Abstract
Hematopoietic multipotent progenitors seed the thymus and then follow consecutive developmental stages until the formation of mature T cells. During this process, phenotypic changes of T cells entail stage-specific transcriptional programs that underlie the dynamic progression towards mature lymphocytes. Lineage-specific transcription factors are key drivers of T cell specification and act in conjunction with epigenetic regulators that have also been elucidated as crucial players in the establishment of regulatory networks necessary for proper T cell development. In this review, we summarize the activity of transcription factors and epigenetic regulators that together orchestrate the intricacies of early T cell development with a focus on regulation of T cell lineage commitment.
1. Introduction
Hematopoiesis, the formation of blood cells, is a well-studied developmental process, during which transcription factors and epigenetic regulators operate together in order to ensure faithful progression toward the production of mature blood cells [1,2]. Lineage restriction events reflect changes in the transcriptional programs and epigenetic regulation [3,4], and defects in these regulatory mechanisms can promote leukemogenesis [5,6]. These intrinsic programs require cell-extrinsic factors, such as signaling pathways, cytokines and growth factors, provided by a specific microenvironment. While most blood cell types develop in the bone marrow [7], specialized white blood cells differentiate in the thymus from multipotent progenitors that pass through different stages until they develop into mature T cells [8,9]. The complex regulatory mechanisms that enable this stepwise differentiation are governed by transcriptional and epigenetic programs [10,11]. Master transcription factors and epigenetic regulators drive the journey from early thymic progenitors to mature T cells by repressing alternative non-T cell fates and enhancing the dynamic progress towards lineage commitment. The roles of transcription and epigenetic factors can be elucidated in a stage-specific manner by using genetically modified mouse models and flow cytometry-based strategies. The interplay between transcriptional programs and epigenetic regulation has been well-characterized for the late differentiation steps (CD4 single positive versus CD8 single positive) and has been reviewed elsewhere [12,13,14,15]. However, less is known about the transcription factors and chromatin modifiers controlling early T cell development. In this review, we discuss the journey of early thymic progenitors toward committed T cells and the lessons learned from murine models about the crucial roles of key transcription factors and epigenetic regulators in the establishment of T cell identity.
2. Introduction of Epigenetics
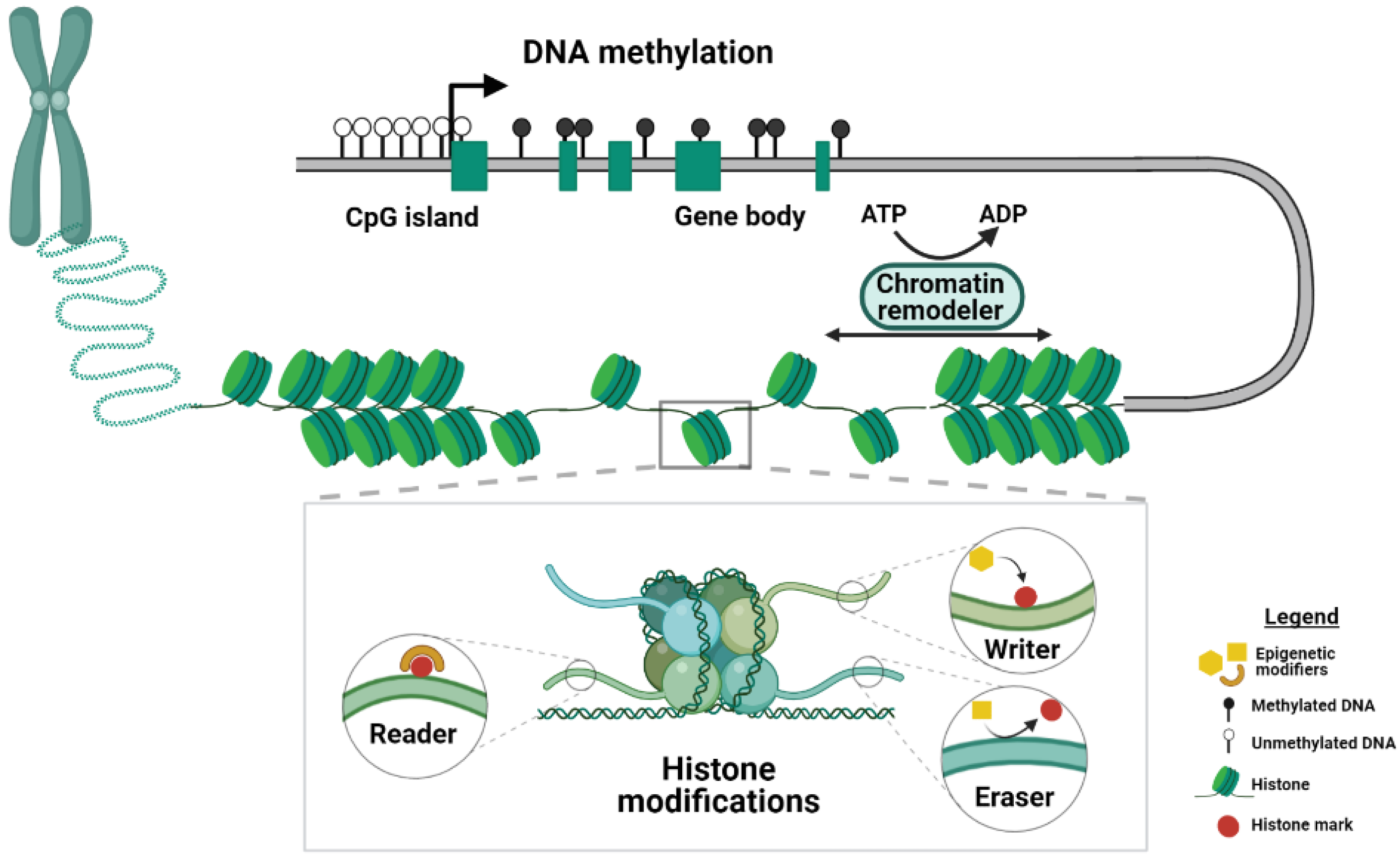
Epigenetic regulation is key to the normal development of an organism because it allows for modulating gene expression levels through the addition of chemical modifications to the DNA and its associated histone proteins, which are referred to as epigenetic marks (Figure 1).
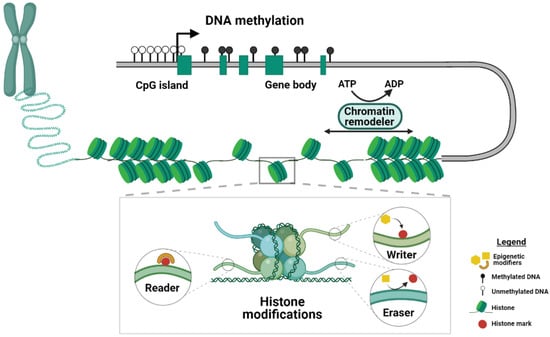
Figure 1.
Schematic of the mechanisms of epigenetic regulation. DNA methylation, histone modifications and chromatin remodeling represent three different kinds of epigenetic mechanisms. Main players in the histone modification machinery are depicted in the inset. Proteins that covalently attach chemical groups to the histone tails are termed writers, whereas the so-called readers can recognize and bind to histone modifications. Enzymes that remove histone marks are termed erasers. DNA methylation is found across different genomic elements. For example, CpG islands, which are often found in the proximity of promoters, are usually depleted of methylation, whereas gene bodies are heavily methylated. The chromatin structure is controlled by chromatin remodeler complexes that use the hydrolysis of ATP to mediate the packaging of the chromatin. Created with BioRender.com.
DNA methylation, which involves the addition of a methyl group to the 5th position of a cytosine in a CpG dinucleotide context, is one of the best-studied epigenetic marks and essential for mammalian development [16]. DNA methylation is mainly associated with gene silencing, and throughout the genome, 70–80% of CpG dinucleotides are methylated [17]. CpG-rich sequences near promoter regions, referred to as CpG islands, usually lack DNA methylation [18]. More recently, DNA methylation has also been associated with active transcription when found across gene bodies [19]. The de novo DNA methyltransferases (DNMTs), DNMT3A and DNMT3B, are responsible for the establishment of DNA methylation early in embryonic development [20], whereas DNMT1 is required for the maintenance of DNA methylation throughout cell divisions, thereby conferring heritable epigenetic memory [21]. In addition to passive loss of DNA methylation during cell divisions, DNA methylation can be actively removed by members of the ten-eleven translocation (TET) enzyme family. These so called “erasers” mediate DNA demethylation by oxidizing 5-methylcytosine to 5-hydroxy-methylcytosine [22].
Histone marks are post-translational modifications found at the amino-terminal tails of histone proteins and include acetylation, lysine and arginine methylation, phosphorylation and ubiquitination [23]. Different genomic features acquire different types of modifications, and histone marks can be associated with transcriptional activation and repression [24]. For example, both mono-/tri-methylation of histone 3 lysine 4 (H3K4me1/3) and H3K27ac are associated with active transcription but get deposited at different genomic elements. Whereas H3K4me1 and H3K27ac mark enhancers, H3K4me3 is mainly found at the promoters of actively transcribed genes [25,26,27]. In contrast, H3K9me3 and H3K27me3 are referred to as repressive marks, and while H3K9me3 is found in repetitive regions of the genome, transcriptionally silenced genes are enriched in H3K27me3 [28]. Additionally, the methylation of arginine residues can be either associated to transcriptional activation or repression [29]. Histone variants H2A and H2B are post-translationally modified, and the ubiquitination is one of the most frequent and studied tags. For example, monoubiquitination of histone H2A lysine 119 (H2AK119ub) is a repressive mark [30], whereas H2BK120ub is associated with actively transcribed genes [31]. The enzymes responsible for the deposition and removal of these marks are referred to as writers and erasers, respectively. In addition, the readers recognize specific modifications and can mediate downstream effects.
Organization and packaging of chromatin can be restructured by so-called chromatin remodeling complexes that use the hydrolysis of ATP molecules to modulate the degree of chromatin compaction [32] (Figure 1). This extra layer of epigenetic regulation allows rapid adjustment of chromatin states and plays a critical role during development [33]. Chromatin remodeling involves nucleosome repositioning, which influences chromatin accessibility and the binding of transcription factors to DNA [34]. Four subfamilies of chromatin remodelers have been identified: imitation switch (ISWI), chromodomain helicase DNA-binding (CHD), switch/sucrose non-fermentable (SWI/SNF) and INO80, according to the similarity between the ATPase domains [35,36]. ISWI and CHD remodeler subfamilies co-operate to assemble the nucleosome on the newly synthetized DNA. SWI/SNF subfamily enzymes use their DNA translocase activity to slide the octamers along the DNA, eject the nucleosome or remove histone dimers. The remodeling mediated by the INO80 subfamily factors is responsible for the exchange of canonical histone proteins and related variants.
3. Overview of Mouse T Cell Development
T cells derive from lymphoid progenitor cells that develop in the fetal liver or adult bone marrow, migrate via the blood and seed the thymus at the cortical medullary junction [37,38]. Specialized thymic epithelial cells provide the necessary stimuli to activate the Notch signaling pathway that drives progenitor cells to develop into mature differentiated cells in a stepwise manner [39,40,41]. Mouse T cell development is marked by changes in the expression of surface molecules, subdividing T cells into double negative (DN), double positive (DP) and single positive (SP) populations, according to the expression of CD4 and CD8 coreceptors [42,43,44]. Mouse and human T cell development are homologous, but different markers distinguish DN stages in humans (compared in Reference [44]). However, here we focus on mouse T cell development because most of the knowledge on epigenetic and transcription factors comes from using genetically modified mouse models. Early T-cell progenitors (ETPs) are the most immature DN subpopulation, characterized by the expression of the receptor tyrosine kinase c-Kit (or CD117) and the hyaluronic acid receptor (CD44), but they lack surface expression of the IL-2Rα chain (CD25) [45,46,47]. This heterogeneous population is multipotent with the potential to generate other lineages, such as myeloid cells, B cells and natural killer (NK) cells [46,47,48]. As ETPs move towards the outer thymic cortex, CD25 expression increases, giving rise to CD44+CD25+ T cells, referred to as DN2. Based on the expression of CD117, DN2 cells can be further subdivided into DN2a and DN2b cells. CD117 is more highly expressed on the surface of DN2a compared to DN2b cells. These two substages define the transition from uncommitted to committed T cells, during which the Notch signaling-induced cascade mediates the repression of alternative lineage development [49,50]. Full T cell lineage commitment occurs upon transition to the CD44−CD25+ DN3 stage. CD27 expression discriminates between two subpopulations, DN3a (CD44−CD25+CD27−) and DN3b cells (CD44−CD25+CD27+), defined as pre- and post-β-selection cells respectively [51]. β-selection is a key developmental checkpoint at the DN3 stage wherein T cell receptor (TCR) rearrangement, mediated by the recombinant activating genes 1 and 2 (RAG1 and RAG2), begins [52,53]. After successful rearrangement of the TCRβ locus, the expressed β chains couple with the invariant pre-Tα receptor, forming the pre-TCR complex [54,55]. Cells with successful β-selection downregulate the expression of CD25 and become DN4 cells, which then progress to the DP stage through the immature single positive (ISP) cells expressing in mouse CD8, but not CD4 nor CD3 [56]. The development of DN3 cells with unsuccessful β-selection stops, and cells undergo apoptosis. At the DP stage, TCRα gene rearrangements initiate, and an αβ-TCR is produced [57]. Subsequently, T cells undergo positive and negative selection in the cortex. Positive selection is intended to select for T cells with a functional TCR, which means they are able to interact with the major histocompatibility complex (MHC) on cortical epithelial cells that function as antigen-presenting cells [58]. During the negative selection, the cells are screened for potential autoreactivity and will undergo apoptosis when a self-peptide is encountered in the MHC [59]. Thymocytes that respond to self-antigens are eliminated. Finally, the cells with a functional TCR differentiate into CD4+ helper or CD8+ cytotoxic T cells. These now-called single positive (SP) cell types can leave the thymus and emigrate to the periphery [60].
4. Key Transcription Factors in Early T Cells
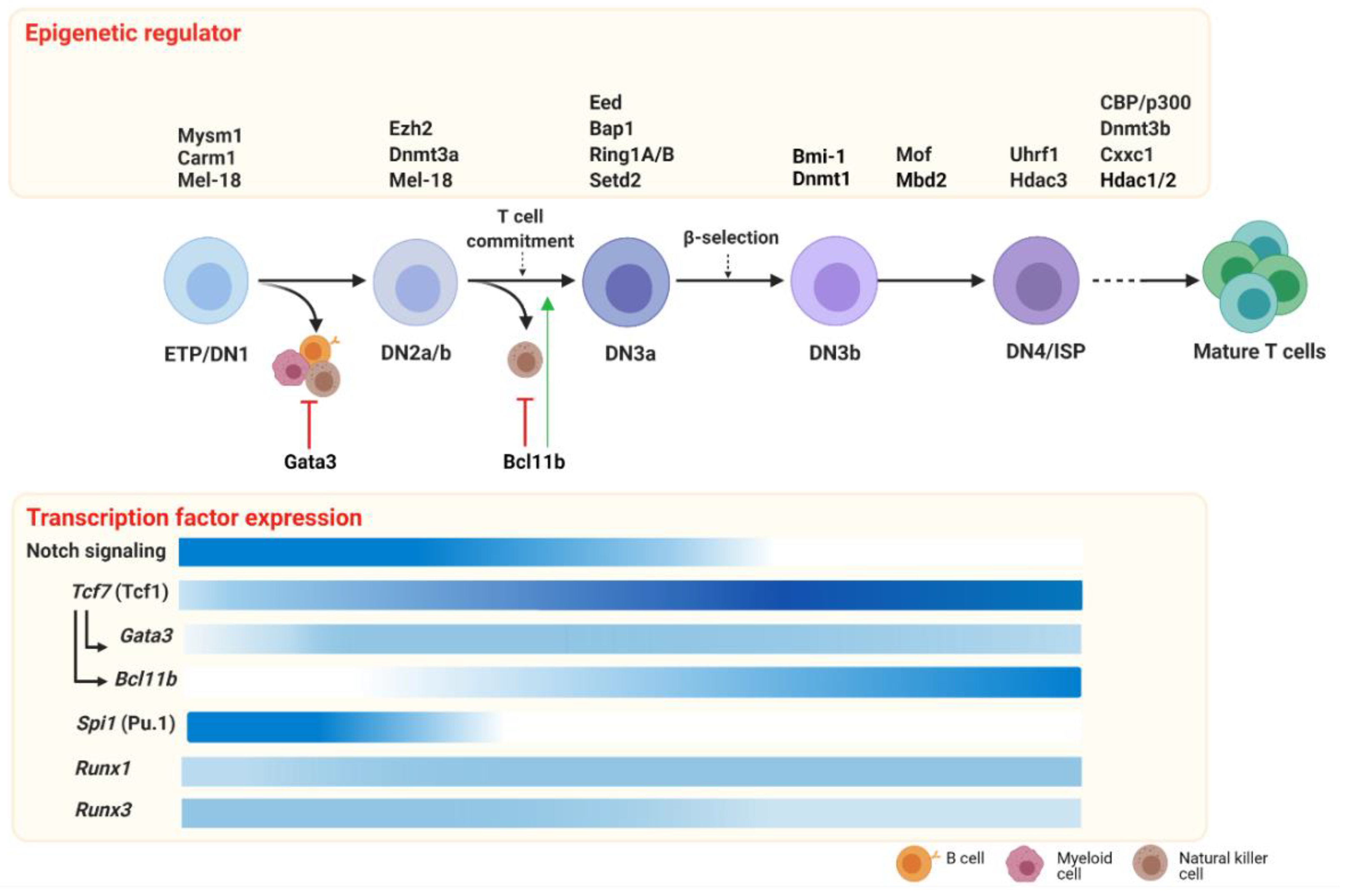
T cell development is dynamically orchestrated by a core set of transcription factors establishing T cell lineage commitment (extensively reviewed in Reference [61]). The critical master regulators for T cell commitment are T cell factor 1 (Tcf1) [62,63], Bcl11b [64], Gata3 [65] and to a lesser extent Pu.1 [66] and the Runx family of transcription factors [67]. The T cell regulatory network is continuously changing throughout all the subsequent developmental stages within the thymus, starting from the most immature cells to the SP cell subsets. The Notch signaling pathway initiates the cascade of transcriptional program changes in multipotent and heterogeneous progenitor cells, which lose their competence for alternative fates and activate the specific T cell regulatory gene network throughout their development [48]. Here, we focus on the key transcription factors Tcf1, Bcl11b, Gata3, Pu.1 and Runx1/3 that have stage-specific patterns of expression (Figure 2) and together create a transcriptional network that regulates early T cell development.
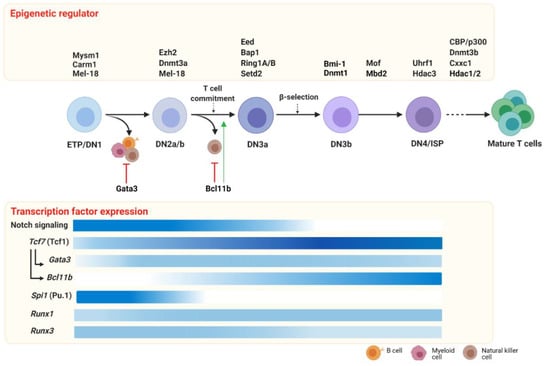
Figure 2.
Epigenetic regulators and transcription factors driving early T cell development. In the thymus, T cell development, triggered by the Notch signaling pathway, requires the expression of key transcription factors (lower panel) and epigenetic modifiers (upper panel). During the first developmental stages, ETP and DN2 cells retain lineage plasticity, most likely sustained by non-T cell transcription factors such as Pu.1 and inhibited by T-cell factors such as Gata3 and Bcl11b (red arrows). The potential to develop myeloid, B and NK cells is shut down at the T cell commitment checkpoint during the transition to the DN2-DN3 stage, which is promoted by Bcl11b (green arrow). DN3b are selected after a successful TCR gene rearrangement (β selection) that, after the last immature stages (DN4/ISP), can progress toward mature CD4+ or CD8+ TCRαβ+ T cells. In the lower panel, the expression patterns of the key transcription factors of early T cells are indicated (the color’s intensity correlates with the gene expression level; darker color indicates a higher expression). Early T cell development is dynamically orchestrated by epigenetic regulators that contribute in shaping T cell survival and specification. In the upper panel, we report the epigenetic regulators at a specific stage or transition, during which the knockout murine models displayed a phenotype. Of note, conditional knockout models have often been employed to study epigenetic regulator function in T cell development. In those cases, it cannot be excluded that the requirement at other stages might exist, but this has not been investigated yet. Abbreviations: ETP: early thymic progenitors; DN: double negative; ISP: immature single positive. Created with BioRender.com.
Tcf1 (encoded by the Tcf7 gene) is a master regulator of T-lineage specification and is activated directly by Notch signaling at the ETP stage [62,63]. The upregulation of Tcf7 upon entry of lymphoid progenitors to the thymus is crucial for the progression toward subsequent developmental stages, and Tcf1 is required for the expression of essential T cells genes such as Bcl11b and Gata3 [62,63]. The functional relationship between these major first transcription factors (Tcf1, Bcl11b and Gata3) that shape the T cell gene program has long been unclear. We recently unraveled the functional hierarchy of this core set of transcription factors in T cell commitment [68]. Our work and others showed that disruption of Tcf7 resulted in abnormal gene expression profiles and chromatin accessibility in DN3 [68] and DP thymocytes [69]. Tcf1 bound to the Gata3 and Bcl11b promoter/enhancer regions, where a more compact chromatin was observed in the absence of Tcf1 [68]. Motif analysis revealed that sites that displayed a differential chromatin opening were enriched for Tcf1 binding sites, suggesting that Gata3 and Bcl11b were two direct Tcf1 targets in thymocytes [68], in accordance with previous studies from Rothenberg and colleagues [64,70]. The ectopic expression of Gata3 and Bcl11b in Tcf7-deficient murine cells suggested a collaboration among the transcription factors: Gata3 suppresses B and myeloid fate in early T cells, whereas Bcl11b rescues impaired T cell development [68]. Interestingly, lack of Tcf1 is also associated with a high chance of developing leukemias [71], as Tcf1 is not only an active transcription factor when associated with the Wnt mediator β-catenin, but also a transcriptional repressor [72]. Collectively, these studies emphasize the role of Tcf1 as essential regulator of T cell development.
Gata3, a member of the Gata transcription factor family, is considered another key member of the T cell transcriptional network. Gata3 plays multiple roles in thymocyte development in a stage-specific manner, affecting T-cell survival, growth, commitment and progression into mature stages [65,73]. Notch signaling and Tcf1 induce the expression of Gata3 that gradually increases from thymic progenitors to mature T cells [74,75,76]. Gata3, in response to Notch signaling, is required for the repression of diversion to non-T cell lineages, such as B cells and mast cells [68,76,77,78]. Gata3 is responsible for driving T cell specification by positively regulating the expression of the essential T cell factor, Bcl11b, at the transition of the T cell commitment stage [77].
The Notch-induced regulatory cascade triggers the activation of Bcl11b, which starts to be expressed at the T cell lineage commitment step occurring at the DN2a–DN2b transition [64,70]. The deficiency of this master regulator affects thymocyte maturation at the commitment checkpoint and impairs T cell receptor rearrangement [79,80]. Bcl11b induces a T cell-specific gene expression program that drives T-lineage commitment by limiting the cells from acquiring non-T cell fates. Specifically, Bcl11b blocks alternative lineages, such as natural killer, innate lymphoid cells and myeloid cells, as genes associated with stem/progenitor and non-T cell lineages are abnormally activated in absence of this master regulator [64,81,82,83]. Therefore, Bcl11b is an essential transcription factor that locks the cells in the T cell lineage by excluding access to other hematopoietic lineages and switching on T cell receptor rearrangement and the T cell program. Bcl11b is also highly expressed in innate lymphoid cells 2 (ILC2s) [84], which share the regulatory network driven by key transcription factors with T cells and thus, effector functionality [85,86]. Interestingly, despite the phenotypical and functional similarity among T cells and ILC2s, Bcl11b controls cell-type specific target genes by binding a distinct set of genomic regions and recruiting different protein complexes [87].
Pu.1 (encoded by Spi1) is a crucial player in the transcriptional regulation in uncommitted T cells, mostly myeloid cells. Its expression starts in the pre-thymic precursors, and it is silenced at the checkpoint of the T cell commitment, when Bcl11b is activated [50]. This hematopoietic transcription factor contributes to the establishment of cell lineage fidelity in myeloid, dendritic and B cells [88,89,90,91]. Nevertheless, Pu.1 is also required for the progression and survival of thymocytes in the early stages [92], and its expression pattern correlates with the transient capability of the cells to give rise to alternative lineages [93,94,95,96,97]. The forced expression of Pu.1 in committed T cells turns on myeloid genes and diverts the cells into macrophage and dendritic cells when Notch signaling is absent [66,98,99,100]. Endogenous Pu.1 mediates the positive and negative regulation of its target genes through a mechanism of re-deployment of other transcription factors such as Runx1 and Satb1 at stage-specific target loci [92,101]. Collectively, Pu.1 coordinates the core transcriptional network of uncommitted T cells by redirecting partner transcription factors.
The Runx transcription factor family regulates the transcriptional program of T cells at different developmental stages [102,103,104,105]. All the family members are initially expressed in the T cell progenitors; however, Runx1 expression increases when Runx2 and Runx3 expression declines [67,106]. The activity of Runx1 has been mainly highlighted in the relationship with other key transcription factors such as Pu.1 and Bcl11b, and together they regulate the dynamic changes of transcriptional signatures before and after T cell commitment, respectively [83,101]. Nevertheless, Runx1 is not the only member of the Runx family to participate in the gene regulatory network of T cell development. Recent work dissected the importance of Runx1 and its paralog Runx3 in stage-specific deletion mouse models, identifying a functional redundancy in activating T cell genes and silencing alternative lineage genes when both factors are co-expressed [67]. Despite their continuous expression levels throughout T cell development, Runx1 and Runx3 control the transcriptome waves in a stage-dependent manner, and the dynamic redistribution of Runx proteins to stage-specific target loci is influenced by their interacting partners, Pu.1 before T cell commitment and Bcl11b in committed T cells [67,83,101]. Therefore, the Runx transcription factor family is required to shape the T cell gene regulatory network in a context-dependent manner.
5. Functions for Epigenetic Regulators in Early T Cell Development
5.1. Histone Modifiers
Transcription factors are not the only drivers of T cell commitment and differentiation, but the acquisition of distinctive, developmental-stage-specific transcriptomes is also dependent on correct epigenetic regulation (Figure 2; Table 1). Many epigenetic regulators are crucial for normal development, and their roles in T cell development have often been studied using conditional models driven by Cre-recombinase that acts at specific stages.

Table 1.
Phenotypes of epigenetic factor-depleted murine models.
Polycomb repressive complex 1 (PRC1) and Polycomb repressive complex 2 (PRC2), which mediate the deposition of repressive histone marks, play important roles during T cell development [130], and mice deficient in Polycomb group genes exhibit T cell defects [131]. Among the PRC1 members, Bmi-1 has been shown to be involved in the regulation of immature T cells. Defective accumulation of thymocytes at the DN3 stage and cell death were observed in Bmi-1 knockout mouse models, and it was shown that during the DN–DP transition, Bmi-1 was critical to regulate the suppression of p19Arf, which was found responsible for the cell cycle arrest [107]. The role of another PRC1 protein, Mel-18, was investigated in T cell development [108], and it was found that Mel-18 supported the expansion of early thymocytes and maintained the expression of Hes1, one of the Notch signaling target genes, in vitro and in vivo. The PRC1 members Ring1A and Ring1B mediate mono-ubiquitination of H2AK119 [132,133], and Ring1A/B double knockout resulted in a block at the DN3 stage and a strong reduction of TCRβ+ cells [113]. Interestingly, chromatin immunoprecipitation experiments identified Pax5, a master regulator of B cell development [134], as one of the main Ring1B targets, and deletion of Pax5 was able to rescue T cell development, suggesting that PRC1 complex members were also involved in the suppression of the B cell lineage during thymocyte development [113].
H2AK119Ub can be removed by deubiquitinating enzymes, including BAP1 [135], MYSM1 [136] and ubiquitin-specific proteases (USP) such as USP16 and USP21 [30]. USP16 and USP21 are not involved in T cell development [137,138,139]. However, critical roles for Bap1 and Mysm1 in early thymocyte development have been reported. Conditional deletion of Bap1 in T cells resulted in reduction of thymic cellularity and loss of DN thymocytes. Through the use of an in vitro differentiation system, it was further shown that Bap1 controlled cell cycle progression at the DN3 stage before the pre-TCR checkpoint through its histone de-ubiquitination function [114]. Severe T cell defects have also been observed in Mysm1-deficient mice [115,116], which was associated with Mysm1 function in the p53 pathway rather than its de-ubiquitinase activity [117].
EED, EZH2 and SUZ12 are components of the PRC2 complex [140], and several studies have evaluated the role of PRC2 proteins in late T cell differentiation and plasticity [141,142,143]. In addition, functions for these factors in early intrathymic lymphopoiesis have been reported. For instance, Eed contributes to the developmental progression at the β-selection checkpoint [109]. Ablation of Ezh2, the H3K27me3 writer, resulted in defective T cell development and abnormal distribution of the repressive mark in Ezh-deficient DN cells [110]. Moreover, the loss of Ezh2 and p53 deregulates the transcriptional program of T cell differentiation, leading to malignant transformation [144]. Interestingly, PRC2 interacts with Ikaros, a key factor that can act as repressor or activator in T cell development [145,146,147,148], and PRC2 binding to specific loci is dependent on Ikaros at the DN3 stage [149], suggesting a functional correlation between epigenetic regulators and transcription factors in early thymocytes.
The readers, writers and erasers of the active histone mark H3K4me are essential epigenetic enzymes with important functions in hematopoiesis [150]. In addition, they can contribute to the control of the T cell lineage. Cxxc1 is a key epigenetic regulator that specifically binds to unmethylated CpG-rich sequences and protects Polycomb-bound genes from DNA methylation in mouse embryonic stem cells [151]. In addition, Cxxc1 is a component of the COMPASS complex that contains the H3K4 methyltransferase Setd1 [152], and this association has been suggested to be critical for Cxxc1′s function in thymocyte development. Loss of Cxxc1 in a conditional mouse model was accompanied by a drastic reduction in thymic cellularity and an arrest at the DN3 stage [111], supporting a critical function of Cxxc1/Setd1 in T cell development.
The histone methyltransferase Setd2 catalyzes methylation at lysine 36 of histone 3 (H3K36me3) [153]. Conditional mouse models with loss-of-function mutations in Setd2 resulted in reduced H3K36me3 and a partial arrest at the DN3 stage, during which Setd2 was required for the TCRβ rearrangement [154]. Consistent with this developmental block at the precursor stage, T cell-specific Setd2 knockout mice showed an expansion of γδ T cells [112], suggesting a function of Setd2 in the early lymphocyte maturation.
Another histone modifier that was identified as a regulator of T lymphocytes was the protein arginine methyltransferase Carm1 [155]. Knockout murine models showed that this epigenetic factor promoted the differentiation at the transition from the DN1 to the DN2 stage [118,119], but the epigenetic changes driven by the arginine methyltransferase activity have not been investigated yet.
The reversible acetylation of lysine residues on histone tails is mediated by histone acetyltransferases (HATs) [156], including the CBP/p300 family of HATs [157]. The function of CBP/p300 has been elucidated in late T cell differentiation [158,159], but not much is known about their function during earlier stages. They acted as transcriptional coactivators, and deletion of either CBP or p300 resulted in a reduction of DP thymocytes, but the double knockout showed a more drastic decrease in the number of mature T cells [120]. Mof is responsible for the global acetylation of H4K16 [160], and its deficiency leads to reduction of DP cells and consequently to a partial block at the DN3-DN4 transition, suggesting a primary role for this HAT in early T cells [121].
Histone deacetylases (HDACs) remove acetyl groups [161], and the roles for these erasers in developing T cells have been reported. While the deletion of Hdac1 or Hdac2 has no impact on thymocyte development, dual conditional deletion results in reduced thymic cellularity with a block at the transition to DP cells, causing a dramatic effect on thymocyte development [122]. This severe phenotype was accompanied by an increased acetylation of H3K9, demonstrating the requirements of Hdac1 and Hdac2 for chromatin regulation in the T cell identity [122]. Hdac3 acts at multiple stages during T cell development [162,163] and Cre-mediated Hdac3 deletion causes an impaired progression of early thymocytes towards the DP stage, resulting in an abnormal accumulation of ISP cells that are not able to produce functional T cells [123].
5.2. DNA Methylation Machinery
Kramer and colleagues established an important role for Dnmt3a in the earliest T cell subsets by showing that its genetic ablation in murine hematopoietic stem cells provoked a significant accumulation of DN2 cells because of reduced apoptosis [124]. On the other hand, it was reported that a T cell-specific Dnmt3b knockout did not have an impact on T cell differentiation in a mouse model of MYC-induced lymphomagenesis [164]. Interestingly, knock-in mice with Dnmt3b mutations corresponding to the mutations found in immunodeficiency, centromeric instability, facial anomalies (ICF) syndrome patients [165] showed an increase of thymocyte apoptosis and an arrest at the DN to DP transition stage [125]. Reduced levels or the absence of serum immunoglobulins are characteristic for ICF syndrome. In addition, T cell defects have been described in ICF patients [166,167,168], but their contribution to the disease is incompletely understood. T cell proliferation and development was also dependent on Dnmt1 because impaired survival of TCRαβ+ cells and lower cell number of DP T cells were observed in Dnmt1 conditional knockout mice [126].
The maintenance of DNA methylation requires UHRF1, which binds to hemi-methylated DNA and recruits DNMT1 [169]. The contribution of Uhrf1 to T cell lineage development was assessed in a conditional mouse model, suggesting a possible functional role in αβ and γδ T cell fate decisions due to its activity as epigenetic regulator [127].
T lymphopoiesis is impaired when the DNA methylation reader, Mbd2, is not expressed [128]. The deficiency in Mbd2 leads to an arrest at the DN3 stage and consequently, a reduction of the DP population. The T cell defect was linked to Mbd2′s function in regulating the expression of key genes of the WNT pathway, including Tcf7. As Wnt signaling has been shown to be crucial for proper T cell development [72,170], these studies have uncovered a link between an epigenetic regulator controlling early T cell development and the Wnt pathway. As reviewed extensively elsewhere [171,172,173], Wnt signaling is required for normal T cell development even though the targeted deletion of beta and gamma-catenin in thymocytes shows no phenotype because these mutations fail to eliminate canonical Wnt signaling. Other studies conclusively have shown the importance of Wnt signaling in early T cell development, as in the combined effects of the nuclear effectors of Wnt signaling, Lef1 and Tcf1 [174].
6. The Interplay of Epigenetic and Transcription Factors in Early T Cell Development
T cell development is built on decisions by progenitor cells that are instructed to differentiate into more mature T cells. The “instructions” that are controlling the development stages are not only external factors such as cytokines and environmental signals. Regulatory networks driven by stage-specific transcription factors constitute a key feature of the developmental changes during lymphopoiesis [10,175].
A specialized class of transcription factors, termed pioneer factors, can bind to their target sites within compacted chromatin [176], and this pioneering activity leads to increased DNA accessibility that allows the recruitment and binding of other transcription factors and chromatin remodelers [177]. The ability to trigger chromatin changes has been observed in lineage-determining transcription factors. For example, Pu.1 is a well-known pioneer transcription factor in myeloid cells [178,179]. A pioneering role of Pu.1 has recently also been described in uncommitted T cells, wherein Pu.1 is able to bind to its closed target sites and initiate the opening of chromatin [176,180]. Pu.1 can recruit two other transcription factors, Satb1 and Runx1, to genomic sites, and it can activate or repress genes by displacement of these transcription factors along the genome [101,181]. Downregulation of Pu.1 expression is associated with the activation of Bcl11b, which defines the transition to committed cells and coincides with the loss of potential to develop toward myeloid lineages [82,92]. Activation of the Bcl11b gene requires the transcription of a non-coding RNA called Thymocyte Differentiation Factor (ThymoD) that facilitates enhancer–promoter communication [182]. Moreover, a computational model based on results from single-cell and bulk studies has been recently used to increase our understanding of the kinetics of T cell lineage commitment, elucidating the complex regulatory mechanism of Bcl11b activation that involves transcriptional and stochastic epigenetic control [183].
The transition from uncommitted to committed T cells also exhibits the largest transformation in chromatin accessibility and topologically associating domain (TAD) conformation [184], and this reorganization of the chromatin landscape correlates with changes in the distribution of genome-wide histone marks [11,185]. DNA sequences that become inaccessible after T cell commitment are highly enriched for a Pu.1 binding motif [11,69], and these sites lose the active histone mark (H3K4me2) and acquire the repressive modification (H3K27me3) [11]. Genomic loci that gain accessibility are bound by Bcl11b, and it seems that Bcl11b itself can contribute to maintaining long-range chromatin interactions of its target genes [184]. The two stage-specific transcription factors Pu.1 and Bcl11b are not the only factors that are able to modulate the chromatin landscape. Indeed, our data and others reveal that the master regulator, Tcf1, can coordinate chromatin accessibility in DN3 and DP T cells, unveiling a role of Tcf1 in the epigenetic landscape of T cells [68,69]. Moreover, Tcf1 exploits its intrinsic HDAC activity to regulate the gene expression in CD8 T cells [186], but it is still undiscovered whether the HDAC domain has a crucial function in the immature stages. Of note, the HDAC activity of Tcf/Lef factors is modest; it is much lower than conventional HDACs and needs high concentrations of inhibitors to be reduced. In addition, many splice variants of Tcf1 exist that do not contain the presumed HDAC activity but play roles in mature CD8 T cell differentiation [187], suggesting the involvement of additional mechanisms.
For instance, it is well established that master transcription factors and chromatin modifiers cooperate in directing B lymphocyte development [188,189]. Additionally, dysregulation of both epigenetic regulators and transcription factors induces immune lineage conversion [190]. While our knowledge about these processes in early T cell development is still limited, it is likely that epigenetic regulation is an essential component of T cell specification.
The progress toward mature T cells is orchestrated by the actions of chromatin modifiers [129], but the functional crosstalk among them and key transcription factors in early T cells remains poorly understood. Mouse models deficient for ATP-dependent remodeling enzymes suggested critical functions of these factors throughout T cell development [191]. In the context of early T cell development, Mbd3, a member of the NuRD complex, has an important impact on thymic lymphopoiesis [192]. Furthermore, it has been shown that a T cell specific knockout of Brg1/SMARCA4, which is the catalytic subunit of the SWI/SNF chromatin remodeling complex, displayed thymic defects associated with a developmental block at the DN stage [193]. Notably, Cd4 and Cd8 gene expression is coordinated by the combined functions of epigenetic regulators and transcription factors that ensure proper silencing in immature T cells [194]. The mechanism of recruitment and redistribution of epigenetic and transcription factors has been elucidated in a DN3-like cell line [83,181]. The master regulator of T cell commitment, Bcl11b, regulates the expression of its target genes by nucleation of chromatin remodeling complexes such as PRC1, NuRD, REST complexes, and Runx1 to specific genomic sites [83,87]. Bcl11b and its partners are able to modulate both activation and repression of genes, collaborating in this way toward the establishment of the T cell identity [83]. Overall, the identification of the complex interaction between chromatin modifiers and transcription factors is crucial for the understanding of the stage-specific transcriptional network and regulation of intrathymic cell fate determination. This is currently an area of intense investigation in several laboratories world-wide.
7. Concluding Remarks
The development of T lymphocytes from uncommitted progenitor cells in the thymus is a highly regulated process. Regulation involves external signals such as cytokines, Wnt, BMP and Notch signals as well as cell-intrinsic mechanisms including epigenetic control. It is therefore not surprising that deletion of fairly ubiquitous epigenetic mediators (HDACs, SWI/SNF factors, etc.) in specific stages of T cell development leads to profound functional defects. In cases where effects are not found, there is likely a molecular redundancy. It should also be noted that in many studies on epigenetic factors, conditional knockout systems are used, most often employing Lck-Cre, which is not active until the DN3 stage. Hence, functional effects in earlier stages of T cell development may not have been discovered yet. More interesting, therefore, are lineage-specific regulators or epigenetic factors that show strong differential expression over the various stages of development, as differential expression may imply an important function. A key issue is the physical and functional interaction between pioneering transcription factors and epigenetic regulators, and especially epigenetic readers. For instance, some major transcription factors may mediate changes in the epigenetic state because they are apparently capable of setting up a specific lineage as was shown for Tcf1, possibly by intrinsic HDAC activity [186]. On the other hand, other examples indicate that epigenetic regulation is superimposed on the actions of lineage-specific transcription factors. A fascinating example is hematopoiesis and T cell development in Wnt3a-deficient mice. In Wnt3a−/− mice, there are fewer hematopoietic stem cells that remain capable of multilineage differentiation but lack self-renewal, as shown in secondary transplantations [170]. Interestingly, transplantation of Wnt3a−/− hematopoietic stem cells (HSCs) in Wnt3a-proficient hosts does not rescue this phenotype. Apparently, there is a window of time in development in the fetal liver during which Wnt3a needs to be present; otherwise, HSCs do not form properly. This is likely due to an epigenetic regulation of the stem cell fate in this time window. Indications for a link to aberrant epigenetic regulation come from RNA-seq experiments on Wnt3a-deficient HSCs, showing sharply reduced expression of genes involved in epigenetic regulation (Staal, unpublished). Thus, this represents an example of an extrinsic effect on epigenetic regulation due to the temporal requirement for Wnt3a signaling by a neighboring cell. How pioneering transcription factors and epigenetic regulators interact and set up lineage specific gene expression programs is currently largely unknown. Given that various developmental checkpoints (T lineage commitment, β selection, positive and negative selection, CD4 vs CD8 division) are well defined functionally and phenotypically, and individual cells can be readily purified for further study, T cell development forms an ideal model to investigate these fascinating questions.
Author Contributions
V.D.C., L.D. and F.J.T.S. discussed and wrote the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
V.D.C. is supported by an NWO-ALW-Open grant to L.D. (ALWOP.358). Work in the Staal laboratory is supported in part by a ZonMW E-RARE grant (40-419000-98-020) and EU H2020 grant RECOMB (755170-2) and has received funding from the European Union Horizon 2020 research and innovation program.
Acknowledgments
We thank Kirsten Canté-Barrett for carefully reading the manuscript and members of the Staal and Daxinger laboratories for helpful discussions.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Zhu, J.; Emerson, S.G. Hematopoietic cytokines, transcription factors and lineage commitment. Oncogene 2002, 21, 3295–3313. [Google Scholar] [CrossRef]
- Rodrigues, C.P.; Shvedunova, M.; Akhtar, A. Epigenetic Regulators as the Gatekeepers of Hematopoiesis. Trends Genet. 2021, 37, 125–142. [Google Scholar] [CrossRef]
- Goode, D.K.; Obier, N.; Vijayabaskar, M.S.; Lie, A.L.M.; Lilly, A.J.; Hannah, R.; Lichtinger, M.; Batta, K.; Florkowska, M.; Patel, R.; et al. Dynamic Gene Regulatory Networks Drive Hematopoietic Specification and Differentiation. Dev. Cell 2016, 36, 572–587. [Google Scholar] [CrossRef]
- Winter, D.R.; Amit, I. The role of chromatin dynamics in immune cell development. Immunol. Rev. 2014, 261, 9–22. [Google Scholar] [CrossRef]
- Hu, D.; Shilatifard, A. Epigenetics of hematopoiesis and hematological malignancies. Genes Dev. 2016, 30, 2021–2041. [Google Scholar] [CrossRef] [PubMed]
- Goyama, S.; Kitamura, T. Epigenetics in normal and malignant hematopoiesis: An overview and update 2017. Cancer Sci. 2017, 108, 553–562. [Google Scholar] [CrossRef]
- Cumano, A.; Godin, I. Ontogeny of the Hematopoietic System. Annu. Rev. Immunol. 2007, 25, 745–785. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Jeremiah Bell, J.; Bhandoola, A. T-cell lineage determination. Immunol. Rev. 2010, 238, 12–22. [Google Scholar] [CrossRef]
- Koch, U.; Fiorini, E.; Benedito, R.; Besseyrias, V.; Schuster-Gossler, K.; Pierres, M.; Manley, N.R.; Duarte, A.; Macdonald, H.R.; Radtke, F. Delta-like 4 is the essential, nonredundant ligand for Notch1 during thymic T cell lineage commitment. J. Exp. Med. 2008, 205, 2515–2523. [Google Scholar] [CrossRef] [PubMed]
- Rothenberg, E.V. Dynamic control of the T-cell specification gene regulatory network. Curr. Opin. Syst. Biol. 2019, 18, 62–76. [Google Scholar] [CrossRef]
- Zhang, J.A.; Mortazavi, A.; Williams, B.A.; Wold, B.J.; Rothenberg, E.V. Dynamic Transformations of Genome-wide Epigenetic Marking and Transcriptional Control Establish T Cell Identity. Cell 2012, 149, 467–482. [Google Scholar] [CrossRef]
- Henning, A.N.; Roychoudhuri, R.; Restifo, N.P. Epigenetic control of CD8+ T cell differentiation. Nat. Rev. Immunol. 2018, 18, 340–356. [Google Scholar] [CrossRef] [PubMed]
- Gray, S.M.; Kaech, S.M.; Staron, M.M. The interface between transcriptional and epigenetic control of effector and memory CD8+T-cell differentiation. Immunol. Rev. 2014, 261, 157–168. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, R.M.; Lopez-Larrea, C.; Suarez-Alvarez, B. Epigenetic dynamics during CD4+ T cells lineage commitment. Int. J. Biochem. Cell Biol. 2015, 67, 75–85. [Google Scholar] [CrossRef] [PubMed]
- Pace, L.; Amigorena, S. Epigenetics of T cell fate decision. Curr. Opin. Immunol. 2020, 63, 43–50. [Google Scholar] [CrossRef]
- Greenberg, M.V.C.; Bourc’His, D. The diverse roles of DNA methylation in mammalian development and disease. Nat. Rev. Mol. Cell Biol. 2019, 20, 590–607. [Google Scholar] [CrossRef] [PubMed]
- Siegfried, Z.; Cedar, H. DNA methylation: A molecular lock. Curr. Biol. 1997, 7, R305–R307. [Google Scholar] [CrossRef]
- Cross, S.H.; Bird, A.P. CpG islands and genes. Curr. Opin. Genet. Dev. 1995, 5, 309–314. [Google Scholar] [CrossRef]
- Jones, P.A. Functions of DNA methylation: Islands, start sites, gene bodies and beyond. Nat. Rev. Genet. 2012, 13, 484–492. [Google Scholar] [CrossRef] [PubMed]
- Okano, M.; Bell, D.W.; Haber, D.A.; Li, E. DNA Methyltransferases Dnmt3a and Dnmt3b Are Essential for De Novo Methylation and Mammalian Development. Cell 1999, 99, 247–257. [Google Scholar] [CrossRef]
- Hermann, A.; Goyal, R.; Jeltsch, A. The Dnmt1 DNA-(cytosine-C5)-methyltransferase Methylates DNA Processively with High Preference for Hemimethylated Target Sites. J. Biol. Chem. 2004, 279, 48350–48359. [Google Scholar] [CrossRef] [PubMed]
- Tahiliani, M.; Koh, K.P.; Shen, Y.; Pastor, W.A.; Bandukwala, H.; Brudno, Y.; Agarwal, S.; Iyer, L.M.; Liu, D.R.; Aravind, L.; et al. Conversion of 5-Methylcytosine to 5-Hydroxymethylcytosine in Mammalian DNA by MLL Partner TET1. Science 2009, 324, 930–935. [Google Scholar] [CrossRef] [PubMed]
- Rothbart, S.B.; Strahl, B.D. Interpreting the language of histone and DNA modifications. Biochim. Biophys. Acta 2014, 1839, 627–643. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Garcia, B.A. Comprehensive Catalog of Currently Documented Histone Modifications. Cold Spring Harb. Perspect. Biol. 2015, 7, a025064. [Google Scholar] [CrossRef] [PubMed]
- Creyghton, M.P.; Cheng, A.W.; Welstead, G.G.; Kooistra, T.; Carey, B.W.; Steine, E.J.; Hanna, J.; Lodato, M.A.; Frampton, G.M.; Sharp, P.A.; et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc. Natl. Acad. Sci. USA 2010, 107, 21931–21936. [Google Scholar] [CrossRef] [PubMed]
- Schübeler, D. Enhancing genome annotation with chromatin. Nat. Genet. 2007, 39, 284–285. [Google Scholar] [CrossRef]
- Heintzman, N.D.; Stuart, R.K.; Hon, G.; Fu, Y.; Ching, C.W.; Hawkins, R.D.; Barrera, L.O.; Van Calcar, S.; Qu, C.; Ching, K.A.; et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat. Genet. 2007, 39, 311–318. [Google Scholar] [CrossRef]
- Trojer, P.; Reinberg, D. Facultative Heterochromatin: Is There a Distinctive Molecular Signature? Mol. Cell 2007, 28, 1–13. [Google Scholar] [CrossRef]
- Di Lorenzo, A.; Bedford, M.T. Histone arginine methylation. FEBS Lett. 2011, 585, 2024–2031. [Google Scholar] [CrossRef]
- Vissers, J.H.; Nicassio, F.; Van Lohuizen, M.; Di Fiore, P.P.; Citterio, E. The many faces of ubiquitinated histone H2A: Insights from the DUBs. Cell Div. 2008, 3, 8. [Google Scholar] [CrossRef]
- Minsky, N.; Shema, E.; Field, Y.; Schuster, M.; Segal, E.; Oren, M. Monoubiquitinated H2B is associated with the transcribed region of highly expressed genes in human cells. Nat. Cell Biol. 2008, 10, 483–488. [Google Scholar] [CrossRef] [PubMed]
- Clapier, C.R.; Iwasa, J.; Cairns, B.R.; Peterson, C.L. Mechanisms of action and regulation of ATP-dependent chromatin-remodelling complexes. Nat. Rev. Mol. Cell Biol. 2017, 18, 407–422. [Google Scholar] [CrossRef]
- Ho, L.; Crabtree, G.R. Chromatin remodelling during development. Nature 2010, 463, 474–484. [Google Scholar] [CrossRef]
- Narlikar, G.J.; Sundaramoorthy, R.; Owen-Hughes, T. Mechanisms and Functions of ATP-Dependent Chromatin-Remodeling Enzymes. Cell 2013, 154, 490–503. [Google Scholar] [CrossRef] [PubMed]
- Längst, G.; Manelyte, L. Chromatin Remodelers: From Function to Dysfunction. Genes 2015, 6, 299–324. [Google Scholar] [CrossRef]
- Clapier, C.R.; Cairns, B.R. The Biology of Chromatin Remodeling Complexes. Annu. Rev. Biochem. 2009, 78, 273–304. [Google Scholar] [CrossRef]
- Schwarz, B.A.; Sambandam, A.; Maillard, I.; Harman, B.C.; Love, P.E.; Bhandoola, A. Selective Thymus Settling Regulated by Cytokine and Chemokine Receptors. J. Immunol. 2007, 178, 2008–2017. [Google Scholar] [CrossRef] [PubMed]
- Schwarz, B.A.; Bhandoola, A. Circulating hematopoietic progenitors with T lineage potential. Nat. Immunol. 2004, 5, 953–960. [Google Scholar] [CrossRef]
- Rothenberg, E.V.; Moore, J.E.; Yui, M.A. Launching the T-cell-lineage developmental programme. Nat. Rev. Immunol. 2008, 8, 9–21. [Google Scholar] [CrossRef]
- Radtke, F.; Wilson, A.; Stark, G.; Bauer, M.; van Meerwijk, J.; MacDonald, H.R.; Aguet, M. Deficient T Cell Fate Specification in Mice with an Induced Inactivation of Notch1. Immunity 1999, 10, 547–558. [Google Scholar] [CrossRef]
- Chen, E.L.Y.; Thompson, P.K.; Zúñiga-Pflücker, J.C. RBPJ-dependent Notch signaling initiates the T cell program in a subset of thymus-seeding progenitors. Nat. Immunol. 2019, 20, 1456–1468. [Google Scholar] [CrossRef]
- Dik, W.A.; Pike-Overzet, K.; Weerkamp, F.; de Ridder, D.; de Haas, E.F.; Baert, M.R.; van der Spek, P.; Koster, E.E.; Reinders, M.J.; van Dongen, J.J.; et al. New insights on human T cell development by quantitative T cell receptor gene rearrangement studies and gene expression profiling. J. Exp. Med. 2005, 201, 1715–1723. [Google Scholar] [CrossRef]
- Scollay, R.; Bartlett, P.; Shortman, K. T Cell Development in the Adult Murine Thymus: Changes in the Expression of the Surface Antigens Ly2, L3T4 and B2A2 during Development from Early Precursor Cells to Emigrants. Immunol. Rev. 1984, 82, 79–103. [Google Scholar] [CrossRef] [PubMed]
- Weerkamp, F.; Baert, M.R.; Brugman, M.H.; Dik, W.A.; De Haas, E.F.; Visser, T.P.; De Groot, C.J.; Wagemaker, G.; Van Dongen, J.J.; Staal, F.J. Human thymus contains multipotent progenitors with T/B lymphoid, myeloid, and erythroid lineage potential. Blood 2006, 107, 3131–3137. [Google Scholar] [CrossRef] [PubMed]
- Godfrey, D.I.; Kennedy, J.; Suda, T.; Zlotnik, A. A developmental pathway involving four phenotypically and functionally distinct subsets of CD3-CD4-CD8- triple-negative adult mouse thymocytes defined by CD44 and CD25 expression. J. Immunol. 1993, 150, 4244–4252. [Google Scholar] [PubMed]
- Ikawa, T.; Kawamoto, H.; Fujimoto, S.; Katsura, Y. Commitment of Common T/Natural Killer (Nk) Progenitors to Unipotent T and Nk Progenitors in the Murine Fetal Thymus Revealed by a Single Progenitor Assay. J. Exp. Med. 1999, 190, 1617–1626. [Google Scholar] [CrossRef] [PubMed]
- Wada, H.; Masuda, K.; Satoh, R.; Kakugawa, K.; Ikawa, T.; Katsura, Y.; Kawamoto, H. Adult T-cell progenitors retain myeloid potential. Nature 2008, 452, 768–772. [Google Scholar] [CrossRef]
- Zhou, W.; Yui, M.A.; Williams, B.A.; Yun, J.; Wold, B.J.; Cai, L.; Rothenberg, E.V. Single-Cell Analysis Reveals Regulatory Gene Expression Dynamics Leading to Lineage Commitment in Early T Cell Development. Cell Syst. 2019, 9, 321–337.e9. [Google Scholar] [CrossRef]
- Masuda, K.; Kakugawa, K.; Nakayama, T.; Minato, N.; Katsura, Y.; Kawamoto, H. T Cell Lineage Determination Precedes the Initiation of TCRβ Gene Rearrangement. J. Immunol. 2007, 179, 3699–3706. [Google Scholar] [CrossRef]
- Yui, M.A.; Feng, N.; Rothenberg, E.V. Fine-Scale Staging of T Cell Lineage Commitment in Adult Mouse Thymus. J. Immunol. 2010, 185, 284–293. [Google Scholar] [CrossRef]
- Taghon, T.; Yui, M.A.; Pant, R.; Diamond, R.A.; Rothenberg, E.V. Developmental and Molecular Characterization of Emerging β- and γδ-Selected Pre-T Cells in the Adult Mouse Thymus. Immunity 2006, 24, 53–64. [Google Scholar] [CrossRef] [PubMed]
- Dudley, E.C.; Petrie, H.T.; Shah, L.M.; Owen, M.J.; Hayday, A.C. T cell receptor β chain gene rearrangement and selection during thymocyte development in adult mice. Immunity 1994, 1, 83–93. [Google Scholar] [CrossRef]
- Wilson, A.; Held, W.; Macdonald, H.R. Two waves of recombinase gene expression in developing thymocytes. J. Exp. Med. 1994, 179, 1355–1360. [Google Scholar] [CrossRef] [PubMed]
- Terhorst, C. Structure and function of the T3/T cell receptor complex. Behring Inst. Mitt. 1985, 77, 22–29. [Google Scholar]
- Aifantis, I.; Feinberg, J.; Fehling, H.J.; Di Santo, J.P.; Von Boehmer, H. Early T Cell Receptor β Gene Expression Is Regulated by the Pre-T Cell Receptor–CD3 Complex. J. Exp. Med. 1999, 190, 141–144. [Google Scholar] [CrossRef]
- Hugo, P.; Waanders, G.A.; Scollay, R.; Shortman, K.; Boyd, R.L. Ontogeny of a novel CD4+CD8−CD3−thymocyte subpopulation: A comparison with CD4− CD8+ CD3−thymocytes. Int. Immunol. 1990, 2, 209–218. [Google Scholar] [CrossRef] [PubMed]
- Villey, I.; Caillol, D.; Selz, F.; Ferrier, P.; De Villartay, J.-P. Defect in Rearrangement of the Most 5′ TCR–Jα Following Targeted Deletion of T Early α (TEA): Implications for TCR α Locus Accessibility. Immunity 1996, 5, 331–342. [Google Scholar] [CrossRef]
- Davis, M.M.; Boniface, J.J.; Reich, Z.; Lyons, D.; Hampl, J.; Arden, B.; Chien, Y. Ligand recognition by alpha beta T cell receptors. Annu. Rev. Immunol. 1998, 16, 523–544. [Google Scholar] [CrossRef]
- Surh, C.D.; Sprent, J. T-cell apoptosis detected in situ during positive and negative selection in the thymus. Nature 1994, 372, 100–103. [Google Scholar] [CrossRef] [PubMed]
- Weinreich, M.A.; Hogquist, K.A. Thymic Emigration: When and How T Cells Leave Home. J. Immunol. 2008, 181, 2265–2270. [Google Scholar] [CrossRef]
- Hosokawa, H.; Rothenberg, E.V. How transcription factors drive choice of the T cell fate. Nat. Rev. Immunol. 2021, 21, 162–176. [Google Scholar] [CrossRef]
- Germar, K.; Dose, M.; Konstantinou, T.; Zhang, J.; Wang, H.; Lobry, C.; Arnett, K.L.; Blacklow, S.C.; Aifantis, I.; Aster, J.C.; et al. T-cell factor 1 is a gatekeeper for T-cell specification in response to Notch signaling. Proc. Natl. Acad. Sci. USA 2011, 108, 20060–20065. [Google Scholar] [CrossRef] [PubMed]
- Weber, B.N.; Chi, A.W.; Chavez, A.; Yashiro-Ohtani, Y.; Yang, Q.; Shestova, O.; Bhandoola, A. A critical role for TCF-1 in T-lineage specification and differentiation. Nature 2011, 476, 63–68. [Google Scholar] [CrossRef]
- Li, L.; Leid, M.; Rothenberg, E.V. An Early T Cell Lineage Commitment Checkpoint Dependent on the Transcription Factor Bcl11b. Science 2010, 329, 89–93. [Google Scholar] [CrossRef] [PubMed]
- Hosoya, T.; Kuroha, T.; Moriguchi, T.; Cummings, D.; Maillard, I.; Lim, K.C.; Engel, J.D. GATA-3 is required for early T lineage progenitor development. J. Exp. Med. 2009, 206, 2987–3000. [Google Scholar] [CrossRef]
- Anderson, M.K.; Weiss, A.H.; Hernandez-Hoyos, G.; Dionne, C.J.; Rothenberg, E.V. Constitutive Expression of PU.1 in Fetal Hematopoietic Progenitors Blocks T Cell Development at the Pro-T Cell Stage. Immunity 2002, 16, 285–296. [Google Scholar] [CrossRef]
- Shin, B.; Hosokawa, H.; Romero-Wolf, M.; Zhou, W.; Masuhara, K.; Tobin, V.R.; Levanon, D.; Groner, Y.; Rothenberg, E.V. Runx1 and Runx3 drive progenitor to T-lineage transcriptome conversion in mouse T cell commitment via dynamic genomic site switching. Proc. Natl. Acad. Sci. USA 2021, 118. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Perez, L.; Famili, F.; Cordes, M.; Brugman, M.; Van Eggermond, M.; Wu, H.; Chouaref, J.; Granado, D.S.L.; Tiemessen, M.M.; Pike-Overzet, K.; et al. Functional definition of a transcription factor hierarchy regulating T cell lineage commitment. Sci. Adv. 2020, 6, eaaw7313. [Google Scholar] [CrossRef]
- Johnson, J.L.; Georgakilas, G.; Petrovic, J.; Kurachi, M.; Cai, S.; Harly, C.; Pear, W.S.; Bhandoola, A.; Wherry, E.J.; Vahedi, G. Lineage-Determining Transcription Factor TCF-1 Initiates the Epigenetic Identity of T Cells. Immunity 2018, 48, 243–257.e10. [Google Scholar] [CrossRef] [PubMed]
- Kueh, H.Y.; Yui, M.A.; Ng, K.K.; Pease, S.S.; Zhang, J.A.; Damle, S.S.; Freedman, G.; Siu, S.; Bernstein, I.D.; Elowitz, M.B.; et al. Asynchronous combinatorial action of four regulatory factors activates Bcl11b for T cell commitment. Nat. Immunol. 2016, 17, 956–965. [Google Scholar] [CrossRef]
- Tiemessen, M.M.; Baert, M.R.; Schonewille, T.; Brugman, M.H.; Famili, F.; Salvatori, D.C.; Meijerink, J.P.; Ozbek, U.; Clevers, H.; Van Dongen, J.J.; et al. The Nuclear Effector of Wnt-Signaling, Tcf1, Functions as a T-Cell–Specific Tumor Suppressor for Development of Lymphomas. PLoS Biol. 2012, 10, e1001430. [Google Scholar] [CrossRef]
- Staal, F.J.; Meeldijk, J.; Moerer, P.; Jay, P.; van de Weerdt, B.C.; Vainio, S.; Nolan, G.P.; Clevers, H. Wnt signaling is required for thymocyte development and activates Tcf-1 mediated transcription. Eur. J. Immunol. 2001, 31, 285–293. [Google Scholar] [CrossRef]
- Wan, Y.Y. GATA3: A master of many trades in immune regulation. Trends Immunol. 2014, 35, 233–242. [Google Scholar] [CrossRef]
- David-Fung, E.-S.; Yui, M.A.; Morales, M.; Wang, H.; Taghon, T.; Diamond, R.A.; Rothenberg, E.V. Progression of regulatory gene expression states in fetal and adult pro-T-cell development. Immunol. Rev. 2006, 209, 212–236. [Google Scholar] [CrossRef] [PubMed]
- Del Real, M.M.; Rothenberg, E.V. Architecture of a lymphomyeloid developmental switch controlled by PU.1, Notch and Gata3. Development 2013, 140, 1207–1219. [Google Scholar] [CrossRef] [PubMed]
- Hozumi, K.; Negishi, N.; Tsuchiya, I.; Abe, N.; Hirano, K.; Suzuki, D.; Yamamoto, M.; Engel, J.D.; Habu, S. Notch signaling is necessary for GATA3 function in the initiation of T cell development. Eur. J. Immunol. 2008, 38, 977–985. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Ojeda, M.E.; Klein Wolterink, R.G.; Lemaitre, F.; Richard-Le Goff, O.; Hasan, M.; Hendriks, R.W.; Cumano, A.; Di Santo, J.P. GATA-3 promotes T-cell specification by repressing B-cell potential in pro–T cells in mice. Blood 2013, 121, 1749–1759. [Google Scholar] [CrossRef] [PubMed]
- Taghon, T.; Yui, M.A.; Rothenberg, E.V. Mast cell lineage diversion of T lineage precursors by the essential T cell transcription factor GATA-3. Nat. Immunol. 2007, 8, 845–855. [Google Scholar] [CrossRef]
- Ikawa, T.; Hirose, S.; Masuda, K.; Kakugawa, K.; Satoh, R.; Shibano-Satoh, A.; Kominami, R.; Katsura, Y.; Kawamoto, H. An Essential Developmental Checkpoint for Production of the T Cell Lineage. Science 2010, 329, 93–96. [Google Scholar] [CrossRef]
- Wakabayashi, Y.; Watanabe, H.; Inoue, J.; Takeda, N.; Sakata, J.; Mishima, Y.; Hitomi, J.; Yamamoto, T.; Utsuyama, M.; Niwa, O.; et al. Bcl11b is required for differentiation and survival of αβ T lymphocytes. Nat. Immunol. 2003, 4, 533–539. [Google Scholar] [CrossRef]
- Li, P.; Burke, S.; Wang, J.; Chen, X.; Ortiz, M.; Lee, S.C.; Lu, D.; Campos, L.; Goulding, D.; Ng, B.L.; et al. Reprogramming of T Cells to Natural Killer-Like Cells upon Bcl11b Deletion. Science 2010, 329, 85–89. [Google Scholar] [CrossRef] [PubMed]
- Longabaugh, W.J.R.; Zeng, W.; Zhang, J.A.; Hosokawa, H.; Jansen, C.S.; Li, L.; Romero-Wolf, M.; Liu, P.; Kueh, H.Y.; Mortazavi, A.; et al. Bcl11b and combinatorial resolution of cell fate in the T-cell gene regulatory network. Proc. Natl. Acad. Sci. USA 2017, 114, 5800–5807. [Google Scholar] [CrossRef]
- Hosokawa, H.; Romero-Wolf, M.; Yui, M.A.; Ungerbäck, J.; Quiloan, M.L.G.; Matsumoto, M.; Nakayama, K.I.; Tanaka, T.; Rothenberg, E.V. Bcl11b sets pro-T cell fate by site-specific cofactor recruitment and by repressing Id2 and Zbtb16. Nat. Immunol. 2018, 19, 1427–1440. [Google Scholar] [CrossRef] [PubMed]
- Califano, D.; Cho, J.J.; Uddin, M.N.; Lorentsen, K.J.; Yang, Q.; Bhandoola, A.; Li, H.; Avram, D. Transcription Factor Bcl11b Controls Identity and Function of Mature Type 2 Innate Lymphoid Cells. Immunity 2015, 43, 354–368. [Google Scholar] [CrossRef]
- Yagi, R.; Zhong, C.; Northrup, D.L.; Yu, F.; Bouladoux, N.; Spencer, S.; Hu, G.; Barron, L.; Sharma, S.; Nakayama, T.; et al. The Transcription Factor GATA3 Is Critical for the Development of All IL-7Rα-Expressing Innate Lymphoid Cells. Immunity 2014, 40, 378–388. [Google Scholar] [CrossRef] [PubMed]
- Harly, C.; Kenney, D.; Ren, G.; Lai, B.; Raabe, T.; Yang, Q.; Cam, M.C.; Xue, H.H.; Zhao, K.; Bhandoola, A. The transcription factor TCF-1 enforces commitment to the innate lymphoid cell lineage. Nat. Immunol. 2019, 20, 1150–1160. [Google Scholar] [CrossRef] [PubMed]
- Hosokawa, H.; Romero-Wolf, M.; Yang, Q.; Motomura, Y.; Levanon, D.; Groner, Y.; Moro, K.; Tanaka, T.; Rothenberg, E.V. Cell type–specific actions of Bcl11b in early T-lineage and group 2 innate lymphoid cells. J. Exp. Med. 2020, 217. [Google Scholar] [CrossRef]
- Nerlov, C.; Graf, T. PU.1 induces myeloid lineage commitment in multipotent hematopoietic progenitors. Genes Dev. 1998, 12, 2403–2412. [Google Scholar] [CrossRef]
- Feng, R.; Desbordes, S.C.; Xie, H.; Tillo, E.S.; Pixley, F.; Stanley, E.R.; Graf, T. PU.1 and C/EBPalpha/beta convert fibroblasts into macrophage-like cells. Proc. Natl. Acad. Sci. USA 2008, 105, 6057–6062. [Google Scholar] [CrossRef]
- Houston, I.B.; Kamath, M.B.; Schweitzer, B.L.; Chlon, T.M.; DeKoter, R.P. Reduction in PU.1 activity results in a block to B-cell development, abnormal myeloid proliferation, and neonatal lethality. Exp. Hematol. 2007, 35, 1056–1068. [Google Scholar] [CrossRef]
- Carotta, S.; Dakic, A.; D’Amico, A.; Pang, S.H.; Greig, K.T.; Nutt, S.L.; Wu, L. The Transcription Factor PU.1 Controls Dendritic Cell Development and Flt3 Cytokine Receptor Expression in a Dose-Dependent Manner. Immunity 2010, 32, 628–641. [Google Scholar] [CrossRef]
- Champhekar, A.; Damle, S.S.; Freedman, G.; Carotta, S.; Nutt, S.L.; Rothenberg, E.V. Regulation of early T-lineage gene expression and developmental progression by the progenitor cell transcription factor PU.1. Genes Dev. 2015, 29, 832–848. [Google Scholar] [CrossRef]
- Wu, L.; Li, C.L.; Shortman, K. Thymic dendritic cell precursors: Relationship to the T lymphocyte lineage and phenotype of the dendritic cell progeny. J. Exp. Med. 1996, 184, 903–911. [Google Scholar] [CrossRef]
- Lee, C.K.; Kim, J.K.; Kim, Y.; Lee, M.K.; Kim, K.; Kang, J.K.; Hofmeister, R.; Durum, S.K.; Han, S.S. Generation of Macrophages from Early T Progenitors In Vitro. J. Immunol. 2001, 166, 5964–5969. [Google Scholar] [CrossRef] [PubMed]
- Balciunaite, G.; Ceredig, R.; Rolink, A.G. The earliest subpopulation of mouse thymocytes contains potent T, significant macrophage, and natural killer cell but no B-lymphocyte potential. Blood 2005, 105, 1930–1936. [Google Scholar] [CrossRef] [PubMed]
- Luc, S.; Luis, T.C.; Boukarabila, H.; Macaulay, I.C.; Buza-Vidas, N.; Bouriez-Jones, T.; Lutteropp, M.; Woll, P.S.; Loughran, S.J.; Mead, A.J.; et al. The earliest thymic T cell progenitors sustain B cell and myeloid lineage potential. Nat. Immunol. 2012, 13, 412–419. [Google Scholar] [CrossRef]
- Bell, J.J.; Bhandoola, A. The earliest thymic progenitors for T cells possess myeloid lineage potential. Nature 2008, 452, 764–767. [Google Scholar] [CrossRef] [PubMed]
- Franco, C.B.; Scripture-Adams, D.D.; Proekt, I.; Taghon, T.; Weiss, A.H.; Yui, M.A.; Adams, S.L.; Diamond, R.A.; Rothenberg, E.V. Notch/Delta signaling constrains reengineering of pro-T cells by PU.1. Proc. Natl. Acad. Sci. USA 2006, 103, 11993–11998. [Google Scholar] [CrossRef] [PubMed]
- Laiosa, C.V.; Stadtfeld, M.; Xie, H.; De Andres-Aguayo, L.; Graf, T. Reprogramming of Committed T Cell Progenitors to Macrophages and Dendritic Cells by C/EBPα and PU.1 Transcription Factors. Immunity 2006, 25, 731–744. [Google Scholar] [CrossRef]
- Lefebvre, J.M.; Haks, M.C.; Carleton, M.O.; Rhodes, M.; Sinnathamby, G.; Simon, M.C.; Eisenlohr, L.C.; Garrett-Sinha, L.A.; Wiest, D.L. Enforced Expression of Spi-B Reverses T Lineage Commitment and Blocks β-Selection. J. Immunol. 2005, 174, 6184–6194. [Google Scholar] [CrossRef]
- Hosokawa, H.; Ungerbäck, J.; Wang, X.; Matsumoto, M.; Nakayama, K.I.; Cohen, S.M.; Tanaka, T.; Rothenberg, E.V. Transcription Factor PU.1 Represses and Activates Gene Expression in Early T Cells by Redirecting Partner Transcription Factor Binding. Immunity 2018, 49, 782. [Google Scholar] [CrossRef] [PubMed]
- Sato, T.; Ohno, S.; Hayashi, T.; Sato, C.; Kohu, K.; Satake, M.; Habu, S. Dual Functions of Runx Proteins for Reactivating CD8 and Silencing CD4 at the Commitment Process into CD8 Thymocytes. Immunity 2005, 22, 317–328. [Google Scholar] [CrossRef]
- Egawa, T.; Tillman, R.E.; Naoe, Y.; Taniuchi, I.; Littman, D.R. The role of the Runx transcription factors in thymocyte differentiation and in homeostasis of naive T cells. J. Exp. Med. 2007, 204, 1945–1957. [Google Scholar] [CrossRef] [PubMed]
- Taniuchi, I.; Osato, M.; Egawa, T.; Sunshine, M.J.; Bae, S.C.; Komori, T.; Ito, Y.; Littman, D.R. Differential Requirements for Runx Proteins in CD4 Repression and Epigenetic Silencing during T Lymphocyte Development. Cell 2002, 111, 621–633. [Google Scholar] [CrossRef]
- Guo, Y.; Maillard, I.; Chakraborti, S.; Rothenberg, E.V.; Speck, N.A. Core binding factors are necessary for natural killer cell development and cooperate with Notch signaling during T-cell specification. Blood 2008, 112, 480–492. [Google Scholar] [CrossRef]
- David-Fung, E.S.; Butler, R.; Buzi, G.; Yui, M.A.; Diamond, R.A.; Anderson, M.K.; Rowen, L.; Rothenberg, E.V. Transcription factor expression dynamics of early T-lymphocyte specification and commitment. Dev. Biol. 2009, 325, 444–467. [Google Scholar] [CrossRef]
- Miyazaki, M.; Miyazaki, K.; Itoi, M.; Katoh, Y.; Guo, Y.; Kanno, R.; Katoh-Fukui, Y.; Honda, H.; Amagai, T.; Van Lohuizen, M.; et al. Thymocyte Proliferation Induced by Pre-T Cell Receptor Signaling Is Maintained through Polycomb Gene Product Bmi-1-Mediated Cdkn2a Repression. Immunity 2008, 28, 231–245. [Google Scholar] [CrossRef]
- Miyazaki, M.; Kawamoto, H.; Kato, Y.; Itoi, M.; Miyazaki, K.; Masuda, K.; Tashiro, S.; Ishihara, H.; Igarashi, K.; Amagai, T.; et al. Polycomb Group Gene mel-18 Regulates Early T Progenitor Expansion by Maintaining the Expression of Hes-1, a Target of the Notch Pathway. J. Immunol. 2005, 174, 2507–2516. [Google Scholar] [CrossRef] [PubMed]
- Ikawa, T.; Masuda, K.; Endo, T.A.; Endo, M.; Isono, K.; Koseki, Y.; Nakagawa, R.; Kometani, K.; Takano, J.; Agata, Y.; et al. Conversion of T cells to B cells by inactivation of polycomb-mediated epigenetic suppression of the B-lineage program. Genes Dev. 2016, 30, 2475–2485. [Google Scholar] [CrossRef]
- Arenzana, T.L.; Lianoglou, S.; Seki, A.; Eidenschenk, C.; Cheung, T.; Seshasayee, D.; Hagenbeek, T.; Sambandam, A.; Noubade, R.; Peng, I.; et al. Tumor suppressor BAP1 is essential for thymic development and proliferative responses of T lymphocytes. Sci. Immunol. 2018, 3, eaal1953. [Google Scholar] [CrossRef]
- Huang, X.F.; Nandakumar, V.; Tumurkhuu, G.; Wang, T.; Jiang, X.; Hong, B.; Jones, L.; Won, H.; Yoshii, H.; Ozato, K.; et al. Mysm1 is required for interferon regulatory factor expression in maintaining HSC quiescence and thymocyte development. Cell Death Dis. 2016, 7, e2260. [Google Scholar] [CrossRef][Green Version]
- Nijnik, A.; Clare, S.; Hale, C.; Raisen, C.; McIntyre, R.E.; Yusa, K.; Everitt, A.R.; Mottram, L.; Podrini, C.; Lucas, M.; et al. The critical role of histone H2A-deubiquitinase Mysm1 in hematopoiesis and lymphocyte differentiation. Blood 2012, 119, 1370–1379. [Google Scholar] [CrossRef]
- Gatzka, M.; Tasdogan, A.; Hainzl, A.; Allies, G.; Maity, P.; Wilms, C.; Wlaschek, M.; Scharffetterkochanek, K. Interplay of H2A deubiquitinase 2A-DUB/Mysm1 and the p19ARF/p53 axis in hematopoiesis, early T-cell development and tissue differentiation. Cell Death Differ. 2015, 22, 1451–1462. [Google Scholar] [CrossRef]
- Richie, E.R.; Schumacher, A.; Angel, J.M.; Holloway, M.; Rinchik, E.M.; Magnuson, T. The Polycomb-group gene eed regulates thymocyte differentiation and suppresses the development of carcinogen-induced T-cell lymphomas. Oncogene 2002, 21, 299–306. [Google Scholar] [CrossRef]
- Su, I.H.; Dobenecker, M.W.; Dickinson, E.; Oser, M.; Basavaraj, A.; Marqueron, R.; Viale, A.; Reinberg, D.; Wülfing, C.; Tarakhovsky, A. Polycomb Group Protein Ezh2 Controls Actin Polymerization and Cell Signaling. Cell 2005, 121, 425–436. [Google Scholar] [CrossRef]
- Cao, W.; Guo, J.; Wen, X.; Miao, L.; Lin, F.; Xu, G.; Ma, R.; Yin, S.; Hui, Z.; Chen, T.; et al. CXXC finger protein 1 is critical for T-cell intrathymic development through regulating H3K4 trimethylation. Nat. Commun. 2016, 7, 11687. [Google Scholar] [CrossRef]
- Moffitt, A.B.; Ondrejka, S.L.; McKinney, M.; Rempel, R.E.; Goodlad, J.R.; Teh, C.H.; Leppa, S.; Mannisto, S.; Kovanen, P.E.; Tse, E.; et al. Enteropathy-associated T cell lymphoma subtypes are characterized by loss of function of SETD2. J. Exp. Med. 2017, 214, 1371–1386. [Google Scholar] [CrossRef]
- Li, J.; Zhao, Z.; Carter, C.; Ehrlich, L.I.; Bedford, M.T.; Richie, E.R. Coactivator-associated arginine methyltransferase 1 regulates fetal hematopoiesis and thymocyte development. J. Immunol. 2013, 190, 597–604. [Google Scholar] [CrossRef]
- Kim, J.; Lee, J.; Yadav, N.; Wu, Q.; Carter, C.; Richard, S.; Richie, E.; Bedford, M.T. Loss of CARM1 Results in Hypomethylation of Thymocyte Cyclic AMP-regulated Phosphoprotein and Deregulated Early T Cell Development. J. Biol. Chem. 2004, 279, 25339–25344. [Google Scholar] [CrossRef]
- Kasper, L.H.; Fukuyama, T.; Biesen, M.A.; Boussouar, F.; Tong, C.; De Pauw, A.; Murray, P.J.; Van Deursen, J.M.; Brindle, P.K. Conditional Knockout Mice Reveal Distinct Functions for the Global Transcriptional Coactivators CBP and p300 in T-Cell Development. Mol. Cell. Biol. 2006, 26, 789–809. [Google Scholar] [CrossRef]
- Gupta, A.; Hunt, C.R.; Pandita, R.K.; Pae, J.; Komal, K.; Singh, M.; Shay, J.W.; Kumar, R.; Ariizumi, K.; Horikoshi, N.; et al. T-cell-specific deletion of Mof blocks their differentiation and results in genomic instability in mice. Mutagenesis 2013, 28, 263–270. [Google Scholar] [CrossRef]
- Dovey, O.M.; Foster, C.T.; Conte, N.; Edwards, S.A.; Edwards, J.M.; Singh, R.; Vassiliou, G.; Bradley, A.; Cowley, S.M. Histone deacetylase 1 and 2 are essential for normal T-cell development and genomic stability in mice. Blood 2013, 121, 1335–1344. [Google Scholar] [CrossRef]
- Stengel, K.R.; Zhao, Y.; Klus, N.J.; Kaiser, J.F.; Gordy, L.E.; Joyce, S.; Hiebert, S.W.; Summers, A.R. Histone Deacetylase 3 Is Required for Efficient T Cell Development. Mol. Cell. Biol. 2015, 35, 3854–3865. [Google Scholar] [CrossRef] [PubMed]
- Kramer, A.C.; Kothari, A.; Wilson, W.C.; Celik, H.; Nikitas, J.; Mallaney, C.; Ostrander, E.L.; Eultgen, E.; Martens, A.; Valentine, M.C.; et al. Dnmt3a regulates T-cell development and suppresses T-ALL transformation. Leukemia 2017, 31, 2479–2490. [Google Scholar] [CrossRef]
- Ueda, Y.; Okano, M.; Williams, C.; Chen, T.; Georgopoulos, K.; Li, E. Roles for Dnmt3b in mammalian development: A mouse model for the ICF syndrome. Development 2006, 133, 1183–1192. [Google Scholar] [CrossRef] [PubMed]
- Lee, P.P.; Fitzpatrick, D.R.; Beard, C.; Jessup, H.K.; Lehar, S.; Makar, K.W.; Pérez-Melgosa, M.; Sweetser, M.T.; Schlissel, M.S.; Nguyen, S.; et al. A Critical Role for Dnmt1 and DNA Methylation in T Cell Development, Function, and Survival. Immunity 2001, 15, 763–774. [Google Scholar] [CrossRef]
- Zhang, Y.; Chen, Y.; Ma, R.; Jiang, Y.; Liu, J.; Lin, Y.; Chen, S.; Xia, M.; Zou, F.; Zhang, J.; et al. UHRF1 Controls Thymocyte Fate Decisions through the Epigenetic Regulation of EGR1 Expression. J. Immunol. 2020, 204, 3248–3261. [Google Scholar] [CrossRef]
- Cheng, L.; Zhou, K.; Chen, X.; Zhou, J.; Cai, W.; Zhang, Y.; Wang, X.; Wang, C.; Yuan, W.; Zhou, J.; et al. Loss of MBD2 affects early T cell development by inhibiting the WNT signaling pathway. Exp. Cell Res. 2021, 398, 112400. [Google Scholar] [CrossRef]
- Shapiro, M.J.; Shapiro, V.S. Chromatin-Modifying Enzymes in T Cell Development. Annu. Rev. Immunol. 2020, 38, 397–419. [Google Scholar] [CrossRef]
- Onodera, A.; Nakayama, T. Epigenetics of T cells regulated by Polycomb/Trithorax molecules. Trends Mol. Med. 2015, 21, 330–340. [Google Scholar] [CrossRef] [PubMed]
- Raaphorst, F.M.; Otte, A.P.; Meijer, C.J. Polycomb-group genes as regulators of mammalian lymphopoiesis. Trends Immunol. 2001, 22, 682–690. [Google Scholar] [CrossRef]
- Cao, R.; Tsukada, Y.; Zhang, Y. Role of Bmi-1 and Ring1A in H2A Ubiquitylation and Hox Gene Silencing. Mol. Cell 2005, 20, 845–854. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Wang, L.; Erdjument-Bromage, H.; Vidal, M.; Tempst, P.; Jones, R.S.; Zhang, Y. Role of histone H2A ubiquitination in Polycomb silencing. Nature 2004, 431, 873–878. [Google Scholar] [CrossRef] [PubMed]
- Medvedovic, J.; Ebert, A.; Tagoh, H.; Busslinger, M. Pax5: A master regulator of B cell development and leukemogenesis. Adv. Immunol. 2011, 111, 179–206. [Google Scholar] [CrossRef] [PubMed]
- Scheuermann, J.C.; de Ayala Alonso, A.G.; Oktaba, K.; Ly-Hartig, N.; McGinty, R.K.; Fraterman, S.; Wilm, M.; Muir, T.W.; Muller, J. Histone H2A deubiquitinase activity of the Polycomb repressive complex PR-DUB. Nature 2010, 465, 243–247. [Google Scholar] [CrossRef] [PubMed]
- Zhu, P.; Zhou, W.; Wang, J.; Puc, J.; Ohgi, K.A.; Erdjument-Bromage, H.; Tempst, P.; Glass, C.K.; Rosenfeld, M.G. A Histone H2A Deubiquitinase Complex Coordinating Histone Acetylation and H1 Dissociation in Transcriptional Regulation. Mol. Cell 2007, 27, 609–621. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Liu, R.B.; Cao, Q.; Fan, K.Q.; Huang, L.J.; Yu, J.S.; Gao, Z.J.; Huang, T.; Zhong, J.Y.; Mao, X.-T.; et al. USP16-mediated deubiquitination of calcineurin A controls peripheral T cell maintenance. J. Clin. Investig. 2019, 129, 2856–2871. [Google Scholar] [CrossRef]
- Fan, Y.; Mao, R.; Yu, Y.; Liu, S.; Shi, Z.; Cheng, J.; Zhang, H.; An, L.; Zhao, Y.; Xu, X.; et al. USP21 negatively regulates antiviral response by acting as a RIG-I deubiquitinase. J. Exp. Med. 2014, 211, 313–328. [Google Scholar] [CrossRef]
- Pannu, J.; Belle, J.I.; Förster, M.; Duerr, C.U.; Shen, S.; Kane, L.; Harcourt, K.; Fritz, J.H.; Clare, S.; Nijnik, A. Ubiquitin Specific Protease 21 Is Dispensable for Normal Development, Hematopoiesis and Lymphocyte Differentiation. PLoS ONE 2015, 10, e0117304. [Google Scholar] [CrossRef]
- Margueron, R.; Reinberg, D. The Polycomb complex PRC2 and its mark in life. Nature 2011, 469, 343–349. [Google Scholar] [CrossRef]
- Dobenecker, M.W.; Park, J.S.; Marcello, J.; McCabe, M.T.; Gregory, R.; Knight, S.D.; Rioja, I.; Bassil, A.K.; Prinjha, R.K.; Tarakhovsky, A. Signaling function of PRC2 is essential for TCR-driven T cell responses. J. Exp. Med. 2018, 215, 1101–1113. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Kinkel, S.; Maksimovic, J.; Bandala-Sanchez, E.; Tanzer, M.C.; Naselli, G.; Zhang, J.G.; Zhan, Y.; Lew, A.M.; Silke, J.; et al. The polycomb repressive complex 2 governs life and death of peripheral T cells. Blood 2014, 124, 737–749. [Google Scholar] [CrossRef] [PubMed]
- Gray, S.M.; Amezquita, R.A.; Guan, T.; Kleinstein, S.H.; Kaech, S.M. Polycomb Repressive Complex 2-Mediated Chromatin Repression Guides Effector CD8+ T Cell Terminal Differentiation and Loss of Multipotency. Immunity 2017, 46, 596–608. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Oshima, M.; Sato, D.; Matsui, H.; Kubota, S.; Aoyama, K.; Nakajima-Takagi, Y.; Koide, S.; Matsubayashi, J.; Mochizuki-Kashio, M.; et al. Ezh2 loss propagates hypermethylation at T cell differentiation–regulating genes to promote leukemic transformation. J. Clin. Investig. 2018, 128, 3872–3886. [Google Scholar] [CrossRef]
- Georgopoulos, K.; Moore, D.D.; Derfler, B. Ikaros, an early lymphoid-specific transcription factor and a putative mediator for T cell commitment. Science 1992, 258, 808–812. [Google Scholar] [CrossRef]
- Koipally, J.; Heller, E.J.; Seavitt, J.R.; Georgopoulos, K. Unconventional Potentiation of Gene Expression by Ikaros. J. Biol. Chem. 2002, 277, 13007–13015. [Google Scholar] [CrossRef]
- Georgopoulos, K.; Winandy, S.; Avitahl, N. The role of the Ikaros gene in lymphocyte development and homeostasis. Annu. Rev. Immunol. 1997, 15, 155–176. [Google Scholar] [CrossRef] [PubMed]
- Koipally, J.; Renold, A.; Kim, J.; Georgopoulos, K. Repression by Ikaros and Aiolos is mediated through histone deacetylase complexes. EMBO J. 1999, 18, 3090–3100. [Google Scholar] [CrossRef]
- Oravecz, A.; Apostolov, A.; Polak, K.; Jost, B.; Le Gras, S.; Chan, S.; Kastner, P. Ikaros mediates gene silencing in T cells through Polycomb repressive complex 2. Nat. Commun. 2015, 6, 8823. [Google Scholar] [CrossRef] [PubMed]
- Gu, B.; Lee, M.G. Histone H3 lysine 4 methyltransferases and demethylases in self-renewal and differentiation of stem cells. Cell Biosci. 2013, 3, 39. [Google Scholar] [CrossRef]
- Boulard, M.; Edwards, J.R.; Bestor, T.H. FBXL10 protects Polycomb-bound genes from hypermethylation. Nat. Genet. 2015, 47, 479–485. [Google Scholar] [CrossRef]
- Shilatifard, A. The COMPASS Family of Histone H3K4 Methylases: Mechanisms of Regulation in Development and Disease Pathogenesis. Annu. Rev. Biochem. 2012, 81, 65–95. [Google Scholar] [CrossRef] [PubMed]
- Wagner, E.J.; Carpenter, P.B. Understanding the language of Lys36 methylation at histone H3. Nat. Rev. Mol. Cell Biol. 2012, 13, 115–126. [Google Scholar] [CrossRef] [PubMed]
- Ji, Z.; Sheng, Y.; Miao, J.; Li, X.; Zhao, H.; Wang, J.; Cheng, C.; Wang, X.; Liu, K.; Zhang, K.; et al. The histone methyltransferase Setd2 is indispensable for V(D)J recombination. Nat. Commun. 2019, 10, 3353. [Google Scholar] [CrossRef] [PubMed]
- Bedford, M.T.; Richard, S. Arginine methylation an emerging regulator of protein function. Mol. Cell 2005, 18, 263–272. [Google Scholar] [CrossRef]
- Roth, S.Y.; Denu, J.M.; Allis, C.D. Histone Acetyltransferases. Annu. Rev. Biochem. 2001, 70, 81–120. [Google Scholar] [CrossRef]
- Shiama, N. The p300/CBP family: Integrating signals with transcription factors and chromatin. Trends Cell Biol. 1997, 7, 230–236. [Google Scholar] [CrossRef]
- Fukuyama, T.; Kasper, L.H.; Boussouar, F.; Jeevan, T.; Van Deursen, J.; Brindle, P.K. Histone Acetyltransferase CBP Is Vital To Demarcate Conventional and Innate CD8+ T-Cell Development. Mol. Cell. Biol. 2009, 29, 3894–3904. [Google Scholar] [CrossRef]
- Castillo, J.; Wu, E.; Lowe, C.; Srinivasan, S.; Mccord, R.; Wagle, M.C.; Jayakar, S.; Edick, M.G.; Eastham-Anderson, J.; Liu, B.; et al. CBP/p300 Drives the Differentiation of Regulatory T Cells through Transcriptional and Non-Transcriptional Mechanisms. Cancer Res. 2019, 79, 3916–3927. [Google Scholar] [CrossRef]
- Smith, E.R.; Pannuti, A.; Gu, W.; Steurnagel, A.; Cook, R.G.; Allis, C.D.; Lucchesi, J.C. The Drosophila MSL Complex Acetylates Histone H4 at Lysine 16, a Chromatin Modification Linked to Dosage Compensation. Mol. Cell. Biol. 2000, 20, 312–318. [Google Scholar] [CrossRef]
- Seto, E.; Yoshida, M. Erasers of Histone Acetylation: The Histone Deacetylase Enzymes. Cold Spring Harb. Perspect. Biol. 2014, 6, a018713. [Google Scholar] [CrossRef]
- Philips, R.L.; Lee, J.H.; Gaonkar, K.; Chanana, P.; Chung, J.Y.; Romero Arocha, S.R.; Schwab, A.; Ordog, T.; Shapiro, V.S. HDAC3 restrains CD8-lineage genes to maintain a bi-potential state in CD4+CD8+ thymocytes for CD4-lineage commitment. eLife 2019, 8, e43821. [Google Scholar] [CrossRef] [PubMed]
- Hsu, F.-C.; Belmonte, P.J.; Constans, M.M.; Chen, M.W.; McWilliams, D.C.; Hiebert, S.W.; Shapiro, V.S. Histone Deacetylase 3 Is Required for T Cell Maturation. J. Immunol. 2015, 195, 1578–1590. [Google Scholar] [CrossRef]
- Hlady, R.A.; Novakova, S.; Opavska, J.; Klinkebiel, D.; Peters, S.L.; Bies, J.; Hannah, J.; Iqbal, J.; Anderson, K.M.; Siebler, H.M.; et al. Loss of Dnmt3b function upregulates the tumor modifier Ment and accelerates mouse lymphomagenesis. J. Clin. Investig. 2012, 122, 163–177. [Google Scholar] [CrossRef] [PubMed]
- Hansen, R.S.; Wijmenga, C.; Luo, P.; Stanek, A.M.; Canfield, T.K.; Weemaes, C.M.; Gartler, S.M. The DNMT3B DNA methyltransferase gene is mutated in the ICF immunodeficiency syndrome. Proc. Natl. Acad. Sci. USA 1999, 96, 14412–14417. [Google Scholar] [CrossRef]
- Weemaes, C.M.; van Tol, M.J.; Wang, J.; van Ostaijen-ten Dam, M.M.; van Eggermond, M.C.; Thijssen, P.E.; Aytekin, C.; Brunetti-Pierri, N.; van der Burg, M.; Graham Davies, E.; et al. Heterogeneous clinical presentation in ICF syndrome: Correlation with underlying gene defects. Eur. J. Hum. Genet. 2013, 21, 1219–1225. [Google Scholar] [CrossRef] [PubMed]
- Pezzolo, A.; Prigione, I.; Facchetti, P.; Castellano, E.; Viale, M.; Gimelli, G.; Pistoia, V. T-cell apoptosis in ICF syndrome. J. Allergy Clin. Immunol. 2001, 108, 310–312. [Google Scholar] [CrossRef]
- Kamae, C.; Imai, K.; Kato, T.; Okano, T.; Honma, K.; Nakagawa, N.; Yeh, T.-W.; Noguchi, E.; Ohara, A.; Shigemura, T.; et al. Clinical and Immunological Characterization of ICF Syndrome in Japan. J. Clin. Immunol. 2018, 38, 927–937. [Google Scholar] [CrossRef]
- Bostick, M.; Kim, J.K.; Estève, P.O.; Clark, A.; Pradhan, S.; Jacobsen, S.E. UHRF1 Plays a Role in Maintaining DNA Methylation in Mammalian Cells. Science 2007, 317, 1760–1764. [Google Scholar] [CrossRef]
- Luis, T.C.; Weerkamp, F.; Naber, B.A.; Baert, M.R.; De Haas, E.F.; Nikolic, T.; Heuvelmans, S.; De Krijger, R.R.; Van Dongen, J.J.; Staal, F.J. Wnt3a deficiency irreversibly impairs hematopoietic stem cell self-renewal and leads to defects in progenitor cell differentiation. Blood 2009, 113, 546–554. [Google Scholar] [CrossRef]
- Staal, F.J.; Chhatta, A.; Mikkers, H. Caught in a Wnt storm: Complexities of Wnt signaling in hematopoiesis. Exp. Hematol. 2016, 44, 451–457. [Google Scholar] [CrossRef]
- Famili, F.; Naber, B.A.; Vloemans, S.; De Haas, E.F.; Tiemessen, M.M.; Staal, F.J. Discrete roles of canonical and non-canonical Wnt signaling in hematopoiesis and lymphopoiesis. Cell Death Dis. 2015, 6, e1981. [Google Scholar] [CrossRef]
- Luis, T.C.; Ichii, M.; Brugman, M.H.; Kincade, P.; Staal, F.J. Wnt signaling strength regulates normal hematopoiesis and its deregulation is involved in leukemia development. Leukemia 2012, 26, 414–421. [Google Scholar] [CrossRef]
- Yu, S.; Zhou, X.; Steinke, F.C.; Liu, C.; Chen, S.C.; Zagorodna, O.; Jing, X.; Yokota, Y.; Meyerholz, D.K.; Mullighan, C.G.; et al. The TCF-1 and LEF-1 Transcription Factors Have Cooperative and Opposing Roles in T Cell Development and Malignancy. Immunity 2012, 37, 813–826. [Google Scholar] [CrossRef] [PubMed]
- Kueh, H.Y.; Rothenberg, E.V. Regulatory gene network circuits underlying T cell development from multipotent progenitors. Wiley Interdiscip. Rev. Syst. Biol. Med. 2012, 4, 79–102. [Google Scholar] [CrossRef] [PubMed]
- Zaret, K.S.; Carroll, J.S. Pioneer transcription factors: Establishing competence for gene expression. Genes Dev. 2011, 25, 2227–2241. [Google Scholar] [CrossRef]
- Zaret, K.S. Pioneer Transcription Factors Initiating Gene Network Changes. Annu. Rev. Genet. 2020, 54, 367–385. [Google Scholar] [CrossRef]
- Barozzi, I.; Simonatto, M.; Bonifacio, S.; Yang, L.; Rohs, R.; Ghisletti, S.; Natoli, G. Coregulation of Transcription Factor Binding and Nucleosome Occupancy through DNA Features of Mammalian Enhancers. Mol. Cell 2014, 54, 844–857. [Google Scholar] [CrossRef]
- Minderjahn, J.; Schmidt, A.; Fuchs, A.; Schill, R.; Raithel, J.; Babina, M.; Schmidl, C.; Gebhard, C.; Schmidhofer, S.; Mendes, K.; et al. Mechanisms governing the pioneering and redistribution capabilities of the non-classical pioneer PU.1. Nat. Commun. 2020, 11, 402. [Google Scholar] [CrossRef]
- Ungerbäck, J.; Hosokawa, H.; Wang, X.; Strid, T.; Williams, B.A.; Sigvardsson, M.; Rothenberg, E.V. Pioneering, chromatin remodeling, and epigenetic constraint in early T-cell gene regulation by SPI1 (PU.1). Genome Res. 2018, 28, 1508–1519. [Google Scholar] [CrossRef]
- Hosokawa, H.; Masuhara, K.; Koizumi, M. Transcription factors regulate early T cell development via redeployment of other factors: Functional dynamics of constitutively required factors in cell fate decisions. Bioessays 2021, e2000345. [Google Scholar] [CrossRef] [PubMed]
- Isoda, T.; Moore, A.J.; He, Z.; Chandra, V.; Aida, M.; Denholtz, M.; van Hamburg, J.P.; Fisch, K.M.; Chang, A.N.; Fahl, S.P.; et al. Non-coding Transcription Instructs Chromatin Folding and Compartmentalization to Dictate Enhancer-Promoter Communication and T Cell Fate. Cell 2017, 171, 103–119.e18. [Google Scholar] [CrossRef] [PubMed]
- Olariu, V.; Yui, M.A.; Krupinski, P.; Zhou, W.; Deichmann, J.; Andersson, E.; Rothenberg, E.V.; Peterson, C. Multi-scale Dynamical Modeling of T Cell Development from an Early Thymic Progenitor State to Lineage Commitment. Cell Rep. 2021, 34, 108622. [Google Scholar] [CrossRef]
- Hu, G.; Cui, K.; Fang, D.; Hirose, S.; Wang, X.; Wangsa, D.; Jin, W.; Ried, T.; Liu, P.; Zhu, J.; et al. Transformation of Accessible Chromatin and 3D Nucleome Underlies Lineage Commitment of Early T Cells. Immunity 2018, 48, 227–242.e8. [Google Scholar] [CrossRef]
- Yoshida, H.; Lareau, C.A.; Ramirez, R.N.; Rose, S.A.; Maier, B.; Wroblewska, A.; Desland, F.; Chudnovskiy, A.; Mortha, A.; Dominguez, C.; et al. The cis-Regulatory Atlas of the Mouse Immune System. Cell 2019, 176, 897–912.e20. [Google Scholar] [CrossRef]
- Xing, S.; Li, F.; Zeng, Z.; Zhao, Y.; Yu, S.; Shan, Q.; Li, Y.; Phillips, F.C.; Maina, P.K.; Qi, H.H.; et al. Tcf1 and Lef1 transcription factors establish CD8+ T cell identity through intrinsic HDAC activity. Nat. Immunol. 2016, 17, 695–703. [Google Scholar] [CrossRef]
- Tiemessen, M.M.; Baert, M.R.; Kok, L.; van Eggermond, M.C.; van den Elsen, P.J.; Arens, R.; Staal, F.J. T Cell Factor 1 Represses CD8+ Effector T Cell Formation and Function. J. Immunol. 2014, 193, 5480–5487. [Google Scholar] [CrossRef]
- Gao, H.; Lukin, K.; Ramírez, J.; Fields, S.; Lopez, D.; Hagman, J. Opposing effects of SWI/SNF and Mi-2/NuRD chromatin remodeling complexes on epigenetic reprogramming by EBF and Pax5. Proc. Natl. Acad. Sci. USA 2009, 106, 11258–11263. [Google Scholar] [CrossRef]
- McManus, S.; Ebert, A.; Salvagiotto, G.; Medvedovic, J.; Sun, Q.; Tamir, I.; Jaritz, M.; Tagoh, H.; Busslinger, M. The transcription factor Pax5 regulates its target genes by recruiting chromatin-modifying proteins in committed B cells. EMBO J. 2011, 30, 2388–2404. [Google Scholar] [CrossRef]
- Zhang, M.; Dong, Y.; Hu, F.; Yang, D.; Zhao, Q.; Lv, C.; Wang, Y.; Xia, C.; Weng, Q.; Liu, X.; et al. Transcription factor Hoxb5 reprograms B cells into functional T lymphocytes. Nat. Immunol. 2018, 19, 279–290. [Google Scholar] [CrossRef]
- Wurster, A.L.; Pazin, M.J. ATP-dependent chromatin remodeling in T cells. Biochem. Cell Biol. 2012, 90, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Loughran, S.J.; Comoglio, F.; Hamey, F.K.; Giustacchini, A.; Errami, Y.; Earp, E.; Göttgens, B.; Jacobsen, S.E.W.; Mead, A.J.; Hendrich, B.; et al. Mbd3/NuRD controls lymphoid cell fate and inhibits tumorigenesis by repressing a B cell transcriptional program. J. Exp. Med. 2017, 214, 3085–3104. [Google Scholar] [CrossRef] [PubMed]
- Gebuhr, T.C.; Kovalev, G.I.; Bultman, S.; Godfrey, V.; Su, L.; Magnuson, T. The Role of Brg1, a Catalytic Subunit of Mammalian Chromatin-remodeling Complexes, in T Cell Development. J. Exp. Med. 2003, 198, 1937–1949. [Google Scholar] [CrossRef] [PubMed]
- Gialitakis, M.; Sellars, M.; Littman, D.R. The epigenetic landscape of lineage choice: Lessons from the heritability of CD4 and CD8 expression. Curr. Top. Microbiol. Immunol. 2012, 356, 165–188. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).