossRef]
- Takhar, P.; Corrigan, C.J.; Smurthwaite, L.; O’connor, B.J.; Durham, S.R.; Lee, T.H.; Gould, H.J. Class switch recombination to IgE in the bronchial mucosa of atopic and nonatopic patients with asthma. J. Allergy Clin. Immunol. 2007, 119, 213–218. [Google Scholar] [CrossRef]
- Gould, H.J.; Sutton, B.J. IgE in allergy and asthma today. Nat. Rev. Immunol. 2008, 8, 205. [Google Scholar] [CrossRef]
- Zhang, M.; Murphy, R.F.; Agrawal, D.K. Decoding IgE Fc receptors. Immunol. Res. 2007, 37, 1–16. [Google Scholar] [CrossRef]
- Oh, S.; McCaffery, J.M.; Eichelberger, M.C. Dose-dependent changes in influenza virus-infected dendritic cells result in increased allogeneic T-cell proliferation at low, but not high, doses of virus. J. Virol. 2000, 74, 5460–5469. [Google Scholar] [CrossRef][Green Version]
- Wheeler, M.L.; Limon, J.J.; Bar, A.S.; Leal, C.A.; Gargus, M.; Tang, J.; Brown, J.; Funari, V.A.; Wang, H.L.; Crother, T.R.; et al. Immunological Consequences of Intestinal Fungal Dysbiosis. Cell Host Microbe 2016, 19, 865–873. [Google Scholar] [CrossRef]
- Singanayagam, A.; Ritchie, A.I.; Johnston, S.L. Role of microbiome in the pathophysiology and disease course of asthma. Curr. Opin. Pulm. Med. 2017, 23, 41–47. [Google Scholar] [CrossRef]
- Huang, Y.J.; Charlson, E.S.; Collman, R.G.; Colombini-Hatch, S.; Martinez, F.D.; Senior, R.M. The role of the lung microbiome in health and disease. A National Heart, Lung, and Blood Institute workshop report. Am. J. Respir. Crit. Care Med. 2013, 187, 1382–1387. [Google Scholar] [CrossRef]
- Charlson, E.S.; Diamond, J.M.; Bittinger, K.; Fitzgerald, A.S.; Yadav, A.; Haas, A.R.; Bushman, F.D.; Collman, R.G. Lung-enriched organisms and aberrant bacterial and fungal respiratory microbiota after lung transplant. Am. J. Respir. Crit. Care Med. 2012, 186, 536–545. [Google Scholar] [CrossRef]
- Hilty, M.; Burke, C.; Pedro, H.; Cardenas, P.; Bush, A.; Bossley, C.; Davies, J.; Ervine, A.; Poulter, L.; Pachter, L.; et al. Disordered microbial communities in asthmatic airways. PLoS ONE 2010, 5, e8578. [Google Scholar] [CrossRef]
- Huffnagle, G.B.; Dickson, R.P. The bacterial microbiota in inflammatory lung diseases. Clin. Immunol. 2015, 159, 177–182. [Google Scholar] [CrossRef]
- Van Woerden, H.C.; Gregory, C.; Brown, R.; Marchesi, J.R.; Hoogendoorn, B.; Matthews, I.P. Differences in fungi present in induced sputum samples from asthma patients and non-atopic controls: A community based case control study. BMC Infect. Dis. 2013, 13, 69. [Google Scholar] [CrossRef]
- LeMessurier, K.S.; Iverson, A.R.; Chang, T.C.; Palipane, M.; Vogel, P.; Rosch, J.W.; Samarasinghe, A.E. Allergic inflammation alters the lung microbiome and hinders synergistic co-infection with H1N1 influenza virus and Streptococcus pneumoniae in C57BL/6 mice. Sci. Rep. 2019, 9, 19360. [Google Scholar] [CrossRef]
- Skalski, J.H.; Limon, J.J.; Sharma, P.; Gargus, M.D.; Nguyen, C.; Tang, J.; Coelho, A.L.; Hogaboam, C.M.; Crother, T.R.; Underhill, D.M. Expansion of commensal fungus Wallemia mellicola in the gastrointestinal mycobiota enhances the severity of allergic airway disease in mice. PLoS Pathog. 2018, 14, e1007260. [Google Scholar] [CrossRef]
- Hoffmann, C.; Dollive, S.; Grunberg, S.; Chen, J.; Li, H.; Wu, G.D.; Lewis, J.D.; Bushman, F.D. Archaea and fungi of the human gut microbiome: Correlations with diet and bacterial residents. PLoS ONE 2013, 8, e66019. [Google Scholar] [CrossRef]
- Michael, C.F.; Waters, C.M.; LeMessurier, K.S.; Samarasinghe, A.E.; Song, C.Y.; Malik, K.U.; Lew, D.B. Airway Epithelial Repair by a Prebiotic Mannan Derived from Saccharomyces cerevisiae. J. Immunol. Res. 2017, 2017, 8903982. [Google Scholar] [CrossRef]
- Lew, D.B.; LeMessurier, K.S.; Palipane, M.; Lin, Y.; Samarasinghe, A.E. Saccharomyces cerevisiae-Derived Mannan Does Not Alter Immune Responses to Aspergillus Allergens. Biomed Res. Int. 2018, 2018, 3298378. [Google Scholar] [CrossRef]
- Cooper, P.J.; Chico, M.E.; Vaca, M.G.; Moncayo, A.L.; Bland, J.M.; Mafla, E.; Sanchez, F.; Rodrigues, L.C.; Strachan, D.P.; Griffin, G.E. Effect of albendazole treatments on the prevalence of atopy in children living in communities endemic for geohelminth parasites: A cluster-randomised trial. Lancet 2006, 367, 1598–1603. [Google Scholar] [CrossRef]
- Van den Biggelaar, A.H.; Rodrigues, L.C.; van Ree, R.; van der Zee, J.S.; Hoeksma-Kruize, Y.C.; Souverijn, J.H.; Missinou, M.A.; Borrmann, S.; Kremsner, P.G.; Yazdanbakhsh, M. Long-term treatment of intestinal helminths increases mite skin-test reactivity in Gabonese schoolchildren. J. Infect. Dis. 2004, 189, 892–900. [Google Scholar] [CrossRef] [PubMed]
- Apiwattanakul, N.; Palipane, M.; Samarasinghe, A.E. Immune responses to fungal aeroallergen in Heligmosomoides polygyrus-infected mice vary by age. Cell Immunol. 2017, 317, 26–36. [Google Scholar] [CrossRef] [PubMed]
- Zaiss, M.M.; Rapin, A.; Lebon, L.; Dubey, L.K.; Mosconi, I.; Sarter, K.; Piersigilli, A.; Menin, L.; Walker, A.W.; Rougemont, J.; et al. The Intestinal Microbiota Contributes to the Ability of Helminths to Modulate Allergic Inflammation. Immunity 2015, 43, 998–1010. [Google Scholar] [CrossRef]
- Simon-Nobbe, B.; Denk, U.; Poll, V.; Rid, R.; Breitenbach, M. The spectrum of fungal allergy. Int. Arch. Allergy Immunol. 2008, 145, 58–86. [Google Scholar] [CrossRef]
- Caminati, M.; Menzella, F.; Guidolin, L.; Senna, G. Targeting eosinophils: Severe asthma and beyond. Drugs Context 2019, 8, 212587. [Google Scholar] [CrossRef]
- Rosenberg, H.F.; Dyer, K.D.; Domachowske, J.B. Respiratory viruses and eosinophils: Exploring the connections. Antiviral Res. 2009, 83, 1–9. [Google Scholar] [CrossRef]
- Tiwary, M.; Rooney, R.J.; Liedmann, S.; LeMessurier, K.S.; Samarasinghe, A.E. Eosinophil Responses at the Airway Epithelial Barrier during the Early Phase of Influenza A Virus Infection in C57BL/6 Mice. Cells 2021, 10, 509. [Google Scholar] [CrossRef]
- Ferastraoaru, D.; Hudes, G.; Jerschow, E.; Jariwala, S.; Karagic, M.; de Vos, G.; Rosenstreich, D.; Ramesh, M. Eosinophilia in Asthma Patients Is Protective Against Severe COVID-19 Illness. J. Allergy Clin. Immunol. Pract. 2021, 9, 1152–1162.e3. [Google Scholar] [CrossRef]
- Jacobsen, E.A.; Jackson, D.J.; Heffler, E.; Mathur, S.K.; Bredenoord, A.J.; Pavord, I.D.; Akuthota, P.; Roufosse, F.; Rothenberg, M.E. Eosinophil Knockout Humans: Uncovering the Role of Eosinophils Through Eosinophil-Directed Biological Therapies. Annu. Rev. Immunol. 2021. [Google Scholar] [CrossRef]


| Source | Mediator | Effect | References |
|---|---|---|---|
| Fungi | Serine proteases | Membrane permeability Disruption of tight junctions Airway smooth muscle constriction | [18,19,20,21,22,23,24] |
| Epithelial cells | Interleukins -25 and -33 TSLP TGF-β | Inflammation Leukocyte activation Airway remodeling | [25,26,27,28,29,30] |
| Dendritic cells | Pattern recognition receptors Interleukin-6 TNF-α | Fungal recognition Interleukin-17A production Neutrophil recruitment | [31,32,33,34] |
| TH2 cells | Interleukin-4 Interleukin-5 Interleukin-13 | Inflammation B cell class switching Eosinophil activation Mucus cell activation | [26,27,31,35] |
| TH17 cells | Interleukin-17A | Neutrophil recruitment Epithelial cell activation | [33] |
| Plasma cells | Immunoglobulin E Immunoglobulin A | Mast cell activation Fungal neutralization | [26,27,31,35,36,37] |
| Eosinophils | Interleukins -17 and -23 DNA traps | Inflammation Fungal neutralization | [38,39] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tiwary, M.; Samarasinghe, A.E. Initiation and Pathogenesis of Severe Asthma with Fungal Sensitization. Cells 2021, 10, 913. https://doi.org/10.3390/cells10040913
Tiwary M, Samarasinghe AE. Initiation and Pathogenesis of Severe Asthma with Fungal Sensitization. Cells. 2021; 10(4):913. https://doi.org/10.3390/cells10040913
Chicago/Turabian StyleTiwary, Meenakshi, and Amali E. Samarasinghe. 2021. "Initiation and Pathogenesis of Severe Asthma with Fungal Sensitization" Cells 10, no. 4: 913. https://doi.org/10.3390/cells10040913
APA StyleTiwary, M., & Samarasinghe, A. E. (2021). Initiation and Pathogenesis of Severe Asthma with Fungal Sensitization. Cells, 10(4), 913. https://doi.org/10.3390/cells10040913






